Exploration of the Human Genome by Investigation of Personalized SNPs
Published online:
Abstract
Students often shy away from tedious bioinformatics approaches to exploring their genomes. However, in our expanding digital world these skills are some of the most relevant and valuable. To increase students' interest in their own genomes, I have designed a computer-based laboratory lesson that was coupled with opportunity for the students to be genotyped by the consumer sequencing company, 23andMe. This lesson employs multiple open-access websites through which students explore a health-related single nucleotide polymorphism (SNP) in which they are most interested. Through a series of guided activities, students investigate the genomic region in which their SNP lies, investigate if there are any genome-wide association studies about this SNP, and then determine what model organism would be the best to use if they were to conduct future research about the gene in which the SNP lies. This module could be adapted as a supplement for a variety of Biology lecture or laboratory courses including but not limited to genetics and molecular biology.
Citation
Keller, L.C. 2015. Exploring the Human Genome by Investigation of Personalized SNPs. CourseSource. https://doi.org/10.24918/cs.2015.11Society Learning Goals
Bioinformatics
- DNA - Information Storage [GENOMICS]
- Where are data about the genome found (e.g., nucleotide sequence, epigenomics) and how are they stored and accessed?
- How can bioinformatics tools be employed to analyze genetic information?
Genetics
- Genetic variation
- How do different types of mutations affect genes and the corresponding mRNAs and proteins?
- Genetics of model organisms
- How do the results of molecular genetic studies in model organisms help us understand aspects of human genetics and genetic diseases?
- Genetics & Ethics
- How does genetics impact society and society impact genetics?
Science Process Skills
- Process of Science
- Interpret, evaluate, and draw conclusions from data
- Construct explanations and make evidence-based arguments about the natural world
- Modeling/ Developing and Using Models
- Recognize the important roles that scientific models, of many different types (conceptual, mathematical, physical, etc.), play in predicting and communicating biological phenomena
- Make inferences and solve problems using models and simulations
- Build and evaluate models of biological systems
- Communication and Collaboration
- Share ideas, data, and findings with others clearly and accurately
Lesson Learning Goals
Students will explore the human genome and gain an appreciation for the vast amount of information that is freely available to the public in on-line databases. Additionally, students will demonstrate the ability to use these databases to investigate disease-causing single nucleotide polymorphisms (SNPs) and the genes they affect.Lesson Learning Objectives
Students successfully completing this lesson will be able to:- Effectively use the bioinformatics databases (SNPedia, the UCSC Genome Browser, and NCBI) to explore SNPs of interest within the human genome.
- Identify three health-related SNPs of personal interest and use the UCSC Genome Browser to define their precise chromosomal locations and determine whether they lie within a gene or are intergenic.
- Establish a list of all genome-wide association studies correlated with a particular health-related SNP.
- Predict which model organism would be most appropriate for conducting further research on a human disease.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Since the human genome was published in 2001, we have been in the midst of an explosion of digital information that can be used to determine many aspects about ourselves (1). This recent explosion in available genomic and proteomic data about biological systems has created a high demand for educational resources that effectively teach students how to access, interpret, and analyze these data. In fact, there is a fairly new field called bioinformatics education that aims to instruct students to use computer and information technology to gather, analyze, interpret, and integrate data to solve biological problems (2). There are a large number of previously published articles focusing on the incorporation of bioinformatics and computing skills within laboratory courses. These laboratory-based courses often use bioinformatics databases such as NCBI and the UCSC Genome Browser along with software tools such as BLAST, which provide students with practical computing skills (3-6). There has also been a push to teach more intense bioinformatics through the incorporation of computer programming skills in undergraduate courses (7-8). The goal of this lesson was to introduce students to basic computer skills using freely available databases to explore their personal genomes. Evidence suggests that student learning can be enhanced by integration of personal genome testing into courses (9).
We are rapidly approaching a time when it will be essential for our students to understand and efficiently utilize available databases to learn, not only about genomic architecture, but also about their own personal genetic make-up. To drive home the importance of personalized genetics, students in my human genetics course had the opportunity to opt in for genetic testing from the direct-to-to consumer sequencing company, 23andMe. It is crucial for students to fully understand the implications of personalized genetic testing and understand 23andMe's terms of use and privacy statements prior to opting in. Students should be made aware of the risks of discovering certain information that they are not prepared to handle such as learning that your father is not actually your biological father or that you have an increased risk for breast cancer, Parkinson's disease, or Alzheimer's Disease. My students were encouraged to think about these issues and consult their parents. Everyone in this course had the option to undergo genetic testing or to use the publicly available genetic data provided by 23andMe's education team. This particular lesson uses health-related single-nucleotide polymorphisms (SNPs) that the students wanted to learn more about. My main goal in designing this lesson is to expose students to the vast amount of information freely available at student's fingertips and to reveal that students should not be wary of such information. The ability to learn directly from their own personal genomes made this lesson particularly interesting and exciting for students in this course.
