Translating Co-Translational Translocation
Published online:
Abstract
Understanding the process of proteins targeting to specific locations within a cell is a fundamental part of cell biology. Synthesis of proteins by co-translational translocation, which also determines the resulting protein topology, is a complex process that is often difficult for students to understand. Our goal was to create an engaging lesson to stimulate critical and experimental thinking and help students more effectively learn about co-translational translocation. In the first part of the lesson, students complete a strip sequence activity to order the basic events of co-translational translocation. This activity stimulates peer and class discussion that helps students engage with the material and generate and answer questions. In the second part of the lesson, students complete a four-part guided worksheet that introduces two versions of the protease protection assay with a secretory protein and uses the protease protection assay to evaluate the topology of transmembrane proteins with different types of ER signal sequences. Student responses to in-class clicker questions and exam questions assess student learning. Student survey results indicate the activity was engaging, not confusing, and was helpful for most students.
Citation
Tifft KE, Fisher EJ. 2019. Translating Co-Translational Translocation. CourseSource. https://doi.org/10.24918/cs.2019.40.Society Learning Goals
Cell Biology
- Protein Targeting & Trafficking
- How are cellular components targeted and distributed to different regions and compartments of a cell?
- Methods & Tools of Cell Biology
- How do the methods and tools of cell biology enable and limit our understanding of the cell?
Lesson Learning Goals
Students will understand the:- steps of co-translational translocation.
- role of ER signal sequences in determining protein targeting and topology.
- use of protease protection assays to study co-translational translocation and protein topology.
Lesson Learning Objectives
Students will be able to:- list the steps of co-translational translocation in the correct order.
- describe the key functions of molecules involved in co-translational translocation.
- predict the outcome of co-translational translocation if one of the components is missing.
- identify the characteristics of N-terminal ER signal sequences and internal ER signal sequences.
- predict or interpret the results of a protease protection assay used to assess co-translational translocation or transmembrane protein topology.
- predict the topology of a co-translationally translocated protein when given a description of the ER signal sequence or predict the type of ER signal sequence encoded by the mRNA-based protein topology.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Cells achieve proper localization and topology of proteins, critical factors for proper structure and function, using signals within the protein and other cellular components. Synthesis of most proteins found in the lumen or membranes of the endomembrane system of eukaryotic cells (including the endoplasmic reticulum, Golgi, plasma membrane, and lysosomes) occurs through co-translational translocation. Co-translationally translocated proteins, including luminal proteins, transmembrane proteins, and secreted proteins, cross the membrane of the endoplasmic reticulum (ER) during translation.
The relatively well understood process of co-translational translocation is summarized here, described in textbooks like Molecular Biology of the Cell (1), and reviewed in more detail in the literature (2,3). Co-translational translocation of a protein is specified by translation of an ER signal sequence that binds to the Signal Recognition Particle (SRP). SRP pauses translation by binding to the ribosome and causes the ribosome to associate with the ER by binding to the SRP receptor on the ER membrane. At the ER, the ribosome binds to the translocon, a protein channel that allows the polypeptide chain to cross the ER membrane, and translation resumes. The protein (or parts of the protein) travels through the translocon into the lumen of the ER.
The location of the ER signal sequence within the protein sequence and the presence or absence of additional transmembrane domains determine the topology of the fully-synthesized protein. N-terminal ER signal sequences (also called signal peptides) bind to the translocon so that the N-terminus of the signal sequence is on the cytosolic side of the translocon. Removal of N-terminal ER signal sequences occurs during or after translation by proteolytic cleavage by signal peptidase which is located on the luminal side of the ER membrane. After cleavage of the N-terminal ER signal sequence, the new N-terminus of the protein is located in the ER lumen. Internal ER signal sequences do not undergo cleavage by signal peptidase and become transmembrane domains. The amino acid sequence surrounding internal ER signal sequences determines the orientation of the signal sequence within the translocon and thus the topology of the protein during and after synthesis. The side of the internal ER signal sequence with more positively charged amino acid residues nearby orients on the cytosolic side of the translocon.
A powerful biochemical approach to studying co-translational translocation involves in vitro protein translation in the presence of isolated microsomes, small ER-derived vesicles that support ER functions including co-translational translocation (1,4-6). Protease protection assays reveal the topology of the translation products relative to the microsomal membrane (4-6). The basis of a protease protection assay is the ability of a phospholipid membrane to protect a protein, or part of a protein, from digestion by a protease. Analysis of the protein that remains after digestion (often using SDS-PAGE) provides information about the topology.
A hands-on activity for introductory biology described by Michele Lebonte uses playdough as a way to visualize the changes to proteins during co-translational translocation and the final topology (7). Although there are published problems/questions on co-translational translocation and the protease protection assay (for example, in the Molecular Biology of the Cell textbook and associated problems book) (1), we are not aware of any other published active-learning lessons designed for the classroom. We sought to develop learning activities for our intermediate-level cell biology class that reinforced understanding of the specific steps of co-translational translocation and incorporated the protease protection assay as an experimental approach.
We designed the activities in this lesson to actively engage students in “translating” basic facts about co-translational translocation into a conceptual understanding of the process and the resulting protein topology and stimulate critical thinking. The lesson introduces an experimental approach to study co-translationally translocated proteins and requires evaluation and prediction of experimental data. The lesson incorporates Vision and Change core concepts and competencies including structure and function, information flow, exchange and storage, ability to apply the process of science, and ability to communicate and collaborate with other disciplines (8). The lesson also tightly aligns with learning goals associated with specific cell biology topics endorsed by the American Society for Cell Biology including protein targeting and methods and tools of cell biology (https://qubeshub.org/community/groups/coursesource/courses/cell-biology).
