Forensic Phylogenetics: Implementing Tree-thinking in a Court of Law
Published online:
Abstract
Here we describe an in-class activity developed for Evolutionary Biology and Biodiversity (BioEE1780) at Cornell University, an introductory course required of all biology majors. This activity offers instructors an engaging and real-life framework for teaching challenging concepts in phylogenetics and tree interpretation. The activity is adapted from a study in forensic phylogenetics reported in Scaduto et al. (2010) that showcased the use of gene genealogies in a court of law to infer willful transmission of a deadly disease to multiple victims.
Citation
Ballen C.J., Drake A.G., and Zamudio K.R. 2019. Forensic phylogenetics: Implementing tree-thinking in a court of law. CourseSource. https://doi.org/10.24918/cs.2019.16Lesson Learning Goals
- Students will understand the basic concepts of monophyly, paraphyly, and phylogenetic divergence.
- Students will understand how phylogenies can be used to inform the evolution and transmission of pathogens.
- Students will be able to interpret how evolutionary trees inform the timing and order of transmission.
Lesson Learning Objectives
- Students will be able to infer the topological and temporal relationships expected in an evolutionary tree (phylogeny) of a pathogen in the case of transmission from one host to the next.
- Students will be able to draw trees representing the transmission events from one host (patient zero) to multiple secondary patients.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Phylogenetic trees are tools that biologists use to depict evolutionary relationships and organismal diversification, but they can be challenging for students to conceptualize (1,2). Published articles on teaching in this field introduce students to phylogenetics through tree reconstruction by building a table of synapomorphies (3), or using identifiable traits with which to group similar fictitious organisms (4). Both of these approaches engage students through thought exercises, and are the first step in understanding how trees are built. Here we provide a lesson for the next step in teaching phylogenetics: interpreting and making inference from tree topologies, or ‘tree-thinking.’
We apply ‘tree thinking’ to a real-world question, providing students the opportunity to actively participate in phylogenetic tree interpretation using an engaging case study example from the primary literature (Box 1).

Box 1. Case study example from primary literature
The scenario is based on a real court case where the defendant was accused of purposefully infecting six women with HIV virus (5). Students are instructed to read a news article about the court case before class (https://abcnews.go.com/2020/hiv-criminal-philippe-padieu-busted-women-lied/story?id=14491705), and then guided by the instructor, use phylogenetic information to trace the origin and the direction of transmission of HIV from the source to the multiple victims. Working in groups, students predict what the phylogenetic tree of virus sequences from the defendant and the victims should look like. Once they arrive at a correct prediction, they use the real phylogenetic topology to reveal that the defendant was the source of all virus lineages isolated from the victims, and to identify the order in which transmission occurred to each victim. We used this case study to help students interpret tree topologies, understand how disease transmission will result in paraphyly of host pathogens relative to pathogens of victims, and to understand that the topology can also indicate the order of transmission to multiple victims. This activity appeals to students because they actively participate in an applied case of forensic discovery, and use ‘tree thinking’ in a real court case.
INTENDED AUDIENCE
We taught this lesson to introductory biology students at Cornell University. The course is largely composed of first and second year students. In the class, this activity occurs in the second of four phylogenetic-themed lectures, so the students already know about what trees represent, and are familiar with basic terms in phylogenetics.
REQUIRED LEARNING TIME
The activity can be completed in approximately 50 minutes to one hour.
PRE-REQUISITE STUDENT KNOWLEDGE
Students first learn basic phylogenetic terms and concepts in pre-lecture videos or textbook readings.
Basic terms and concepts:
- Branches and tips
- Nodes
- Clades/monophyletic groups
- Characters
- Taxa
More complicated terms and concepts:
- Synapomorphies are ‘shared derived characters’
- Ancestor-descendent relationships
- Rotating around nodes
- Trees with different shapes
- Molecular clocks
Activities and assessments from the introductory phylogenetics lectures used in this course can be found in the textbook, ‘Life: The science of biology’ (Sadava et al. 2016). See full citation below:
- Ballen CJ. 2016. ‘Reconstruction and using phylogenetic trees: Active learning module’, in Sadava, D.E, Hillis, D.M., Heller, H.C., Hacker, S. D., eds., Life: The science of biology. London, UK: Macmillan Learning.