This lesson was part of a Human Genetics lecture/laboratory course taught at Quinnipiac University during fall 2014. This course explored the nature of the human genome, how chromosomes are organized, what genes look like, and how the instructions therein give rise to a human being. Contemporary topics such as The Human Genome Project and personalized medicine were highlighted and students enrolled were encouraged to consider undergoing personal genome testing as part of the course curriculum. Any student opting in for genetic testing will log onto the 23andMe website, pay $99, and have their personal genomes sequenced for the cost of a textbook approximately 6-8 weeks prior to the start of class. This course utilized the students' experiences with their own genomes as the backdrop for class discussions on DNA sequencing, SNP analysis, bioinformatics, and the ethical and legal debates about personalized genomics. This new course has been taught only once, but is scheduled to be taught again in the spring of 2015.
Each week, the course met for three 50-minute lectures and one three-hour laboratory session. There were 20 students enrolled in the course and 17 of them opted in for genetic testing by 23andMe. By the end of the course, two of the students who opted out of genetic testing at the beginning of the course changed their minds and requested a kit from 23andMe. Students ranged from sophomores to seniors and were majoring in Biology, Biomedical Sciences, Health Sciences, or Behavioral Neuroscience. The laboratory part of this course was composed of approximately 25% wet lab experiments, which included human trait inheritance analysis, mitochondrial haplotyping, and preparation of human chromosome spreads. The remaining 75% of the laboratory involved primarily exploration and analysis of genetic information pertaining to the student's personal genetic information. Students that opted out of genetic testing were given access to example genetic profiles provided directly from 23andMe.
Intended Audience
Life science majors with at least one year of introductory biology.
Learning time
This lesson was conducted within one 3-hour laboratory session. However, a user can modify the timeline in Table 1, shortening the lesson by including only the most relevant information for a particular class.
Pre-requisite student knowledge
Students should have basic computer skills, including proficiency in Microsoft Word and Excel. Additionally, students should understand gene structure and function, single-nucleotide polymorphisms, and some knowledge of the human genome project.
SCIENTIFIC TEACHING THEMES
Active Learning
Students engage in think-pair-share discussions at the beginning of the laboratory to assess their knowledge of scientific databases. After the laboratory session, the whole class discusses the results of their bioinformatics exploration.
Assessment
Pre-assessment: In small group discussion and sharing out to the class, students describe what they think they can discover about a particular SNP based on bioinformatics approaches.
Assignment: Students turn in a screen shot from the UCSC Genome Browser representing the SNP of interest, along with a short description of the genomic region including nearby genes, conservation of the region in other vertebrate models, and citations of three published genome-wide association studies.
Participate in Discussion: After turning in the assignment, students participated in a class-wide discussion of what they learned about on-line genomic information.
Inclusive Teaching
- Discussion of the similarities among all human genomes acknowledges the enormous genetic conservation between all of us.
- Examination of particular health-related SNPs also demonstrates that all of us are at risk for some diseases regardless of age, gender, race, etc.
- Enabling students to choose a particular SNP is inherently inclusive, since each student can pursue an individual interest.
- The diversity of choices across the class will provide a variety of examples that may be more or less common in various backgrounds.
