It's a Substrate... It's a Protein...No - It's an Enzyme! Teaching Using 3D Serine Protease Physical Modeling Activities to Confront Misconceptions.
Editor: Ellis Bell
Published online:
Abstract
Reported misconceptions of enzyme-substrate interactions highlight the necessity for better, targeted instructional tools and assessments. A series of active learning activities with corresponding three-dimensional (3D) physical models were developed to target undergraduate biochemistry students’ conceptual understanding of space, electrostatic interactions, and stereochemistry in enzyme-substrate interactions. This lesson includes two activities utilizing physical models of elastase, chymotrypsin, and trypsin. These enzymes are widely taught in undergraduate biochemistry courses and are exceptional examples of a variety of enzyme paradigms. The Model Exploration activity guides students in an exploration of these models to connect conceptual and visual content. The Problem Solving activity uses two-dimensional representations of the physical models to further build student's understanding of enzyme-substrate interactions. These activities are implemented in two consecutive fifty-minute classes or alternatively combined for a seventy-five-minute class. These lessons are an inclusive, student-centered approach to teaching that enables students to confront misconceptions and promotes mastery of the material.
Primary image: Backbones and Surfaces and Substrates! Oh My! Undergraduate Biochemistry Students Working with the Serine Protease Model Set.
Citation
Terrell CR, Kersten CA. 2021. It's a Substrate... It's a Protein...No - It's an Enzyme! Teaching using 3D Serine Protease Physical Modeling Activities to Confront Misconceptions. CourseSource. https://doi.org/10.24918/cs.2021.33Society Learning Goals
Biochemistry and Molecular Biology
- Macromolecular Structure Determines Function and Regulation
- How are structure and function related?
- What is the role of noncovalent intermolecular interactions?
Lesson Learning Goals
- Students will physically manipulate and explore enzymes in three dimensions.
- Students will understand the importance of shape, electrostatic forces and stereochemistry in enzyme binding and catalysis.
- Students will develop mental models of enzyme binding and catalysis.
- Students will visually compare three serine proteases.
- From the Biochemistry and Molecular Biology Learning Framework (1):
- “How are structure and function related?"
- “What is the role of noncovalent intermolecular interactions?”
- From Threshold Concepts for Biochemistry (2):
- “The physical basis of interactions: Interactions occur because of the electrostatic properties of molecules. These properties can involve full, partial, and/or momentary charges. Correct understanding of noncovalent interactions is essential in integrating structure and function.”
Lesson Learning Objectives
- From Schönborn and Anderson Visual Literacy Skills (3):
- Students will be able to “decode the symbolic language composing an External Representation (ER).”
- Students will be able to “interpret and use an ER to solve a problem.”
- Students will be able to “spatially manipulate an ER to interpret and explain a concept.”
- Students will be able to “translate horizontally across multiple ERs of a concept.”
- From the authors:
- Students will be able to assess the shape (size) and electrostatic properties (polarity) of the enzyme active site to determine suitability for binding and/or catalysis with a substrate.
- Students will be able to assess a substrate’s shape (size), stereochemistry, and electrostatic properties (polarity) to determine suitability for binding and/or catalysis within an enzyme’s active site.
- Students will be able to use structural features of active sites to compare binding and/or catalysis.
- Students will be able to compare the residues of the catalytic site and the specificity (binding) pocket within an enzyme and among enzymes.
- Students will be able to predict the product(s) of a serine protease reaction given the substrate and enzyme.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
Introduction
Our primary aim with this project is to help undergraduate biochemistry students develop mental models for abstract biochemistry concepts. We use physical models to promote student conceptual understanding and visual literacy skills, since two dimensional representations are known to propagate learning difficulties (4). In this paper we refer to physical models as any three-dimensional tactile learning aid that is part of the representation category that includes all models (image, graph, tables, etc.).