We also recommend the following reading from Zimmer and Emlen (2015) Making Sense of Life textbook.
Full reading assignment for both lectures:
- Chapter 4: sections 4.1, 4.2, 4.3, Box 4.3.
- Chapter 9: sections 9.3, 9.5 [for important practice in reading phylogenies but not for the details of those examples]
Suggested textbook questions for both lectures:
- Chapter 4 (pp 123-124): Multiple choice 1, 2, 4, 5, 6, 7; Short Answer 2, 3.
Textbook glossary terms: all purple glossary terms within section 4.1, 4.2, and 4.3
Zimmer and Emlen (2015) is the textbook that was used for the activity, however other options for readings might include: Phylogeny and the Tree of Life, Chapter 26 of Campbell Biology by Urry et al. (2017); Evolutionary Patterns, Phylogeny and Fossils, Chapter 22 of Biology: How life works by Morris et al. (2019); or any textbook that covers concepts from the “Pre-requisite Student Knowledge” section above.
PRE-REQUISITE TEACHER KNOWLEDGE
Instructors who teach this module should have a firm understanding of evolutionary biology and tree interpretation. However, knowledge of viral or organismal taxonomy is not necessary.
SCIENTIFIC TEACHING THEMES
ACTIVE LEARNING
For this activity, students work together in small groups (4-5 students) to solve questions posed by the instructor, and a subset of students come to the front of the class to present their work to their peers. In large lectures, we recommend the use of a document camera, which provides live image capture of documents or three-dimensional objects. If instructors do not have access to a document camera, they can instruct students to swap their responses with adjacent groups and have their peers correct them. Alternatively, instructors can present work from previous years that represents common mistakes and misconceptions, and have the students correct them. The instructor can also use iClickers to gauge the classroom understanding of concepts along the way.
ASSESSMENT
Instructors can use questions from the Tree Thinking Concept Inventory (6) as pre- and post- lesson measures of students' understanding. We used identical 'tracker questions' on exams across semesters to gauge student understanding in semesters with and without this activity. 'Tracker questions' are identical or near-identical questions presented to students each semester that can be used to track student performance in response to pedagogical changes such as new activities or approaches to teaching.
INCLUSIVE TEACHING
Because of the crime described in the content of this activity, we recommend issuing a warning to students sometime before the lecture and on the course website (if applicable) about the content, so students have time to consider whether they are able to participate. We also recommend instructors encourage students to read an ABC News article about the court case prior to class. Though students have never reported discomfort in response to this activity to the TAs or instructors in our experience, it does include data related to sexual offenses perpetrated by and committed against a number of women. If students are unable to participate due to the nature of the data, encourage them to let their TA know with no penalty.
Students work in groups to solve problems that are applied to different contexts. Students are encouraged to articulate their thoughts to one another as they move through the questions assigned. A random number generator can be used to call on one of the groups to share their answer, but each group can designate a 'reporter' so those who are shy are not forced to speak in front of a large classroom. Alternatively, the group reporter can be designated by the instructor who assigns it by asking the group: who woke up earliest that morning, has a birthday coming up, or whose name starts with a letter closest to the letter 'Z' (8). These random assignments may reduce public speaking anxiety and help reduce gender bias in participation (7).
LESSON PLAN
In the last decades, Science, Technology, Engineering, and Math (STEM) disciplines began integrating evidence-based teaching practices into undergraduate biology classrooms. In this lesson, we offer instructors an engaging activity for teaching challenging concepts in phylogenetics, with a focus on tree interpretation. The activity is adapted from the primary literature - the data used is from a forensic phylogenetics case reported in 2010 (5). A number of news sources also covered the story, and students can read about it before class: (https://abcnews.go.com/2020/hiv-criminal-philippe-padieu-busted-women-lied/story?id=14491705). Note that if instructors intend to print slides (see S1. Phylogenetic Forensics - Lecture Slides), they should consider that some information is communicated via animations during the PowerPoint presentation and cannot be seen in print-outs.