LESSON PLAN
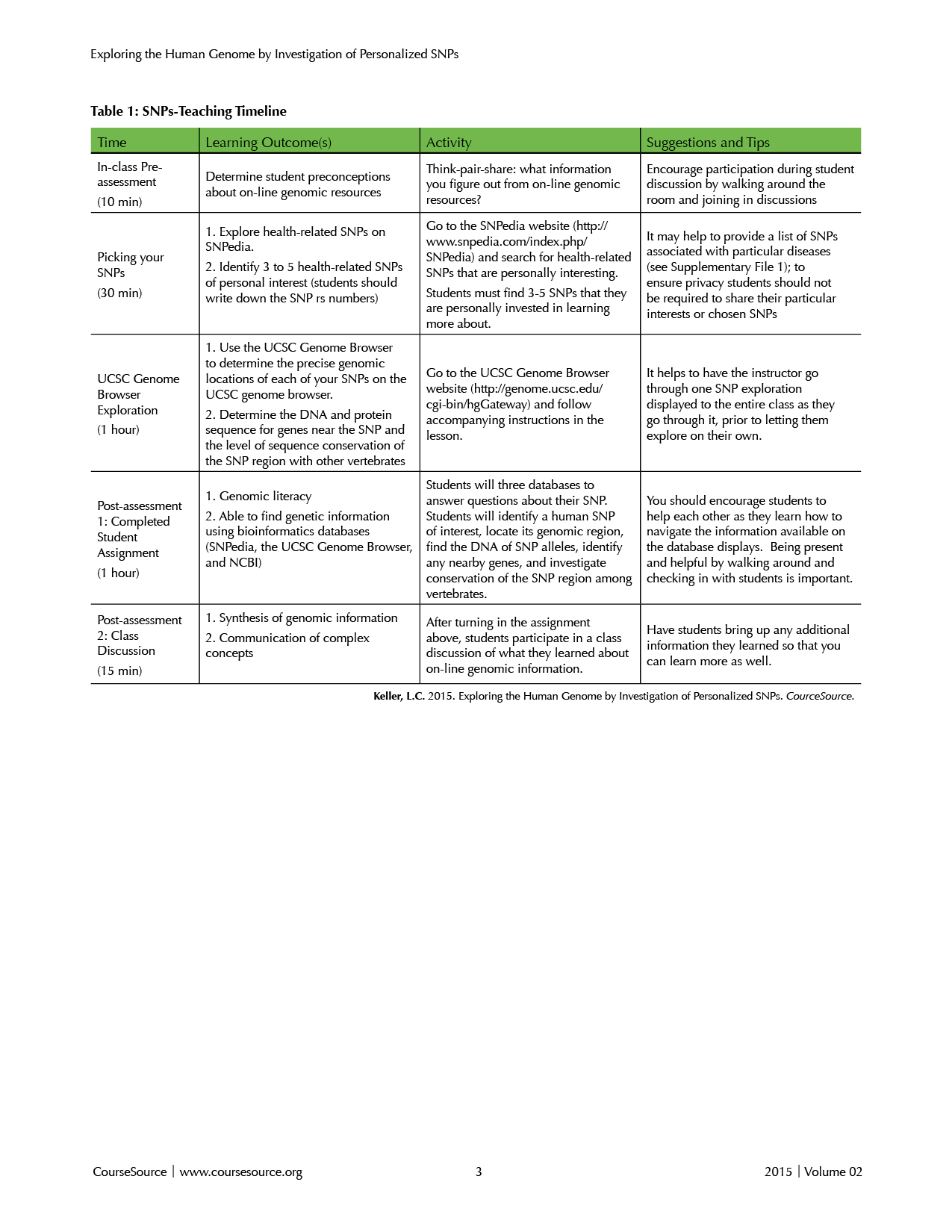
Table 1. SNPs-Teaching Timeline
PRE-CLASS PREPARATION
Approximately 6-8 weeks prior to the start of the semester, students were sent a "welcome to the course" letter in which I described the option for students to undergo direct-to-consumer genetic testing by the company 23andMe. I attempt to describe some of the risks in learning something that you may not really want to know about your health and talk about examples including particular SNPs that dramatically increase the risk of getting breast cancer, Parkinson's disease and Alzheimer's disease. All students were required to read 23andMe's lengthy terms and conditions and privacy statements because I wanted all students to be informed of the issues before they made a decision about getting genetic testing done. I also encouraged students to consult their parents, though parental consent was not required since all my students were 18 or older. I explain that genetic testing by 23andMe is not a requirement for the course or this lesson and that I will provide example genetic data for those students who choose not to undergo personalize genetic testing. Students who decided to get their genome analyzed by 23andMe requested a kit and paid $99, which is less than the majority of textbooks. I did not require a textbook for this course regardless of whether students chose to have their DNA variants determined by 23andMe.
When I teach this lesson, I walk the students through one example as they follow along on their personal computers, using the rs4402960 SNP, which is shown in the figures for this lesson. This particular SNP is associated with type-2 diabetes. I project my computer screen so that all students can watch as I navigate through the exercise. Additionally, I encourage students to help one another out during this demonstration and walk around the class to ensure that students are following along carefully.
Each student must have access to a computer with an internet connection and word processing software such as Microsoft Word during the laboratory session. Familiarize yourself with both SNPedia (12) and the UCSC Genome Browser (11) websites. You should also encourage students to come to lab with a list of health-related traits that they might find particularly interesting. They will be able to use lab time to identify SNPs associated with that health issue. For example, a student who has a family history of diabetes may be interested in investigating SNPs related to diabetes. Clearly, students should not be forced to share their list, to maintain privacy and ensure that students are investigating the things they think are most important. For students who do not complete the pre-lab assignment, it will be useful to provide examples of health-related SNPs that students have looked at in the past as inspiration. A few SNPs that students have been particularly interested in learning more about are those responsible for alcohol craving (SNP rs1799971), the ability to taste cilantro (SNP rs7107418), and the speed of caffeine metabolism (SNP rs762551). A list of SNPs that students from my own class used is available as Supplemental File S3.