During our literature review we learned about student misconceptions related to how enzymes and substrates interact and the Enzyme Substrate Interaction Concept Inventory (ESICI) that measures these misconceptions (5-7). The nineteen misconceptions measured by the ESICI are grouped into five categories: Role of shape and charge in selectivity, How the enzyme interacts with the substrate, Competitive versus noncompetitive inhibition, Conformational change, and Enzyme and substrate characteristics (5). Consistent with our aim, representations of enzymes can be manipulated with molecular modeling software and printed in three dimensions. This generates highly accurate physical models that can be used as learning tools in the classroom. We chose to develop a 3D physical model set of the serine proteases with corresponding activities, because these enzymes are commonly taught in biochemistry courses and the comparison of elastase, trypsin, and chymotrypsin can address several misconceptions documented by Linenberger and Bretz (5). In particular, we designed the activities and physical models to enhance student understanding of how electrostatic properties, stereochemistry and geometry influence enzyme-substrate interactions (Table 1).
Table 1. Enzyme-substrate Misconceptions Targeted with the Model Exploration and Problem Solving Activities
| Misconception (5) | Model Exploration Activity | Problem Solving Activity |
|---|---|---|
| Role of shape and charge in selectivity | ||
| Charged amino acid interacts with OH regardless of sterics | ||
| Students consider charge but not shape | ||
| Similar amino acid will bind in pocket | ||
| How the enzyme interacts with the substrate | ||
| Disregard of relationship between scissile bond and specificity interaction | ||
| Enzyme will bind most tightly to the substrate | ||
| Active site is the only place of interaction | ||
| Allosteric site is not a binding site | ||
| Binding only occurs at transition state | ||
| Active site is on the substrate | ||
| Specificity pocket is not a binding site | ||
| Enzyme will bind most tightly to the active site | ||
| Competitive vs. noncompetitive inhibition | ||
| Inhibitors can bind to the substrate | ||
| Inhibitors interact only via competitive inhibition | ||
| Conformational change | ||
| Allosteric effecter must change enzyme conformation | ||
| An enzyme must change conformation prior to interacting with substrate | ||
| Enzyme and substrate characteristics | ||
| Solvent cannot be a substrate | ||
| Enzyme is a protein therefore a protein cannot be a substrate | ||
| The ‘‘key’’ images represent the enzyme | ||
| Nucleotides cannot be substrates | ||
While our activities are unique in using physical models designed to address identified misconceptions in biochemistry, others have used physical models to engage students with abstract concepts in the molecular life sciences. Oliver-Hoyo and colleagues presented two studies where students utilized physical models of proteins and small molecules to explore noncovalent interactions in structure-function relationships (8,9). Better learning gains and retention were noted when students engaged in a series of learning activities with the physical models rather than a single activity (9). When exploring how physical models can be used to promote better understanding, students who engaged in a “model-dissecting” activity performed better on the post-test assessment than students who did a “model-building” activity (10). Others have investigated the impact of biomolecular physical models on learning gains with respect to gender. One study using a physical model of the Cdc42-interacting protein 4 (CIP4) reported better learning gains for female students, whereas another study demonstrated learning gains for all genders when students used a set of physical models on the flow of genetic information (11,12). The latter study further reported the highest learning gains for lower achieving students (12). In another study by Newman and colleagues, physical models were particularly beneficial for the deaf/hard of hearing (D/HH) student population within their courses (13). This further demonstrates that physical models can support learning for all students.
A few studies have looked at the impact of combining virtual and physical models in the classroom. Students not only prefer the physical models but also rate them as the better tool for aiding in conceptual understanding, compared to other active learning tools (14-16). In one of these studies, the control and intervention groups performed similarly on course assessments; however, when a subset of students from both groups were interviewed, the intervention students answered the “higher order” questions better than the control students (14).
The Enzymes module of our biochemistry course follows the canonical flow of content in undergraduate biochemistry courses (See Figures 1 and 2). The module begins with how enzymes bind substrates with specificity and how enzymes lower the transition state by making optimal shape and electronic interactions with the transition state molecule. Following this, we cover enzyme kinetics, with an emphasis on Michaelis-Menten kinetics. This leads to the serine protease lessons. We use these enzymes at this point in the module, because they are excellent examples of the content covered in the prior enzyme binding and catalysis lessons. We end the module with enzyme regulation content. For our Enzymes module each lesson includes an active learning activity with accompanying physical or virtual model (See Figures 1 and 2).