Before the activity begins, the instructor polls the students with one or two iClicker questions that cover material from the pre-lecture reading/video podcast (‘vodcast’). For example:
iClicker: Which statement is TRUE about phylogenetic relationships? Answer: B
- A. A paraphyletic group includes all the descendants, but not the common ancestor.
- B. A monophyletic group includes the common ancestor and all the descendants.
- C. Taxa in a paraphyletic group are never closely related.
- D. A monophyletic group can include some, but not all, of the descendants.
Then, students are introduced to the field of forensic phylogenetics, or the inference of pathogen phylogenies to infer transmission and trace pathogen origins. Students are also introduced to viral evolution, including their rate of genetic evolution, as well as transmission dynamics. Viral genomes mutate so quickly that they create a population of related lineages in each host. By sequencing highly variable virus genes, scientists can create a phylogenetic tree that shows how those viral lineages are related. The relatedness of the viral lineages in different hosts can support (or not) that one patient infected the other. Specifically, because new viral infections only happen by one or few of the virus particles from any one host, we expect that the population of virus particles from the original host will be paraphyletic relative to the newly infected host, because more descendant lines of the virus will be evolving in the original host. The diversity of the viral pools in different patients can also be used to infer relative order of infection, using a molecular clock.
Students are told about the court case, and about the subsequent forensic data. Virus pools were sequenced from blood samples of all six victims and the defendant, and a phylogenetic tree reconstructed for all viral sequences.
Students are charged with three objectives:
- Draw a phylogeny that provides evidence that the defendant was the index case (first identified case of outbreak)
- Explain how that tree shows evidence for the direction of transmission (source → recipients)
- Provide evidence that the victims were infected at the times/in the order they actually had relationships with the defendant
First the instructor presents questions 1 and 2 (above). The instructor provides little instruction while students are drawing their phylogenies, but reiterates salient points about the case itself (e.g. ‘think about depicting the origin of pathogens and what happens after their transmission’). Using a random number generator, the instructor calls up 2-3 groups to present their work on the document camera. Or, if a document camera is not available, iClicker questions can serve to poll the students about their opinion about which tree topology is correct (e.g. S1. slides 21-24). After discussion, and some re-evaluation, students arrive at a tree that includes all victim viral sequences embedded within the defendant viral pool (paraphyly) (S1. slide 20). While students are generally good at depicting Mr. Paddieu’s viral sequences at the root of the tree, a few student misconceptions one might encounter include students who do not embed victim sequences within the defendant pool (i.e., they are all together on a single branch); or the victims’ viral sequences are depicted as nodes rather than tree tips. While all student trees aren’t collected and students are not provided feedback on them, the instructor walks through the correct answer (S1. slide 27), with an emphasis on why a paraphyletic relationship is observed in the phylogenetic tree of viral sequences.
Next, the instructor introduces the concept of an outgroup which are the “controls” in this case. We compare the victim and defendant sequences (CC01-CC07) to sequences of other HIV patients randomly selected from Texas (R01-R04 samples – control cases). The instructor then presents students with the actual phylogeny reconstructed from HIV-1 pol gene for the seven blood samples in the blind test performed by the forensic lab (CC01-CC07) and the controls (Figure 1). The phylogeny shows repeated paraphyly of sequences derived from CC01, the signature expected of an “index patient”, identifying the defendant as CC01. Instructor polls all groups for who the index case is and based on what evidence (S1. slide 30).