There are a number of available tutorials that are helpful for instructors that may not be familiar with some of the databases used in this lesson and/or that can be posted for students. For example, NCBI has a large number of tutorials of interest for this lesson that can be accessed at http://www.ncbi.nlm.nih.gov/education/tutorials/. Additionally, there is a great YouTube tutorial for the NCBI database at https://www.youtube.com/watch?v=bxx5uaKjMa8. Zweig et al., 2008, have published a UCSC genome browser tutorial paper, which provides detailed instructions for using the UCSC Genome Browser website (10). Additionally, a good YouTube tutorial for the UCSC genome browser website walks the user through the basics of navigating their website: https://www.youtube.com/watch?v=DNXI-M9oQl8.
LAB ACTIVITIES AND TEACHER SCRIPT
Introduction to the Lab: When students arrive in laboratory, have them immediately get out their computers (or log into university computers) and ensure that everyone is connected to the internet. Once all students have arrived and are connected to the internet, give a short explanation of the laboratory activity explaining the importance of students being able to navigate on-line genomic resources. For example,
"Today we will be exploring single-nucleotide polymorphisms associated with aspects of human health that you personally care about. We will not only identify the precise genomic regions in which these SNPs lie, but we will also delve deeper into learning about the genes associated with human disease-causing SNPs. You are learning about biology in an amazing time, when the amount of genetic information is exploding and our knowledge of human disease is growing exponentially. I want each of you to be able to peer into the human genome and have a greater appreciation and understanding of the vast amount of information that is freely accessible to you, or anyone else with an internet connection."
Pre-Assessment Discussion (Think-Pair-Share): Have students think independently about what kind of information they might learn about a particular health-related SNP based on bioinformatics approaches. After two minutes, have the students break into pairs or small groups to share their thoughts with one another. After another two minutes, have each group share out their ideas with the rest of the class. This activity helps the professor assess the student's knowledge about genome browsers and may help launch a discussion about misconceptions about freely available genomic data.
SNPedia Exploration: Explain that SNPedia is a user-created wiki-type website that examines human genetics based on SNPs and genome-wide association studies. Have students open the SNPedia website (http://www.snpedia.com/index.php/SNPedia) and point out that they can find some fun and interesting SNPs to explore, if they scroll down and click on the "Popular" section. Students should then start searching for diseases, health-related traits, or specific SNPs of interest by typing key words into the "Search" box at the top right. As students proceed through this lesson, it is important for them to focus on health-related traits. For example, you can type in "type-2 diabetes" and find dozens of SNPs that have been associated with increased risk of type-2 diabetes. All SNPs are listed as blue Reference SNP identification numbers (rs numbers) within little gray rectangles (i.e. rs4402960). Clicking on one of these rectangles will bring you to a page with more information about that particular SNP. In the box on the right side of the page, you will find genomic variations, a summary of the risk associated with having a particular genotype, as well as links to many other databases that mention this SNP.
Students who have opted in for genetic testing can then link directly to their personal genotype by clicking the 23andMe link farther down in this boxed region. It is always important to remind students that just because they have a particular genotype does not mean that they will get a disease. A few genetic diseases, such as sickle cell anemia and cystic fibrosis, are caused by mutations in a single gene. However, the causes of the large majority of human genetic disorders are much more complex. These disorders do not have a single genetic cause and are likely associated with the effects of multiple genes in combination with lifestyle and environmental factors. These multifactorial diseases, such as heart disease, diabetes, and obesity are known to have a genetic component and often cluster in families. However, there is no precise pattern of inheritance, complicating our ability to determine the risk of inheriting or passing on these disorders. Again, I always stress that it is very rare to see a 100% correlation between having a particular SNP and being predisposed to have a human genetic disease.
Students should generate a list of 3 to 5 SNPs to explore.
UCSC Browser Exploration: Once the students have created their list of SNPs, have them open the UCSC (University of California, Santa Cruz) genome browser website (http://genome.ucsc.edu/cgi-bin/hgGateway) shown in Figure 1. Students should look up one particular SNP (e.g., rs4402960) by entering the rs number within the search term box at the top right (red circle in Figure 1) and pressing return or clicking on the "submit" button.
This search will lead to a page (Figure 2) that lists links to the SNP, depending on the study in which that SNP was identified (i.e., via genomic location; genome-wide association). Have students just click again on their rs number of choice usually located on the far left side of the page (red circle in Figure 2).