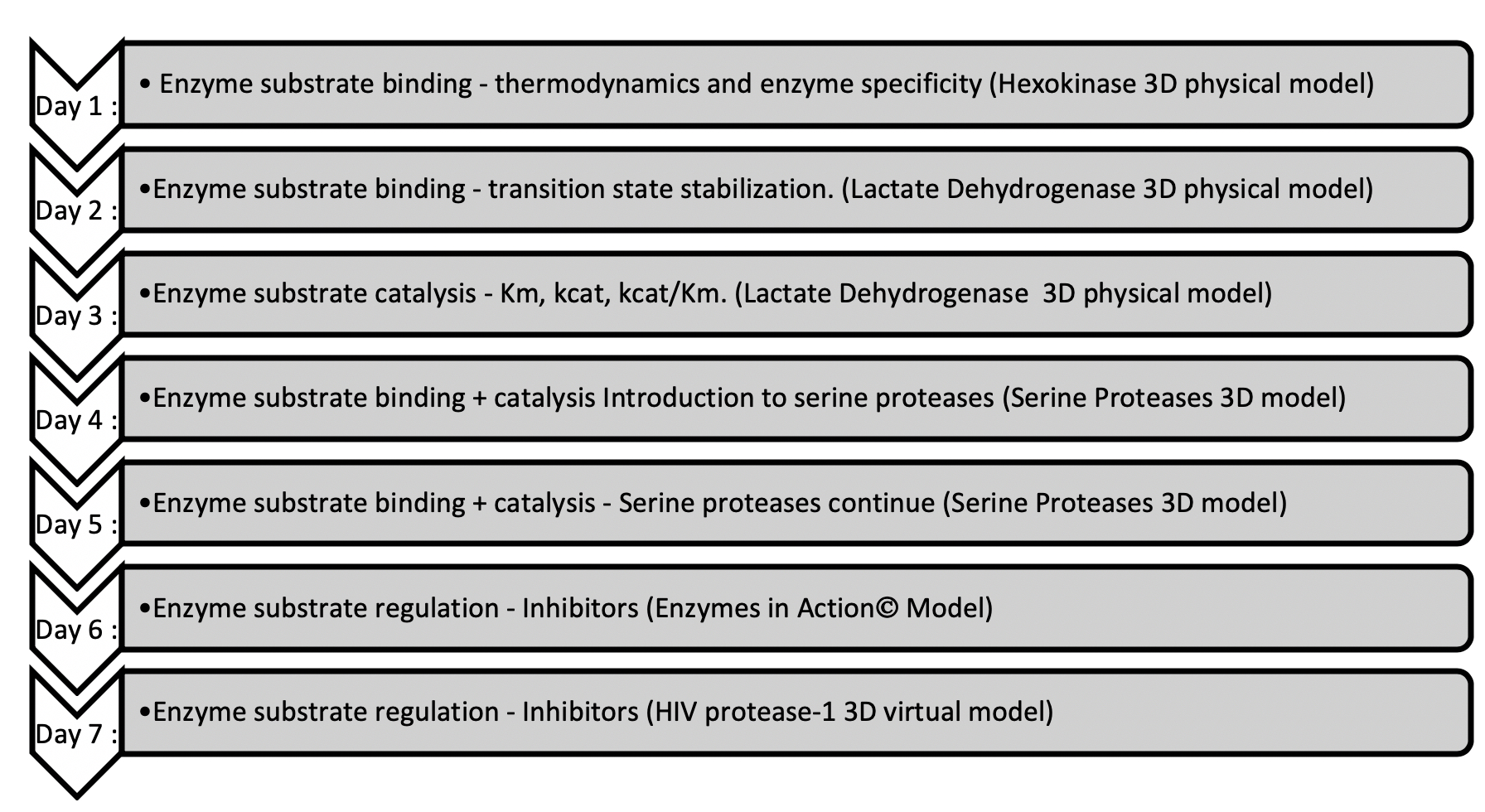
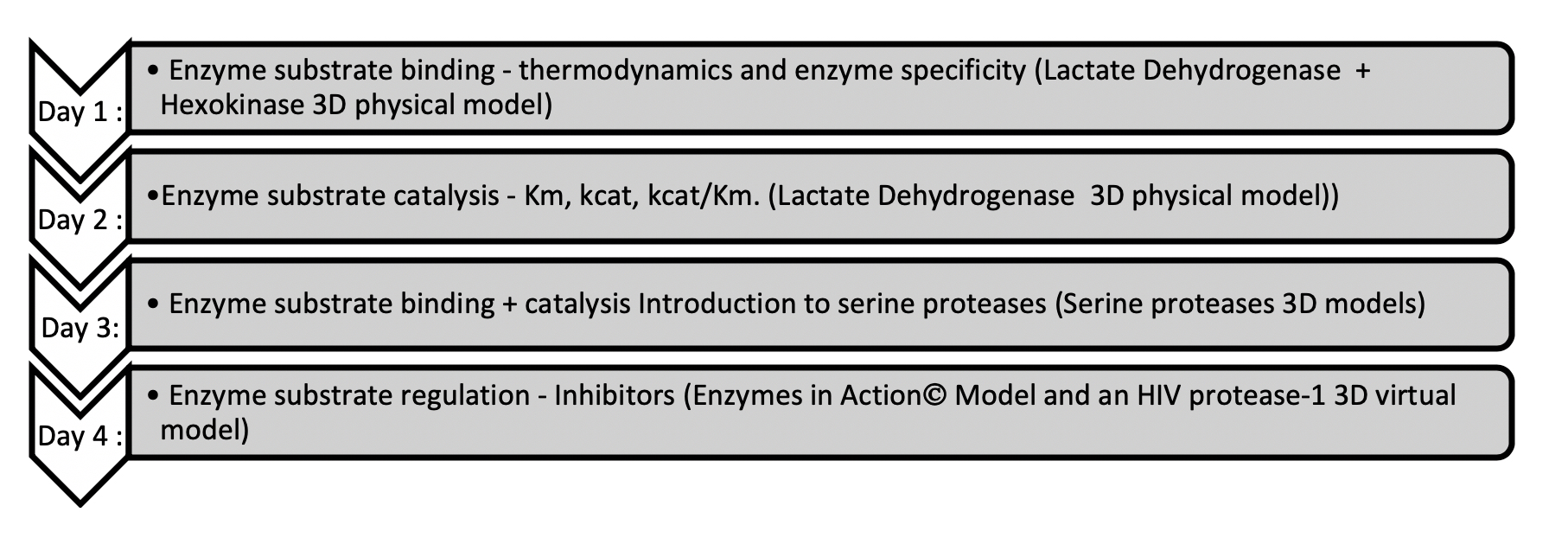
The two activities presented here use the serine protease model set and occur in the middle of our Enzymes module (See Figures 1 and 2). Students complete the Model Exploration activity first and the Problem Solving activity second. Depending on the structure of the course, these activities happen either on consecutive days or are combined into one day. In either case, our biochemistry course is conducted in a flipped-classroom model, with students watching the content video and completing an individual pre-class activity prior to class. At the beginning of class, we begin with student-posed questions from the video and/or pre-class activity. Also, we typically practice the content and representations with recall question(s) from the instructor. After this, students work through the activity in groups using the physical models while the instructor moves through the room answering questions. Each group shares one serine protease physical model set (Figure 3). Groups are guided through the activity with a paper worksheet or learning management system (LMS) quiz. Each student receives a copy of the activity and submits individual answers. A class-wide discussion can occur during the lesson as needed and always at the end. This is a time to go over any questions that students struggle with, pose thought-provoking questions, bring all the concepts and skills together, and allow for reflection.