This lesson is composed of two major activities: a strip sequence activity that challenges students to put the steps of co-translational translocation in the correct order and a worksheet that introduces in vitro translation and a protease protection assay to explore protein topology and ER signal sequences (Table 1). We used this lesson during four semesters of a large enrollment (>200 students) cell biology course. We collected student data and survey responses during Spring 2018 and Spring 2019 with exempt status approval from the Homewood Institutional Review Board (HIRB00006962).
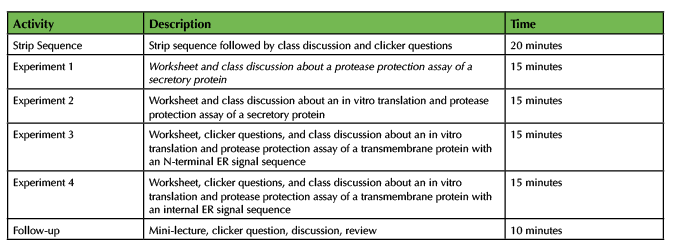
Table 1. Teaching Timeline for Translating Co-translational Translocation
Intended Audience
The intermediate-level cell biology class we designed this lesson for consisted predominantly of sophomores and juniors majoring in biology (or other natural sciences) at a large research university.
Required Learning Time
The in-class time required for these activities as described is about 90 minutes (one or two class sessions).
Prerequisite Student Knowledge
The topics that students need to know before the lesson starts are listed in Table 2 in the "Pre-lesson" section. Our course covered protein translation, biological membranes, and SDS-PAGE prior to the section on co-translational translocation. We created a video introducing the steps of co-translational translocation for a secretory protein and assigned a few homework questions designed to assess basic knowledge (Supporting File S4: Co-translational Translocation- Assessment Questions) due before class started. As an alternative to making your own video, we suggest covering the steps of co-translational translocation by a assigning textbook or other reading, using an existing video, or as a lecture during class (see Table 2 for resources).
Pre-requisite Teacher Knowledge
The topics covered in the lesson and suggested resources for instructors (or students) are listed in Table 2.
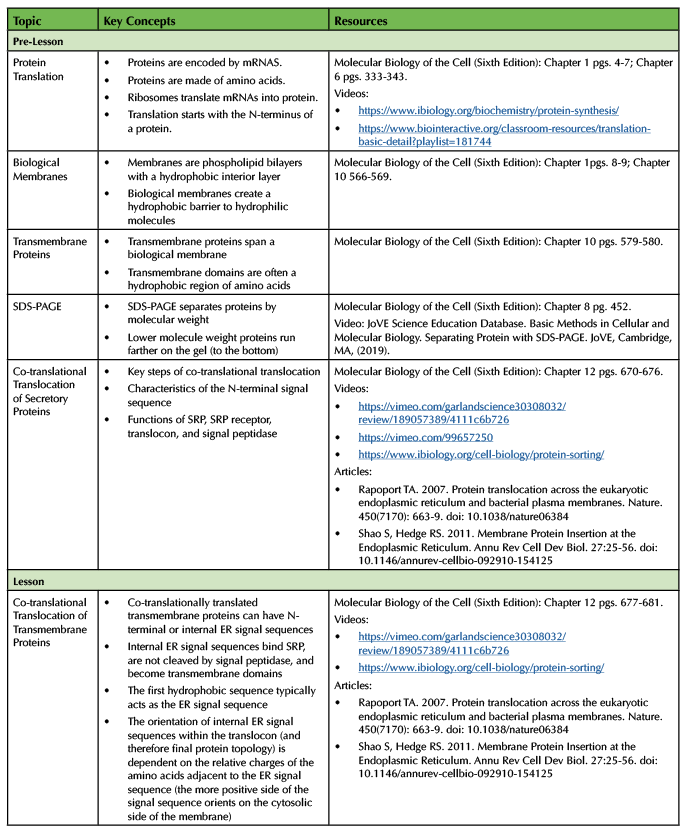
Table 2. List of Lesson Topics and Resources

Table 2. List of Lesson Topics and Resources continued
SCIENTIFIC TEACHING THEMES
Active Learning
During this lesson, students will engage in clicker questions, a strip sequence activity, a worksheet, peer discussion, and group discussion.
Assessment
Learning is assessed by online homework questions, in-class clicker questions, and exam questions.
Inclusive Teaching
This lesson is inclusive because it provides multiple different types of opportunities for students to engage with the material and other members of the class. The strip sequences provide a physical and visual way to interact with the material. The worksheets require students to draw representations of proteins. The peer and group discussions allow the contribution of many student perspectives. Anonymous clicker questions allow everyone, even students who don't want to work in groups or speak aloud, to respond to questions.
LESSON PLAN
Pre-Class Preparation
Strip Sequence Activity
The materials required for the strip sequence activity are a set of twelve paper strips for each group or student. Each paper strip contains a description of one step of co-translational translocation and each set contains all of the steps. The steps within the sets start in random order and students will rearrange the strips in one set into the correct order of events. The strip sequence document (Supporting File S1: Co-translational Translocation- Strip Sequence Document) contains 12 pages, each containing 12 steps of co-translational translocation in a different order, which will generate 12 sets of strips. Print one copy of the document single-sided to make a stack of 12 pages, each with the text in a different order (Figure 1A). Print enough copies of the document to generate enough packets for the desired number of groups. We had students work in groups of three and printed several extras to accommodate students who preferred to work alone or in pairs. Staple each stack of 12 pages 12 times on the left side with a staple aligned with each set of text (Figure 1B). Shuffle the papers to mix up the order of the strips. Cut the stapled stack of papers horizontally (using a paper cutter if possible) between the lines of text to generate sets of 12 strips in random order (Figure 1C). During class, give each group one set of strips. Students will rip the set of strips apart generating 12 individual strips to put in order (Figure 1D).

