Reading primary scientific literature enhances students’ understanding of material, increases their self-efficacy, and critical thinking skills. However, scientific articles often present multiple challenges to the students, the first among them is the unfamiliar nature of scientific texts: their high information density, formal language, and abundance of scientific jargon. In this lesson, students are taught the Talking to the Text method to metacognitively read scientific texts. The Talking to the Text method, which is part of the Reading Apprenticeship approach, enables students to have a metacognitive conversation with a scientific text, annotating it with their own questions, connections to previous knowledge, predictions, and drawings. The method is introduced through instructor’s modeling, individual practice, peer interactions, and class discussions. The collaborative approach of this lesson normalizes the struggles that students face when reading complex scientific texts. We have successfully used the Talking to the Text method in an introductory biology course with diverse students who often do not have experience in reading dense scientific texts. We find that this method promotes inclusion and holds students accountable to address what they do not understand in the text. In a survey, half of the students indicated that they were still using the method at the end of the semester. Students commented that the method improved their understanding of concepts, built their metacognitive skills, and helped them connect new information with what they already know. We believe that this method can be valuable for all students who are starting to read complex scientific texts.
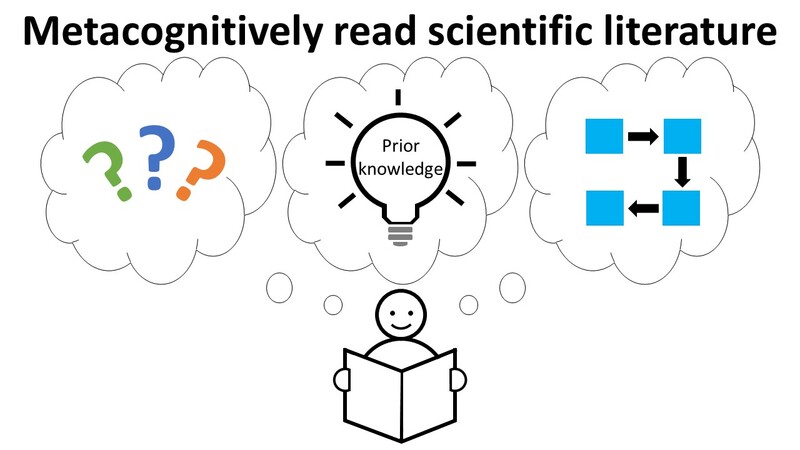
Primary Image: Using Talking to the Text to metacognitively read scientific literature. A student reads a scientific text using the Talking to the Text method. The student engages in metacognitive conversation with the text by identifying the confusing parts of the text, making connections between new and existing knowledge, and drawing pictures of concepts or experiments.