The page that opens should provide the genomic region in which their SNP lies (Figure 3). The information on this page can look very daunting to students. It is important to walk through the information with them, using an example SNP of your choice. You should point out the chromosomal region shown at the top under the move and zoom buttons (i.e. chr3:185,511,437-195,511,937, red rectangle in Figure 3). To the right of this number, the number of base pairs currently being displayed is listed (i.e. 501 bp, red rectangle in Figure 3). Additionally, the page shows a diagram of the entire chromosome and indicates the approximate location of the SNP by a red line (red circle in Figure 3). Notice that this particular SNP is near the end of chromosome three in the human genome.
To determine if their SNP is within a gene or to see if there are any nearby genes, students should look for lines below the scale markers (see Figure 4). These lines represent genetic sequences of genes, with the gene names on the far left (red circle in Figure 4). Thick lines represent exons (coding region of DNA), whereas the thin lines represent introns and arrows indicate the direction of transcription. Medium-width lines represent untranslated regions, which are usually at the 3' and 5' ends of a gene.
The SNP of interest can be found by looking below the genes under the section titled, "Single Nucleotide Polymorphisms." Notice the boxed region in Figure 4, which indicates the presence of two SNPs (rs763450 and rs4403960). The SNP that you searched for is automatically placed in the center of the page and is indicated by a black box with white letters showing the SNP number.
To look more carefully at a particular SNP, students can click the SNP name, which opens a page that provides the precise location of the SNP and the nucleotides that have been observed in human alleles at this position. Sometimes both the Chimpanzee and human alleles are included.
To learn more about the gene in which the SNP resides, or the gene nearest to the SNP, you can return to the Genome Browser Assembly page and click the gene name on the far left hand side (red circle in Figure 4). This selection will bring you to a description and page index for this gene with a lot of information about the gene.
To gain access to the sequence of the gene itself, have students write down the RefSeq Summary number (i.e. NM_001007225). Students can then search this RefSeq number by entering it into the National Center of Biotechnology Information nucleotide search page (http://www.ncbi.nlm.nih.gov/nucleotide/). Some students become quite proficient at navigating the NCBI website and discover that they can link directly to the genomic, mRNA, and protein sequences from this browser. I encourage students to explore the website but to write down the RefSeq Summary number so they can always refer back to it. Students can learn the nucleotide number and sequence in the gene associated with their SNP of interest.
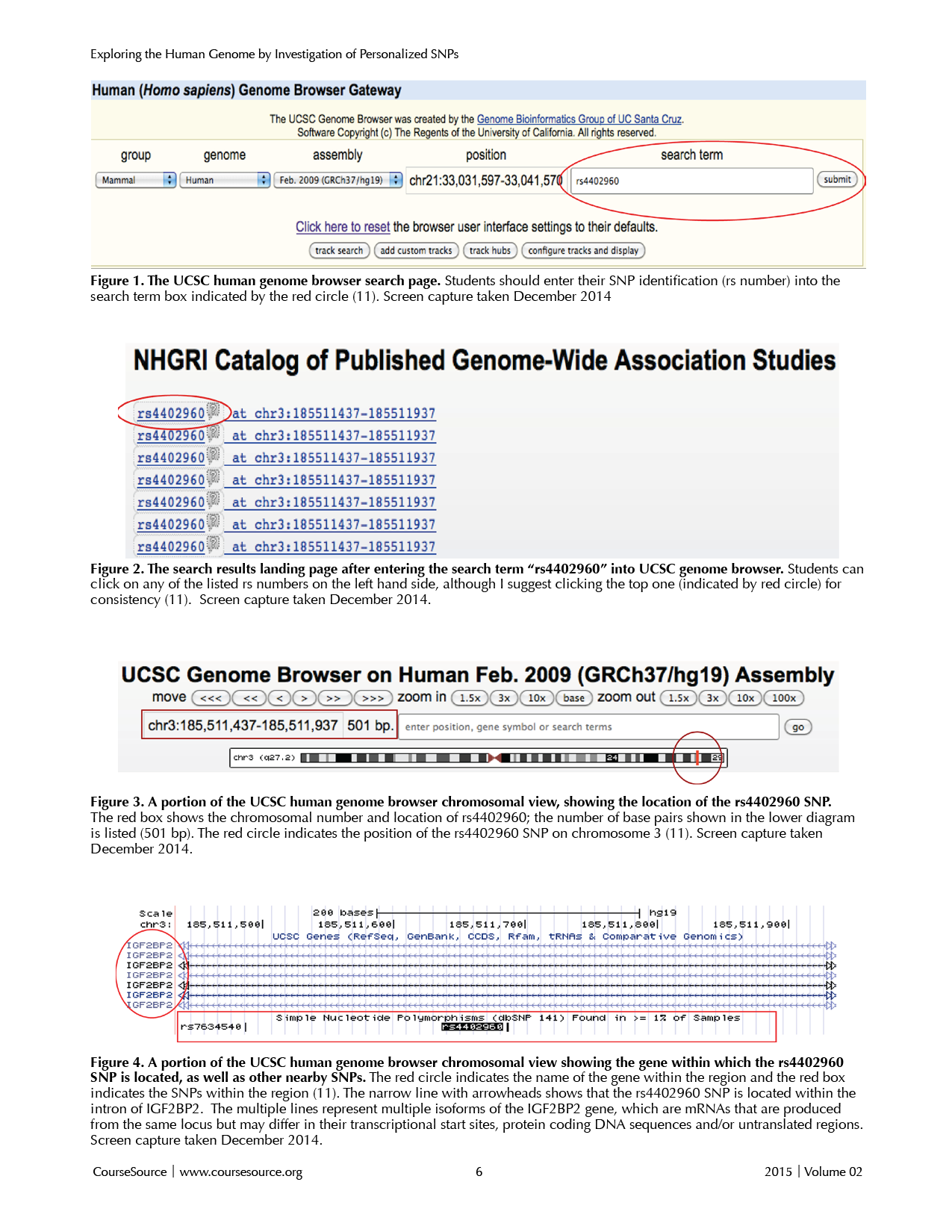
Figures 1-4. SNPs