Figure 1: Strip Sequence Preparation. The photos show the stages of preparation for the strip sequences used in the strip sequence activity. A stack of 12 printed pages (A) is stapled on the left side (B) and cut between lines of text (C) creating stapled stacks of strips that students will rip apart and use to complete the strip sequence activity (D).
Protease Protection Assay Worksheet
Print one copy of the protease protection assay worksheet (Supporting File S2: Co-translational Translocation- Protease Protection Assay Worksheet) for each student.
Class slides
Download the class slides PowerPoint file (Supporting File S3: Co-translational Translocation- Class Presentation Slides). Remove, add, or modify slides. Remove text from the notes section if desired.
In-class Activities
Pre-lesson Clicker Questions
If desired, measure student knowledge before the lesson by asking some assessment questions before the lesson and comparing the answers to the same questions after the lesson as an indicator of learning gains. We asked several of the clicker questions (#1, 2, 3, 8, and 9; Supporting File S4: Co-translational Translocation- Assessment Questions) both before and after the activities (see Assessment of Student Learning section).
Strip Sequence Activity
Briefly introduce the ordering activity by explaining that students should work in groups of three to put the steps on the paper strips in the right order (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 2). We walk around the room to hand out the packets of strips to groups of students while encouraging them to form groups of three. While the students work, we walk around the room answering questions and monitoring progress. The correct order of events is shown on Slide 3 of the class slides (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 3). Cleavage of most signal peptides by signal peptidase (step 11) is thought to occur co-translationally, but the timing varies and cleavage can occur after the completion of translation (9). The order of the other steps is unambiguous based on the current canonical understanding of the process as is presented in textbooks, but this is a simplified and generalized view. When the majority of students finish (assessed by observing or by asking students to click in), bring the class back together to discuss the answers and any questions as a whole class. We tried several different formats for this discussion (see teaching discussion) and recommend asking students to report on the order of events in some format, confirming the correct order of events, and answering any questions. In our experience this activity leads students to ask questions about the exact order of some events for which the answer may or may not be known or may vary depending on the specific protein or organism.
Following the discussion, use clicker questions 1-3 (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slides 4-6) as formative assessment to give students practice thinking about co-translational translocation and feedback on their learning. Clicker questions, answers, and explanations are provided in the notes section of the slides (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slides 4-6). We read each clicker question and the answers out loud before allowing students some time to discuss with their neighbors and vote which typically takes about one and a half minutes in our experience. After revealing the results of the vote, we choose how to proceed based on the results. If the majority of students (>70-80%) answer correctly, we either confirm the right answer and move on or ask a student to describe their reasoning. If a majority of students do not answer correctly, we facilitate an extended discussion (in groups or as an entire class) and/or ask for a revote. After each clicker question, we confirm the correct answer to the question, provide an explanation, and answer any student questions. In the case of questions 1-3, the majority of students answered correctly and did not have many questions so the follow-up was limited and took less than one minute per question.
Protease Protection Assay Worksheet
Start by introducing rough microsomes and the protease protection assay (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 7). The worksheet contains diagrams of the experimental set-up and the SDS-PAGE results for four different experiments. Table 3 summarizes the four experiments and lists key points and important notes.
Experiment 1
In experiment 1, an existing secretory protein inside microsomes is treated with a protease and/or a detergent to demonstrate the principle of the protease protection assay. Introduce experiment 1 (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 8) and instruct students to draw a diagram of the protein they expect in tubes 2-4 based on the representation in tube 1 and the results of the SDS-PAGE. After allowing enough time for the students to work, ask students to describe what they drew in each tube and explain how to use the SDS-PAGE results as evidence. The next slide provides an example of what the students should draw and explanations are in the notes section (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 9). Make sure to explicitly state the correct answers, address key points (Table 3), and answer any student questions.
Experiment 2
In remaining experiments, protein translation occurs in vitro before or after the addition of microsomes. Co-translational translocation of proteins occurs only if microsomes are present during translation. Addition of a protease to the products of translation enables evaluation of the protein topology relative to the microsomal membrane. In experiment 2, the protein of interest is a secretory protein. Introduce experiment 2 (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 10) and instruct students to draw a diagram of the protein they expect in tubes 5-9 and draw the corresponding bands on the diagram of the SDS-PAGE gel. After allowing enough time for the students to work, ask students to describe what they drew in each tube and on the SDS-PAGE gel and explain why. The next slide provides an example of what the students should draw and explanations are in the notes section (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 11). Make sure to explicitly state the correct answers, address key points (Table 3), and answer any student questions.
Experiment 3
In experiment 3 the protein of interest is a transmembrane protein with an N-terminal ER signal sequence. Begin by stating that transmembrane proteins also have ER signal sequences and undergo synthesis by co-translational translocation (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 12). Introduce experiment 3 (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 13). Ask students to focus on the SDS-PAGE results provided and ask clicker question 4 (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 14) which took a total of about 2-3 minutes in our classes. Next, proceed to slide 15 and ask students to draw a diagram of the protein present in each tube and label the N-terminus and C-terminus on each diagram (Supporting File S3: Co-translational Translocation- Class Presentation Slides). When students finish, ask clicker question 5 (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 16). In our experience, this clicker question required time for significant discussion (~5-10 minutes in our experience). To conclude, show the key version provided (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 17) and make sure to explicitly state the correct answers, address key points (Table 3), and answer any student questions. Next, show a diagram or video representing the co-translational translocation of a transmembrane protein with an N-terminal ER signal sequence to help students visualize the process (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 18).