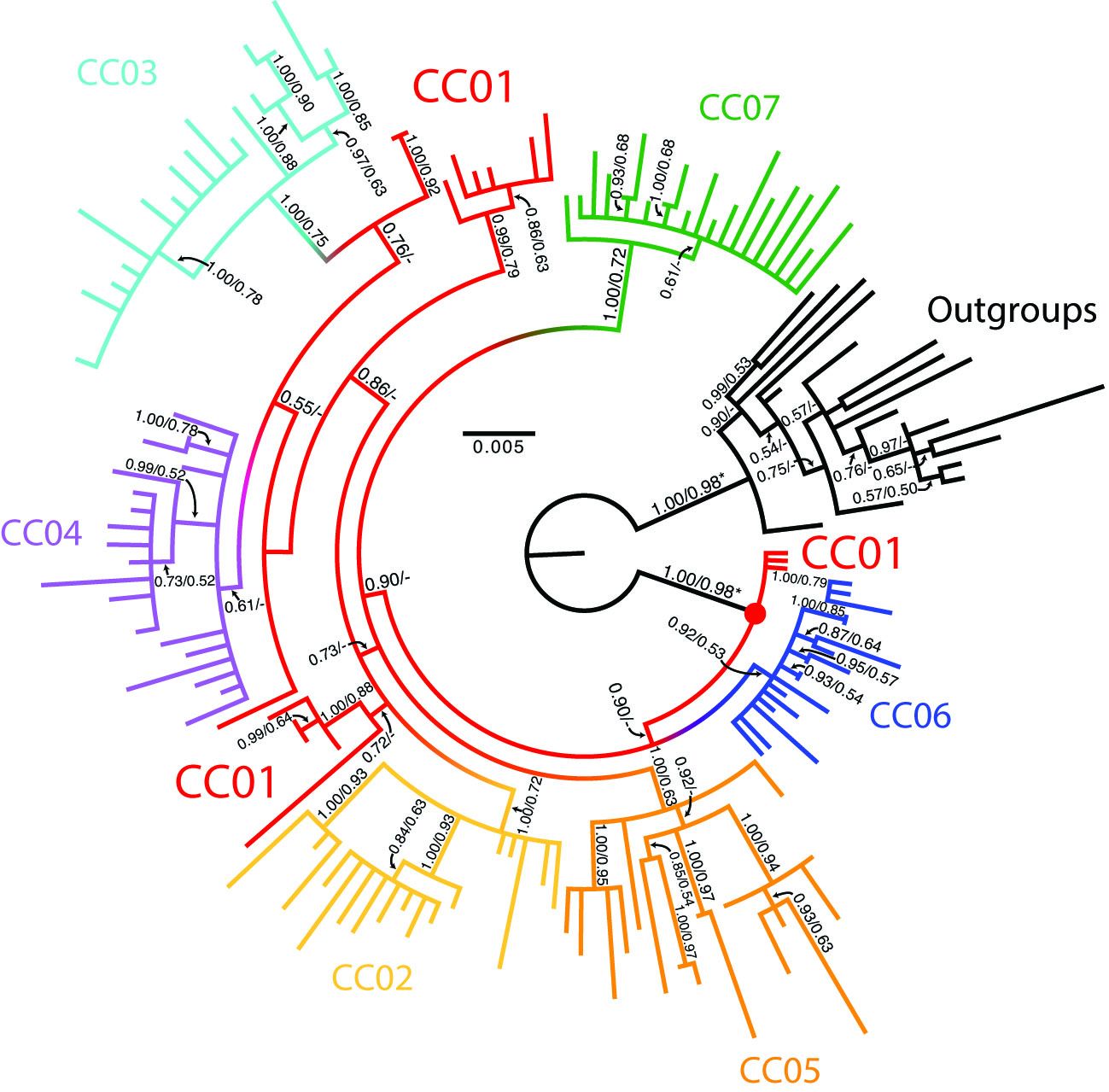
Figure 1. Phylogeny of HIV1 pol genes sequenced anonymously from viral pools of six victims and the defendant (CCO1-CCO7), plus control samples. Used with permission from Scaduto et al. 2010 (5).