On the NCBI page (shown in Figure 5), students can scroll down to examine the "Features" (top red circle in Figure 5) where it lists the gene, the exons (including the base pairs for each exon), and the coding region (CDS). Students can click on the CDS title on the left hand side (bottom red circle in Figure 5), which will highlight the coding region within the sequence at the bottom of the page and will also provide students with the translated protein sequence. Figure 5 shows the translated protein sequence on the right hand side.
Students should now return to the UCSC Genome Browser page for their SNP and play around with the zoom functions shown in Figure 6. To zoom out have students press the zoom out "10X" button until they can see any nearby genes (red circle in Figure 6). Students may have to press this several times to view additional genes. This scale can be disorienting to students, so encourage students to keep an eye on the scale bar to determine the size of the genome region that they are viewing. If students ever get confused, or lose their particular SNP, they can always type the SNP number into the search term box at the top to re-center their SNP. Give them a few minutes to play around with this feature while you walk around the room helping students out. Some SNPs are not located within genes and are therefore referred to as intergenic, a term that you should define during this laboratory activity if students are not already familiar with it. Some genomic regions will contain a high density of genes, while other regions will be rather barren. Some genes will be very large, covering hundreds of thousands of nucleotides.
Have students return to their original SNP by either zooming back in or by entering their SNP number back into the search term box at the top. Then have the students click on the "zoom in to base" button (red arrow in Figure 6), which will zoom in all the way to display the actual nucleotide sequence of this genomic region.

Figures 5-6. SNPs
Scrolling down the page will allow students to examine if a particular SNP has been identified as a point of interest in at least one genome-wide association study (indicated by green title "NHGRI Catalog of Published Genome-Wide Association Studies) or within other publications (indicated by black title "SNPs in Publications"). The red arrows in Figure 7 highlight these features. Clicking on these titles will expand the display to include links to each genome-wide association study (see green boxes in Figure 7).
Students can then click each green box to bring them to a new page that contains a link to the PubMed reference for that particular published genome-wide association study in which their SNP was identified (red box in Figure 8). Students should be encouraged to explore the various publications associated with their SNP to select the SNP that may be most important for future research regarding the human health trait in which they were interested.
Students can then examine the conservation of that genomic region with other vertebrate animals. Have students find and click on the blue "Multiz Alignment of 100 Vertebrates" title (Figure 9) to display the DNA alignment of this region among various vertebrates. For example, the particular genome region within the gene in Figure 9 is conserved between humans, Rhesus monkeys, and dogs, but not in mice or elephants. In addition, the gene is not present in chicken, Xenopus, Zebrafish, or Lampreys.