Experiment 4
Introduce experiment 4, in which the protein of interest in a transmembrane protein with an internal ER signal sequence, using slide 19 (Supporting File S3: Co-translational Translocation- Class Presentation Slides). Ask students to answer clicker question 6 (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 20) and facilitate the follow-up discussion (1-2 minutes in our experience). After concluding that the protein does not have an N-terminal ER signal sequence, describe the characteristics of internal ER signal sequences (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 21 and 22). Return to the worksheet diagrams (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 23) and ask students to draw the proteins present in each tube based on the SDS-PAGE results. When a majority of students finish working, ask clicker question 7 (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 24) (1-3 minutes in our experience). The next slide provides an example of what the students should draw and explanations are in the notes section (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 25). Make sure to explicitly state the correct answers, address key points (Table 3), and answer any student questions. Next, describe the insertion of multi-pass membrane proteins based on slide 26 (Supporting File S3: Co-translational Translocation- Class Presentation Slides). Use clicker questions 8 and 9 as formative assessment to test understanding of internal ER signal sequences (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slides 27-28). In our experience, these clicker questions took between 2-4 minutes each. We suggest wrapping up the lesson with a summary of key characteristics of co-translational translocation (Supporting File S3: Co-translational Translocation- Class Presentation Slides, slide 29).
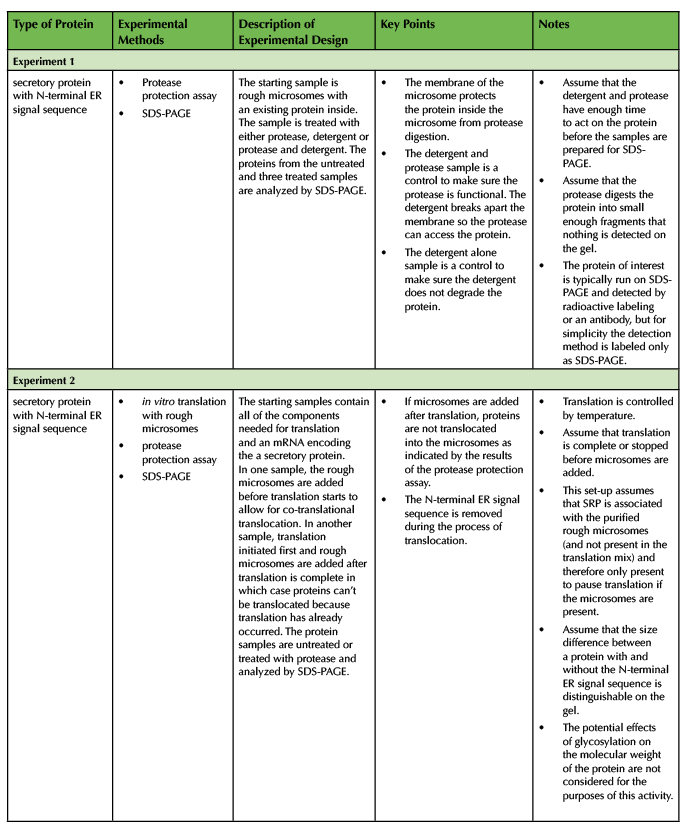
Table 3. Summary of Experiments
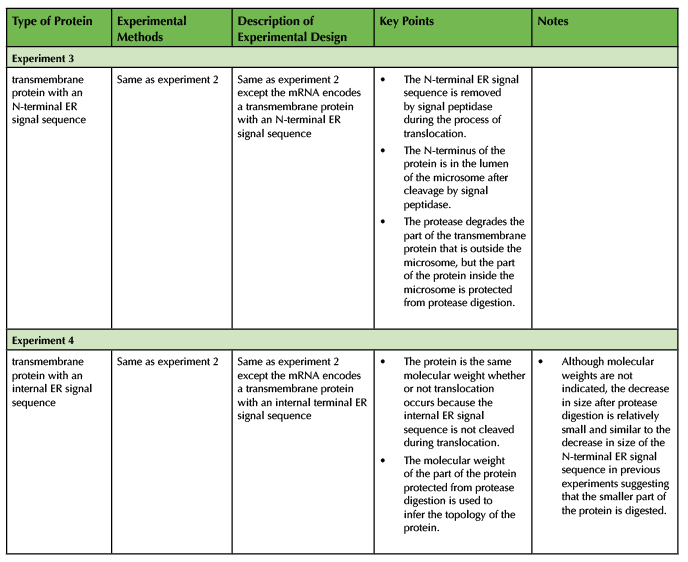
Table 3. Summary of Experiments continued
TEACHING DISCUSSION
Assessment of Student Learning
We assigned online homework questions (Supporting File S4: Co-translational Translocation- Assessment Questions) linked to the material presented in the pre-class video and due before the class in which we used the lesson. Students tend to do very well answering these basic homework questions and in Spring 2019 at least 93% of students correctly answered each question (n=264).
We used clicker questions (formative assessment) and exam questions (summative assessment) to assess student learning. Students earned credit for responding to clicker questions and the credit was not tied to getting the answer correct, thus eliminating any extrinsic incentive for students to get the answers correct. During the Spring 2018 semester we asked five clicker questions (#1, 2, 3, 8, and 9; Supporting File S4: Co-translational Translocation- Assessment Questions) before the lesson, then repeated the relevant questions after the related parts of the class. The purpose of asking the questions twice was to compare student knowledge before the lesson and after the lesson as an indicator for learning gains. We recommend asking the pre-lesson questions only if it is important to measure learning gains because we felt that the questions take up valuable time, the students did not benefit directly from the pre-lesson questions, and the format (asking clicker questions before covering the content and without discussion) was inconsistent with our typical clicker question implementation. During the pre-lesson clicker questions, we read the question and answers and instructed students to answer independently without talking to their classmates which took about one minute per question. We did not reveal responses or provide any feedback. In contrast, for the clicker questions asked during the lesson, we read the question and answers, encouraged students to discuss answers with peers during the voting, and confirmed the correct answer. We also provided feedback, facilitated class discussion, and answered questions as necessary.