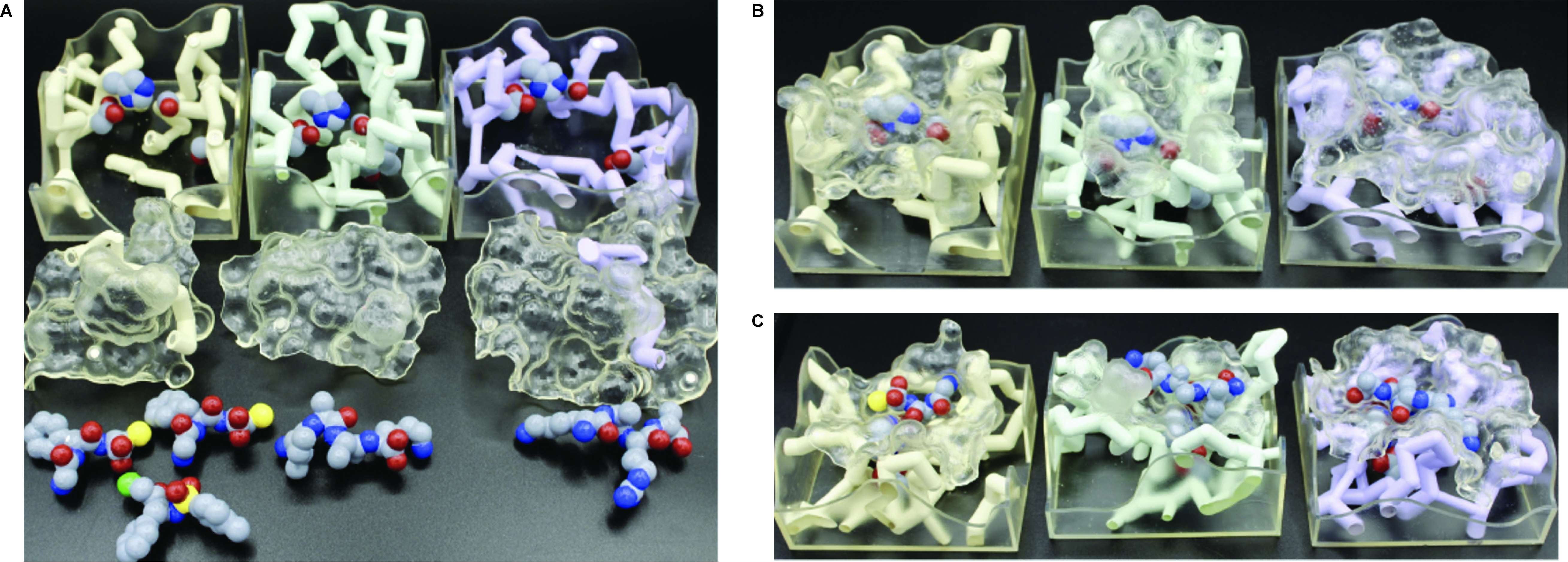
Intended Audience
The intended audience for these activities is third- and fourth-year undergraduate students enrolled in an upper-level biochemistry course. Our students are pursuing a Bachelor of Science in Health Sciences degree at a small, public, primarily undergraduate institution. Prior to enrollment in our biochemistry course, students have completed four semesters of college-level chemistry (general chemistry I and II, organic chemistry I and II), at least one semester of college-level biology, and at least pre-calculus, with a minimum grade of C- in all prerequisite coursework.
Required Learning Time
The Model Exploration and Problem Solving activities can be completed in two consecutive 50 minute course periods. Alternatively, they can be combined for longer class times or laboratory times. These models could also be used in the laboratory setting for more in-depth discussion and to offer students more time to build their visual literacy skills. Prior to class, students individually watch a content video (~37 minutes) and complete a pre-class assignment (~20 minutes).
Prerequisite Student Knowledge
Prior to the Enzymes module, students complete the Introduction to Biochemistry and Protein Structure and Function modules. Students should also be familiar with CPK (Corey, Pauling, and Koltun) colors and the various renderings of macromolecules (e.g., ball and stick, surface, ribbon diagram, etc.). Our students are required to memorize the one and three letter amino acid codes, along with biologically relevant functional groups. Also, they can draw these functional groups and amino acids in skeletal and Lewis structure form at any pH between 0 – 14. Our students are also able to draw and identify non-covalent interactions between functional groups.