Last, the instructor uses the “molecular clock” as a means to judge the relative date of infections for the six victims and compares them to the dates of relationships between defendant and victims known from testimony. Armed with the dates of relationships, students are asked to debate whether the order in which victims were infected matches the order they would infer from the phylogeny, and if not, they are asked to explain why not. Students are presented with the dates that the defendant was in a relationship with each of the victims, and have to examine whether those dates are concordant with the tree. The order of infection as inferred from the tree roughly parallels the order in which the victims dated the defendant, with some exceptions. Students quickly identify that many of the victims had relationships with the defendant overlapping in time, and can follow up with discussions of infection probabilities, and contact rate to explain the temporal pattern inferred from the topology.
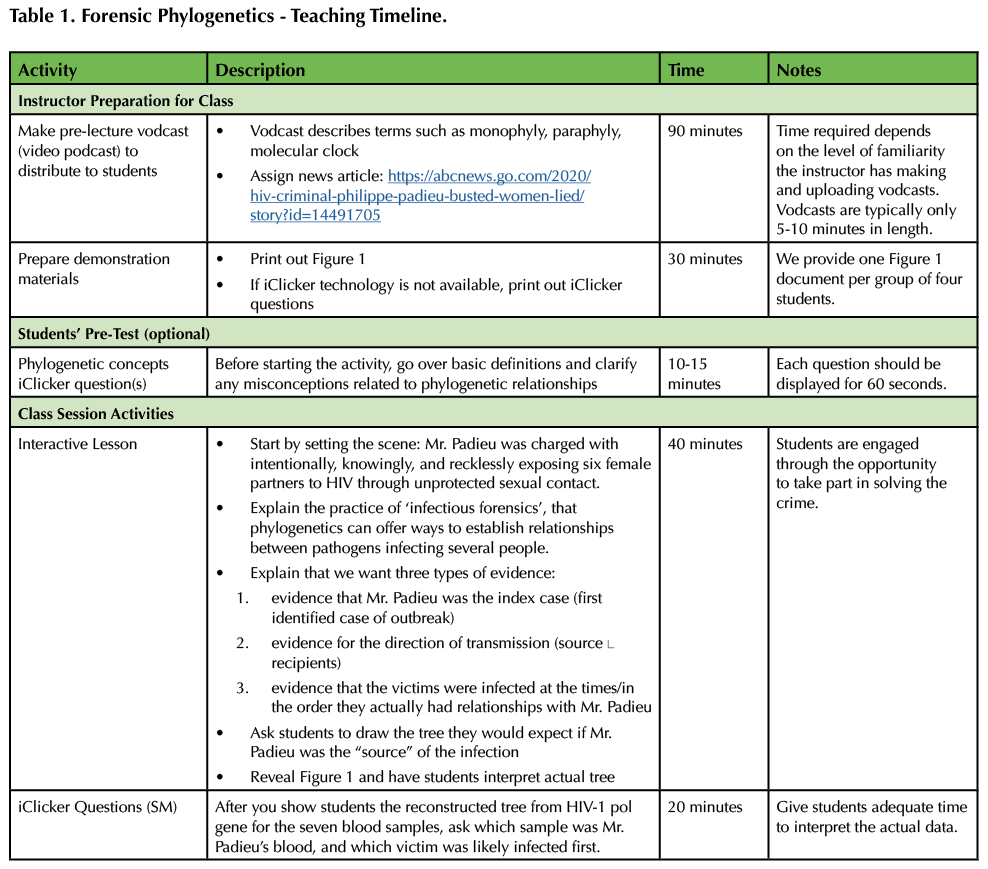
Table 1. Forensic Phylogenetics - Teaching Timeline
TEACHING DISCUSSION
Based on our observations, students particularly enjoyed participating in an activity that was based on a real court case. Students appeared engaged and learned about other aspects in evolution such as rapid genomic evolution in viruses, molecular clocks, HIV transmission, and the principles of forensic phylogenetics.