Clicker questions 1-3 addressed learning objectives associated with the strip sequence activity (Supporting File S4: Co-translational Translocation- Assessment Questions). The responses to the pre-lesson questions 1, 2, and 3 ranged from 46% correct to 63% correct (Figure 2A). Students had enough information to answer these questions based on the pre-class material, so this reflects students' knowledge after some instruction, but before the activity. When we asked the same questions after the strip sequence activity, the percent of correct responses increased significantly (73%-85%) consistent with student learning during the activity.
Clicker questions 8 and 9 addressed transmembrane proteins and internal ER signal sequences (Supporting File S4: Co-translational Translocation- Assessment Questions). Responses to question 8 before the lesson were about 10% correct, which is even lower than expected by random guessing (Figure 2A). We think that the question format may lead students to think that answer choices c-e are more likely correct than answers a or b. Pre-lesson answers for question 9 were about 28% correct, only slightly higher than expected by random guessing (Figure 2A). We expected a low correct response rate as students had no previous instruction on internal ER signal sequences. The post-worksheet scores were 73% and 63% correct for questions 8 and 9 respectively suggesting a substantial increase in student choice of the correct answer during the activity.
Additional clicker questions (4-7) asked during the worksheet activity related to the protease protection assay and provided feedback (formative assessment) to students and instructors during the learning process (Supporting File S4: Co-translational Translocation- Assessment Questions). Over 50% of students responded correctly to these clicker questions (Figure 2B). The results indicate the majority of the students did the work necessary to come to correct conclusions, but the fact that not all students got the answer correct indicates to us this activity was not too easy and thus likely helpful for learning this complex material.
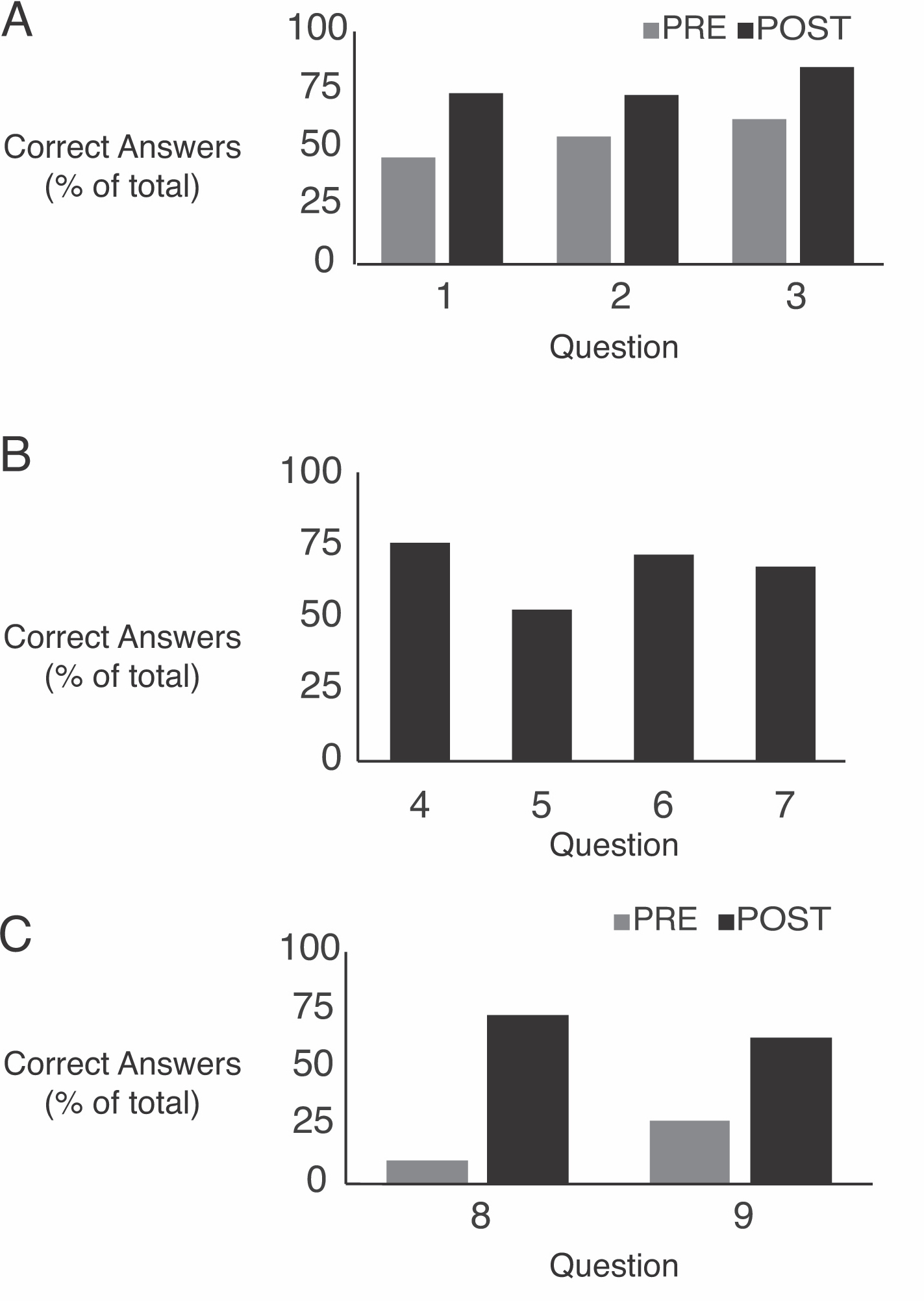
Figure 2: Clicker Question Responses. (A) Percent of correct responses to clicker questions asked pre-lesson or post-lesson. n= 181-193. (B) Percent of correct responses to clicker questions asked during the protease protection assay worksheet activity. n=191-202
We used exam questions on mid-term and final exams as summative assessment. We used different exam questions each semester and therefore cannot make a direct comparison between different semesters. In Spring 2019 the midterm exam (one week after the lesson) contained exam questions 1 and 2 and the comprehensive final exam (12 weeks after the lesson) contained exam questions 3-5 (Supporting File S4: Co-translational Translocation- Assessment Questions). The graph of the results shows the percent of students within each score range for the set of questions on the midterm and final exam (Figure 3). On exam 1, 58% of students scored within the 90% or above and 82% of students scored 70% or above which demonstrates substantial understanding of the concepts. On the final exam 30% of students scored 90% or above and 68% of students scored 70% or above indicating satisfactory understanding and retention of the material. Possible explanations for the lower scores on the final exam include a higher difficulty of questions or the fact that the exam was comprehensive and 12 weeks after the lesson.