Prerequisite Teacher Knowledge
The instructor needs knowledge of elastase, trypsin and chymotrypsin with respect to enzyme specificity and catalytic mechanism. We created an instructor’s guide to using the models (Supporting File S3: Enzyme Substrate Interaction - Instructor Guide Video on Teaching using Serine Protease models) and an accompanying description of the timeline in Table 2.
Table 2. Descriptive Timeline of Topics in the Instructor Guide Video.
| Timeline of Topics in Instructor Guide Video (S3) | Time in video |
|---|---|
| Introduction and considerations for what to say at beginning of class time, how to orient the students with the models, whether to use these in-class or in-lab. | 0 – 3:32 min |
| Discussion as to whether or not students learn better with the serine protease models. | 3:32 – 4:17 min |
| Re-capping the pieces of the model, the renderings, and the meanings of each piece. | 4:17 – 5:02 min |
| Role-playing the first task in the Model Exploration Activity – matching the surface plates to the backbone models. | 5:02 – 6:20 min |
| Role-playing the second task in the Model Exploration Activity – make observations and identifying which model is which protease. Discussion points to have with students for this task. | 6:20 – 9:14 min |
| Role-playing the third task in the Model Exploration Activity – comparing the catalytic and binding site of the enzymes | 9:14 – 10:44 min |
| Classroom management and common student behaviors to notice. | 10:44 – 11:12 min |
| Role-playing the fourth task in the Model Exploration Activity – placing the CPK stickers on the surface plate, making observations and common student struggles with this task. | 11:12 – 13:36 min |
| Role-playing the fifth task in the Model Exploration Activity – matching the substrate to the enzyme, making observations and common student struggles with this task. | 13:36 – 21:10 min |
| Discussion points to have with students at this point in the Model Exploration Activity | 21:10 – 21:50 min |
| Role-playing the sixth task in the Model Exploration Activity – placing gem stickers on catalytic site and correct peptide bond to help students understand the specificity of hydrolysis. | 21:50 – 23:14 min |
| Role-playing the seventh task in the Model Exploration Activity – observing the impact of stereochemistry on binding and/or catalysis. Using the epimer substrates. | 23:14 – 24:40 min |
| Discussion around the inhibitor model and how it can be used in class, either in addition to this activity or in the enzyme regulation class session. | 24:40 – 25:37 min |
| Common student misconceptions, struggles, and classroom management | 25:37 – 28:10 min |
Scientific Teaching Themes
Active Learning
Activities outside of class:
-
Students watch a content video prior to class.
-
Students complete a pre-class activity prior to class.
Activities used during class:
-
Problem-based learning within the activity.
-
Group discussion of the activity questions.
-
Group use of the physical models.
-
Group and individual reflection.
-
Class-wide discussion.
Assessment
Learning was measured using these methods:
-
Assessment of student responses to the activity questions, using either rubric analysis or scores generated from the LMS.
-
Pre/post analysis of the Enzyme Substrate-Interaction Concept Inventory.
-
Assessment of student responses to exam questions pertaining to the serine proteases.
-
Overall student performance on exam 2 (pertains to the Enzyme and Lipids modules).
-
Students self-evaluate learning by reviewing answer keys that correspond to pre-class and in-class activities. Students also engage with practice problems and study guide questions related to the materials. Additionally, each class session ends with a reflection.
Inclusive Teaching
These activities use all five strategies for developing a culture of inclusivity and equity in the classroom described by Tanner (17): (1) we know our students’ names, which helps facilitate the conversations that arise during the lesson; (2) relevant examples of treatments of peptidic ulcers and acid reflux using protease inhibitors are incorporated into the class discussion; (3) the majority of class time is spent with students collaborating in small groups; (4) the lessons use various active learning strategies (i.e., physical models, collaborative work, reflective writing, interactive lecture, pre/post questions); and (5) we explicitly talk about how the physical models and activities promote access and equity for all students. In class, we talk about how spatial abilities are typically lower in female students due in part to the toys and games females are introduced to as children. This slow development of spatial ability has been linked to lower retention and success in STEM (18, 19). Ideally these models will help those with low spatial ability train their brains to build a more robust mental model and therefore increase their success and retention in STEM programs. Additionally, the models are in CPK colors which should be accessible to those with color blindness.