In fall 2014, this section of the course was taught using similar material, walking students through reading trees and its application in the broader biology literature, but it was presented through traditional lecturing rather than active learning. To gauge student understanding of material after the implementation of active learning in the phylogenetics module, we tracked student performance with the use of ‘tracker questions’ on the first exam. To see more results from the transition to active learning in this course, please see [9]. This work was approved by IRB protocol number 1410005010.
Student groups were comparable in fall of 2014 and fall 2015. In Fall 2014 the course was 60.7% female and 39.2% male; 35.9% Caucasian, 34.9% Asian-American, and 21.4% underrepresented minority (here we define ‘underrepresented’ students as those who are of African or African American, Latino, Pacific Islander, and Native American origin, and majority students as those who are not underrepresented in STEM fields, including white students who are not of Hispanic origin and Asian American students), with 8.1% of students who did not declare their ethnicity. In fall 2015 the course was 55.7% female and 44.3% male; 38.2% Caucasian, 28.1% Asian- American, and 25.4% URM with 7.0% of students who did not declare their ethnicity. Students in fall 2015 entered the course with comparable, but slightly lower, incoming academic preparation according to their math SAT score (fall 2014 average = 715.37, SD = 67.67; fall 2015 average = 710.12, SD = 70.35, 2-tailed t-test = 0.48).
In the two examples below, we show how students improved in their understanding of interpreting phylogenetic trees.