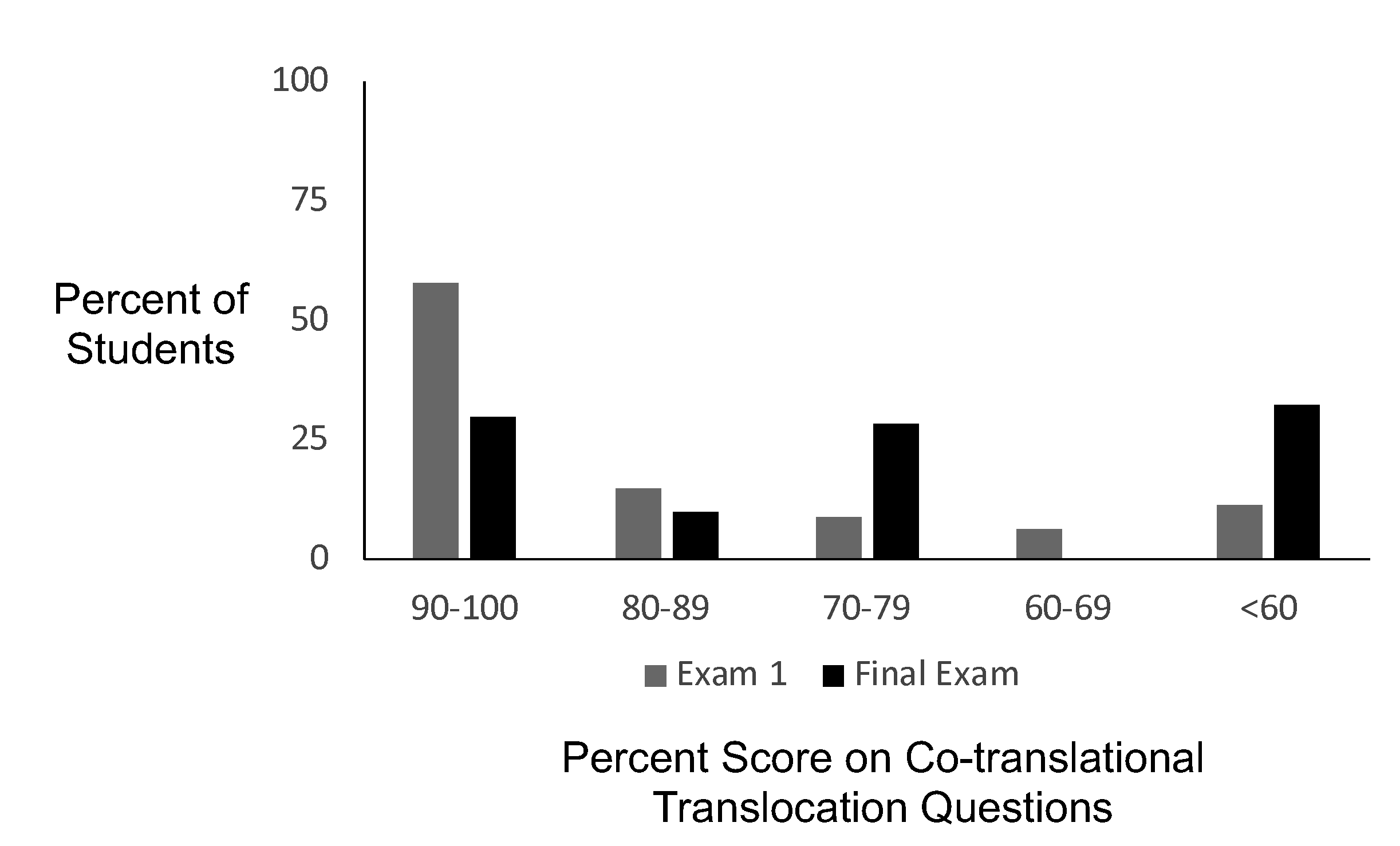
Figure 3: Exam Question Analysis. Percent of students who earned a score within each score range on the co-translational translocation questions on exam 1 (n=292) and the final exam (n=280).