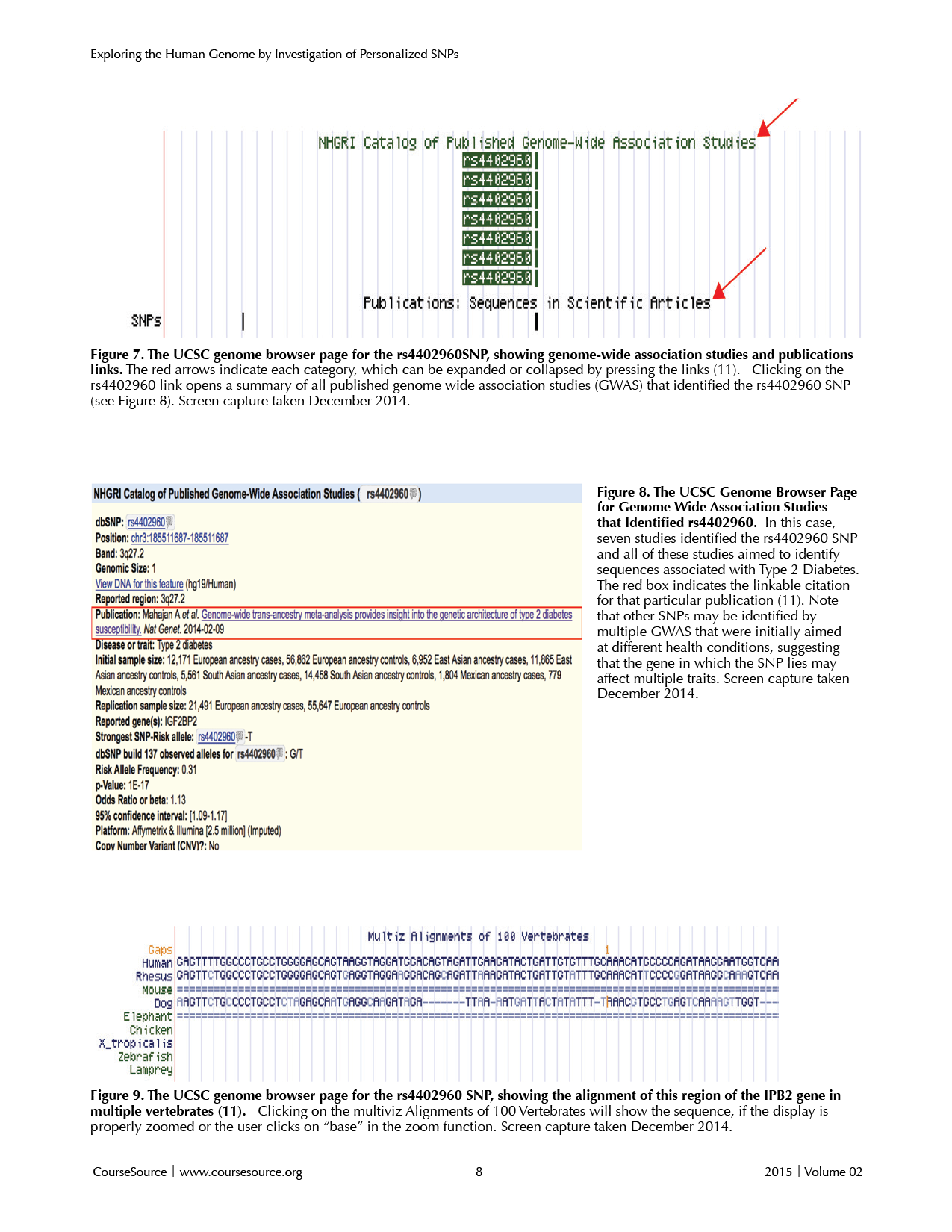
Figures 7-9. SNPs
Student Assignment: Students will need to explore one particular SNP in detail and create a document that describes information about this SNP to turn in by the end of the laboratory. The assignment that I have given in my course is available in within Supplemental Files S1. SNPs-Student Assignment and the corresponding assignment key S2. SNPs-Example Student Assignment.
TEACHING DISCUSSION
Students need to take ownership: This laboratory exercise allows students to develop some of the practical skills necessary to navigate human genetic database information. To get students really engaged in learning about the human genome, I gave students the opportunity to get genotyped by 23andMe, instead of purchasing a textbook. This option allowed students the ability to truly connect with the information that they were studying. However, even if your students do not have their own genetic information, allowing them choose SNPs in which they are particularly interested does seem to give them some ownership of the data analysis. All students were able to find the genomic location of a SNP of interest and dissect the genomic region in relation to genes, genome-wide association studies, and DNA sequence conservation.