Tracker question 1:
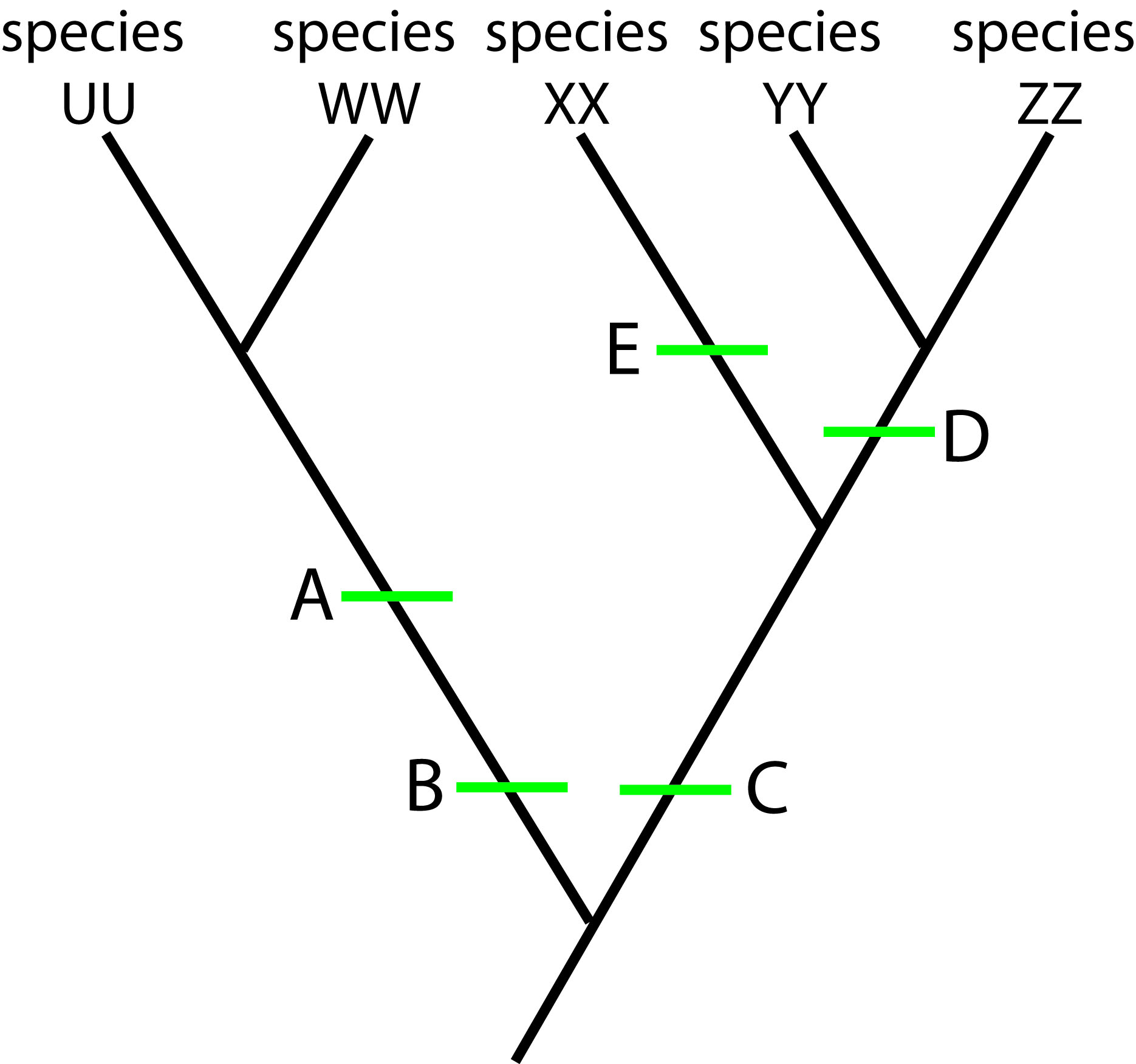
Tracker Question 1 Phylogenetic Tree graphic
In the phylogenetic tree above, species WW is:
- a. more closely related to XX than to ZZ.
- b. more closely related to YY and ZZ than to UU.
- c. more closely related to YY and ZZ than to XX.
- d. equally closely related to XX, YY, and ZZ.
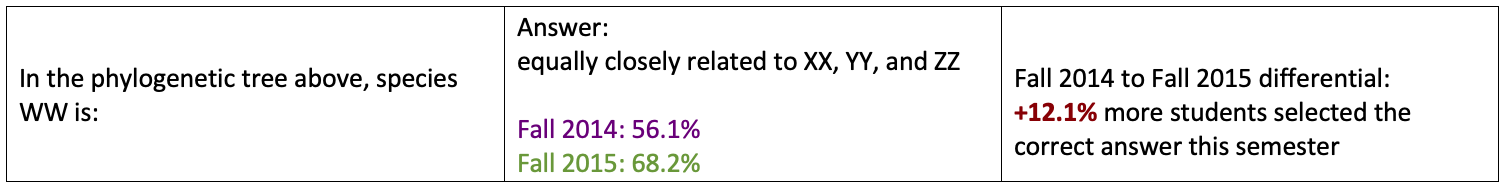
Tracker Question 1: student results
Tracker question 2:
Evolutionary biologists have recently made some exciting discoveries by applying new methods to sequence and analyze Neanderthal DNA [use Figure 2 from Briggs et al (10)].
The figure shows phylogenies constructed from genomic DNA sequences. The modern human DNA samples came from living individuals, while the Neanderthal DNA was extracted from preserved tissue of ages shown on the map in Figure A. Figure B and C show a molecular genealogy of the sequenced genes, so the branch lengths represent sequence divergence (see scale bar), and are NOT calibrated to time. Based on these figures (A-C), answer the question below.
Question: Which of the following statements is most strongly supported by the figures above?
- a. Neanderthals probably migrated among the Feldhofer, Vindija, and El Sidron.
- b. Modern humans occasionally bred with Neanderthals, although certainly less often than they bred with other modern humans.
- c. Modern humans probably evolved from Neanderthal ancestors living in Africa.
- d. Bonobos are more closely related to Chimpanzees than Neanderthals are to Modern Humans.
- e. Modern humans are more closely related to chimpanzees than to Neanderthals.