Assessment of Student Reactions
To collect student feedback on the lesson, we included some specific questions on an anonymous and optional survey. In Spring 2019 we deployed the survey questions (Supporting File S4: Co-translational Translocation- Assessment Questions) about 11 weeks after the lesson as part of an end of the semester survey which also included questions about other aspects of the course. Figure 4 summarizes the results of 183 responses from 293 enrolled students (62.8% response rate). The survey question contained six statements, three positive and three negative, for which students indicated their level of agreement with the statement. Over 70% of students agreed or strongly agreed with that statements that the activities were interesting and engaging, improved understanding of the topics, and helped to retain and apply the information (Figure 4A). In contrast, only 20% or less of students agreed or strongly agreed with the statements that the instructions were unclear/confusing, the activities took too much time, and they would prefer lecture instead of the activities (Figure 4A). We also asked "How would you prefer to spend class time for these topics?" to assess the balance of information delivery and activities in this lesson. Over half of students selected "no change in the amount of information delivery" (Figure 4B). The remainder of the responses were almost evenly distributed between students who wanted more information delivery and less in-class activities or less information delivery and more in-class activities suggesting that the balance during this lesson was appropriate for this student population.
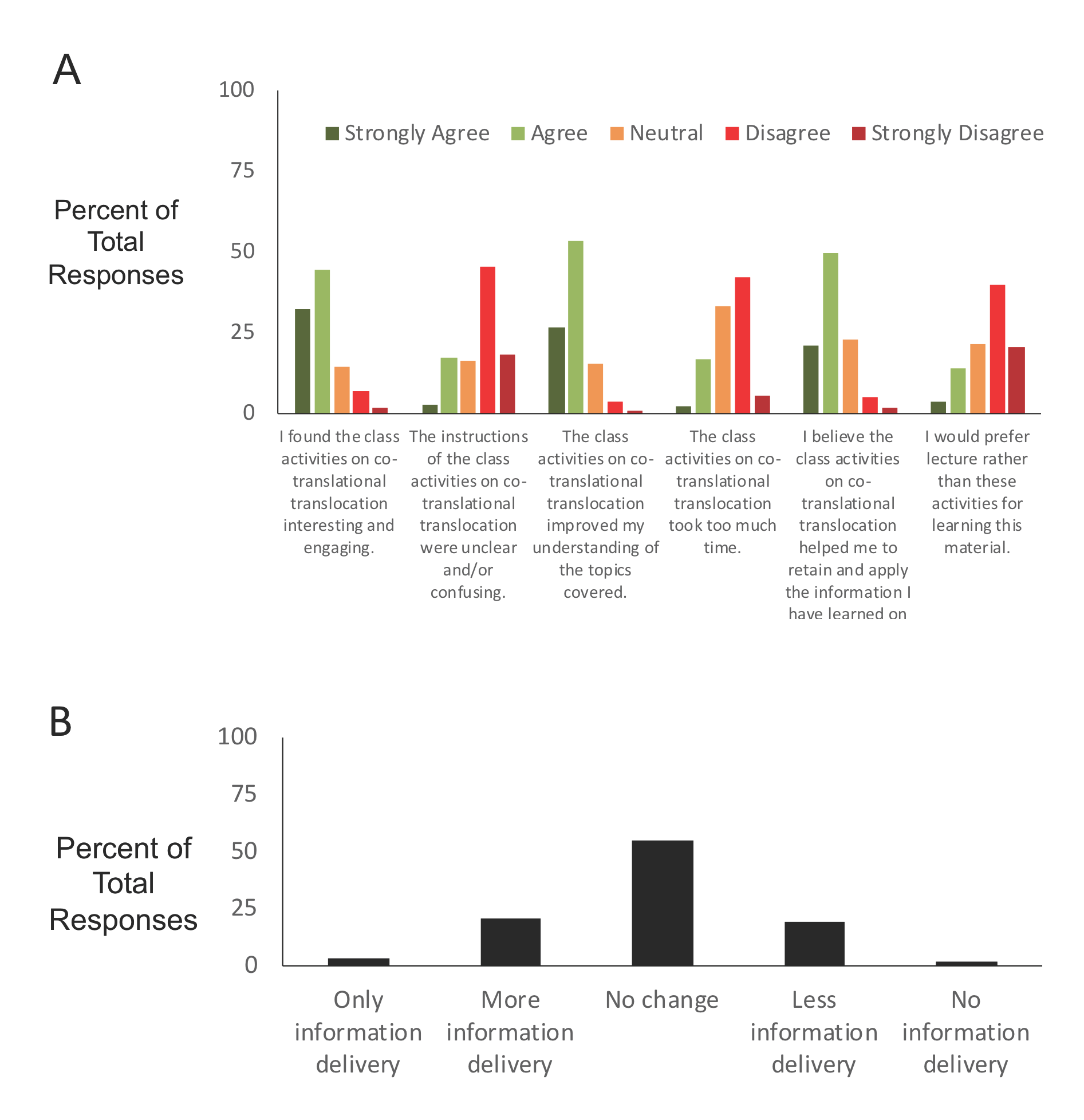
Figure 4: Student Survey Results. (A) Student levels of agreement with statements about the lesson. (B) Student feedback on the amount of information delivery (vs. activities) they would prefer compared to what was done during the lesson. n=183
Improvements/Adaptations/Extensions
In our class, this lesson takes part of two 80-minute classes. Sometimes the class time ends in the middle of a section. We like flexible timing to allow maximum responsiveness to the students, but suggest trying to adjust the timing of the activities to align well with the scheduled class time. Removing any of the sections or using fewer protease protection examples would reduce the time required for the lesson. Potential ways to expand the lesson include spending more time summarizing and reviewing major concepts during the lesson, adding more clicker questions or discussion questions, or adding examples or extensions.