The teacher must be well prepared: Professors need to go through the entire exercise themselves prior to teaching this lesson. Although this lesson lays out a detailed plan, students can easily get confused. It will greatly help your effectiveness to have a firm grasp of all of the relevant websites. Instructors should keep in mind that the freely available databases are constantly being updated and, therefore, the instructor may need to update some of the specific instructions within this lesson as well. Additionally, because human genetics and personalized medicine is a rapidly evolving field, it is very likely that students will come across websites with which the instructor is not familiar, but that have the potential to display the information in new and interesting ways. As the instructor, I encourage students to explore and share any novel information that they find with me and, if it is relevant, I pass this information on to the rest of the class, giving credit to the student who discovered it. This is just one example in how this lesson can be very engaging and interactive.
Students appear to be highly engaged: Students in my human genetics course were very excited to explore the various SNPs that were associated with human health and disease. Even the students who opted out of genetic testing revealed that they appreciated learning how to look up a particular genomic region and dissect out some of the important information. Many of my students were surprised by the lack of homology between many disease SNPs in humans and mice. Mice models of human disease have long been used in order to study and learn more about human diseases, however, my students began to question and think more deeply about the validity of this research after visualizing the lack of homology. This brought up a good opportunity to discuss animal models in research along with the complexity of human diseases.
This exercise is easily adaptable: This laboratory exercise is perfect for adapting to many other courses. For example, it could be used within a molecular biology course to determine the precise location and sequence of a particular gene rather than a SNP. Additionally, the UCSC genome browser contains dozens of features that I did not highlight in this lesson, including the position of promoters and histone variants. Exploring such features would be highly appropriate for an upper-level course.
SUPPLEMENTAL MATERIAL
- Supplemental File S1. SNPs-Student Assignment
- Supplemental File S2. SNPs-Student Example Assignment
- Supplemental File S3. SNPs-Table listing previously examined SNPs
ACKNOWLEDGMENTS
I would like to acknowledge the Quinnipiac University Human Genetics course in the fall of 2014 for participating in this laboratory lesson.
References
- Baltimore D. 2001. Our genome unveiled. Nature 409(6822):814-6.
- Magana AJ, Taleyarkhan M, Alvarado DR, Kane M, Springer J, and Clase K. 2014. A survey of scholarly literature describing the field of bioinformatics education and bioinformatics educational research. CBE- Life Sci Educ 13: 607-623.
- Brame CJ, Pruitt WM, and Robinson LC. 2008. A molecular genetics laboratory course applying bioinformatics and cell biology in the context of original research. CBE Life Sci Educ 7(4): 410-421.
- Banta LM, Crespi EJ, Nehm RH, Schwarz JZ, Singer S, Manduca CA, Bush EC, Collins E, Constance CM, Dean D, Esteban D, Fox S, McDaris J, Paul CA, Quinan G, Raley-Susman KM, Smith ML, Wallace CS, Withers GS, and Caporale L. 2012. Integrating genomics research throughout the undergraduate curriculum: a collection of inquiry-based genomics lab modules. CBE Life Sci Edcu 11(3): 203-208.
- Baumler DJ, Banta LM, Hung KF, Schwarz JA, Cabot EL, Glasner JD, and Perna NT. 2012. Using comparative genomics for inquiry-based learning to dissect virulence of Escherichia coli O157:H7 and Yersinia pestis. CBE Life Sci Educ 11(1): 81-93.
- Carson S and Miller H. 2012. A contemporary, laboratory-intensive course on messenger RNA transcription and processing. Biochem Mol Biol Educ 40(2): 89-99.
- Cooper S. 2001. Integrating bioinformatics into undergraduate courses. Biochem Mol Biol Educ 29: 167-168.
- Wrightman B and Hark AT. 2012. Integration of bioinformatics into an undergraduate biology curriculum and the impact on development of mathematical skill. Biochem Mol Biol Educ 40: 310-319.
- Salari K, Karczewski KJ, Hudgins L, and Ormong KE. 2013. Evidence that personal genome testing enhances students learning in a course on genomics and personalized medicine. PLOS One 8(7): e68853.
- Zweig AS, Karolchik D, Kuhn RM, Haussler D, and Kent WJ. 2008. UCSC genome browser tutorial. Genomics 92(2): 75-84.
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12(6):996-1006.
- Cariaso M and Lennon G. 2012. SNPedia: a wiki supporting personal genome annotation, interpretation and analysis. Nucleic Acids Res 40(Database issue):D1308-12.
Article Files
Login to access supporting documents
Exploring the Human Genome by Investigation of Personalized SNPs(PDF | 4 MB)
Supplemental File S1. SNPs-Student assignment.docx(DOCX | 18 KB)
Supplemental File S2. SNPs-Student Example Assignment assignment.docx(DOCX | 71 KB)
Supplemental File S3. SNPs-Table listing previously examined SNPs explored.docx(DOCX | 23 KB)
- License terms

Comments
Comments
There are no comments on this resource.