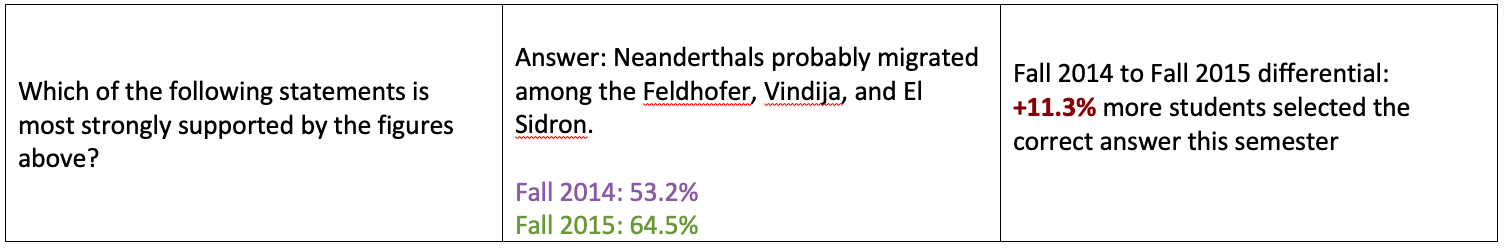
Tracker Question 2: Student results
SUPPORTING MATERIALS
- S1. Forensic Phylogenetics - Lecture Slides. Slides were used in Cornell University’s BioEE1780 Evolutionary Biology & Biodiversity course to teach the activity, Forensic phylogenetics: implementing tree-thinking in a court of law.
ACKNOWLEDGMENTS
We thank P. Lepage, L. Sanfilippo, A. Godert, E. Balko, and Cornell’s Office of Undergraduate Biology, the Center for Teaching Innovation, and Cornell’s Active Learning Initiative for course support. We thank all teaching assistants (particularly R. Petipas, K. Vanderberg, and F. Jóhannesdóttir) and the following instructors of BioEE 1780 for their willingness to experiment with teaching: B. Reed, J. Searle, W. Bemis, A. McCune, I. Lovette, C. Gilbert, and B. Lazzarro. This work was funded by the College of Arts and Sciences and the Active Learning Initiative, Cornell University.
References
- Novick LR, Catley KM. 2007. Understanding phylogenies in biology: the influence of a Gestalt Perceptual Principle. Journal of Experimental Psychology: Applied 13:197.
- Sandvik H. 2008. Tree thinking cannot taken for granted: challenges for teaching phylogenetics. Theory in Biosciences 127:45-51.
- Eddy SL, Crowe AJ, Wenderoth MP, Freeman S. 2013. How should we teach tree-thinking? An experimental test of two hypotheses. Evolution: education and outreach 6:13.
- Goldsmith DW. 2003. The great clade race: presenting cladistic thinking to biology majors & general science students. The American Biology Teacher 65:679-682.
- Scaduto DI, Brown JM, Haaland WC, Zwickl DJ, Hillis DM, Metzker ML. 2010. Source identification in two criminal cases using phylogenetic analysis of HIV-1 DNA sequences. Proceedings of the National Academy of Sciences 107:21242-21247.
- Kummer TA. 2017. PhD Thesis. Brigham Young University, Provo, Utah, United States.
- Eddy SL, Brownell SE, Wenderoth MP. 2014. Gender gaps in achievement and participation in multiple introductory biology classrooms. CBE-Life Sciences Education 13:478-492.
- Tanner, KD. 2013. Structure matters: twenty-one teaching strategies to promote student engagement and cultivate classroom equity. CBE-Life Sciences Education, 12(3), 322-331.
- Ballen CJ, Wieman C, Salehi S, Searle JB, Zamudio KR. 2017. Enhancing diversity in undergraduate science: Self-efficacy drives performance gains with active learning. CBE-Life Sciences Education 16:ar56.
- Briggs AW, Good JM, Green RE, Krause J, Maricic T, Stenzel U, Lalueza-Fox C, Rudan P, Brajković D, Kućan Ž. 2009. Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science 325 (5938), 318-321
Article Files
Login to access supporting documents
Forensic Phylogenetics: Implementing Tree-thinking in a Court of Law(PDF | 598 KB)
S1.Forensic Phylogenetics - Lecture slides.pptx(PPTX | 4 MB)
- License terms

Comments
Comments
There are no comments on this resource.