As a follow-up to the strip sequence activity, we used a document camera or PowerPoint to move the strips in real time based on student input to generate the correct order and addressed any questions that arose. In one class, we asked students to talk about any steps that were challenging to put in order or any questions that came up during their discussions (without showing the correct order). Our intention was to encourage students to take a more active role in the learning process by asking appropriate questions and improving the discussion by focusing specifically on the most challenging parts. The discussions seemed similar in time spent and content, but students preferred that we explicitly show the correct order of events in class.
We see many opportunities for extending the material in this lesson and linking it to other related topics. The experiments could use Western blot or autoradiography instead of SDS-PAGE to visualize proteins after the protease protection assay. The protease protection assay could be used to answer different types of questions including post-translational translocation or identifying the topology of proteins in other membrane-bound compartments. A fluorescence protease protection assay using fluorescently-tagged proteins assesses the topology of proteins in a cellular context (10). Microsomes could be used in other in vitro experiments to answer questions about ER functions (like glycosylation) or specific proteins. Hydropathy plots predict hydrophobic regions of a protein that could correspond to ER signal sequences or transmembrane domains. Real proteins could selected for the protease protection assay or follow-up questions. CFTR is an interesting and medically relevant transmembrane protein to use as an example for co-translational translocation, protein folding, ER-associated degradation, vesicle transport, and endocytosis (11).
For a smaller class, instructors could replace the clicker questions and multiple-choice question presented here with open-ended questions. Slight modifications of the questions describe here would provide additional formative or summative assessment. We envision many possibilities for additional new questions including providing diagrams or data and asking students to make conclusions about topology or co-translational translocation or predict experimental results.
SUPPORTING MATERIALS
- S1. Co-translational Translocation - Strip Sequence Document. A PDF file to print to generate the packets of strip sequences to hand out to students.
- S2. Co-translational Translocation - Protease Protection Assay Worksheet. A PowerPoint file to print as a worksheet for each student.
- S3. Co-translational Translocation - Class Presentation Slides. A PowerPoint file containing slides for presenting in class.
- S4. Co-translational Translocation - Assessment Questions. A word file containing homework questions, clicker questions, exam questions, and survey questions.
ACKNOWLEDGMENTS
We would like to thank members of the Department of Biology who provided support and valuable feedback during the development of this lesson. Special thanks to Beverly Wendland for igniting our interest in co-translational translocation and protease protection experiments and encouraging us to develop active methods for teaching this fundamental material. We greatly appreciate the significant contributions of Carolyn Norris and Christov Roberson during the planning and initial implementation of the lesson and Christov Roberson who provided thoughtful comments on the manuscript. We would also like to thank our students for actively engaging in the lesson, asking questions, and providing feedback.
References
- Alberts B, Johnson AD, Lewis J, Morgan D, Raff M, Roberts K, Walter P. 2014. Molecular Biology of the Cell Sixth Edition. New York, NY: Garland Science.
- Rapoport TA. 2007. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 450(7170): 663-9. doi: 10.1038/nature06384
- Shao S, Hedge RS. 2011. Membrane Protein Insertion at the Endoplasmic Reticulum. Annu Rev Cell Dev Biol. 27:25-56. doi: 10.1146/annurev-cellbio-092910-154125
- Blobel G, Dobberstein B. 1975. Transfer of proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J Cell Biol. 67(3): 852-62. doi: 10.1083/jcb.67.3.852
- Nicchitta CV, Blobel G. 1990. Assembly of translocation-competent proteoliposomes from detergent-solubilized rough microsomes. Cell 60(2): 259-69. doi: 10.1016/0092-8674(90)90741-v
- Sharma A, Mariappan M, Appathurai S, Hegde RS. 2010. In vitro dissection of protein translocation into the mammalian endoplasmic reticulum. Methods Mol Biol. 619: 339-63. doi: 10.1007/978-1-60327-412-8_20.
- LeBonte ML. 2013. A Hands-on Approach to Teaching Protein Translation and Translocation into the ER. The American Biology Teacher 75(3):. 211-213. doi: 10.1525/abt.2013.75.3.10
- American Association for the Advancement of Science. 2011. Vision and Change in Undergraduate Biology Education: A Call to Action, Final Report. Available from: http://visionandchange.org/finalreport/
- Land A, Zonneveld D, Braakman I. 2003. Folding of HIV-1 Envelope glycoprotein involves extensive isomerization of disulfide bonds and conformation-dependent leader peptide cleavage. FASEB J. 17(9): 1058-67. doi: 10.1096/fj.02-0811com
- Lorenz H, Hailey DW, Wunder C, Lippicott-Schwartz J. 2006 The fluorescence protease protection (FPP) assay to determine protein localization and membrane topology. Nat Protoc. 1(1):276-9. doi: 10.1038/nprot.2006.42
- Farinha CM, Canato S. From the endoplasmic reticulum to the plasma membrane: mechanisms of CFTR folding and trafficking. Cell Mol Life Sci. 74:39-55. doi: 10.1007/s00018-016-2387-7.
Article Files
Login to access supporting documents
Translating Co-Translational Translocation(PDF | 574 KB)
S1. Co-translational Translocation - Strip Sequence Document.pdf(PDF | 38 KB)
S2. Co-translational Translocation - Protease Protection Assay Worksheet.pptx(PPTX | 693 KB)
S3. Co-Translational Translocation - Class Presentation Slides.pptx(PPTX | 1 MB)
S4. Co-Translational Translocation - Assessment Questions.docx(DOCX | 588 KB)
- License terms

Comments
Comments
There are no comments on this resource.