Lesson Plan
Classroom Seating Arrangement
The University of Minnesota Rochester classrooms are called “Learning Labs;” with tables arranged in a circle around the room or on wheels that can be moved at the instructor’s discretion. Students sit at tables with up to six classmates. For these activities, students work in groups of six, each sharing one model set. Students are assigned to groups randomly using the course LMS.
Pre-Semester Preparation
While we have our own set of models, the Milwaukee School of Engineering (MSOE) Center for BioMolecular Modeling has a set to borrow through their Lending Library. We administer the Enzyme-Substrate Concept Inventory (ESICI) as a method of assessment at the beginning and end of the semester. For access and permission, contact Stacy Lowery-Bretz (bretzsl@miamioh.edu).
Preparation for Serine Protease Model Exploration Class Session
Prior to the Model Exploration in-class activity, students watch a content video and complete the corresponding pre-class assignment (Supporting File S1: Enzyme Substrate Interaction - Pre-class Activity Serine Proteases). The content video, available on YouTube or by request, compares the active sites of three serine proteases: chymotrypsin, trypsin, and elastase. The role of each catalytic triad residue and the role of key binding pocket residues are described. Additionally, the arrow-pushing serine protease catalyzed mechanism for chymotrypsin and a hypothetical substrate is drawn and talked through in detail. The video also ties in prior knowledge from the previous activities in the Enzyme module. The pre-class assignment is due at the beginning of class and the answer key (Supporting File S2: Enzyme Substrate Interaction - Pre-class Activity Serine Proteases Answer Key) is provided to students after the due date. Students complete the pre-class activity worksheet individually and by hand, so the assignment is posted as a Word and PDF document for accessibility. The pre-class activity and content video are available to students at least one week prior to the Model Exploration in-class activity.
Also, a week prior to the in-class session we acquire the stickers, check over the models and prepare a cart for ease of transport (see Table 3. Lesson Plan for the Model Exploration and Problem Solving Activities). The in-class activity worksheet (Supporting File S4: Enzyme Substrate - Model Exploration Activity) is either printed so that each student has a copy or created in our LMS for individual students to complete.
Table 3. Lesson Plan for the Model Exploration and Problem Solving Activities
| Activity | Description | Estimated Time | Notes |
|---|---|---|---|
| Preparation for Serine Protease Model Exploration Class Session | |||
| Request the physical models |
Send request to the MSOE Lending Library for a set of the serine protease physical models |
10-15 minutes to fill out the request form. | |
| Administer the Concept Inventory (Optional) |
|
20 minutes to administer the ESICI |
|
| Pre-class preparation (At least two – three days prior class periods) |
|
Several class periods to cover prerequisite enzyme content. About 20 minutes to set-up the LMS activity. |
|
| Pre-class preparation (At least one day prior to class) |
|
About 15 minutes to prepare 12 sets. About 30 minutes to set-up the LMS activity. |
|
| Pre-class preparation (10-15 minutes prior to class) |
For each group, lay out the models Note that we have groups of 6-9 students share one model set. |
10-15 minutes prior to class |
Lay out all pieces individually:
|
| Serine Protease Model Exploration Activity Class Session | |||
| At beginning of class, instructor-led Q&A |
|
~ 10 minutes |
These are some questions we cover using the white board drawing (see Figure 5):
Then orient the students to the models as such:
|
| Students begin the activity in small groups | Hand out the printed group activities (one per person) OR open the LMS activity. Remind students to share and to use soft hands. The models are breakable. | ~ 20-40 minutes | |
| Class-wide discussion | Time to wrap- up class with a reflection. | ~5-10 minutes | “Which model (surface plate, backbone, small molecule) best represents electrostatic features? Which model (surface plate, backbone, small molecule) best represents geometric complementarity, and which model (surface plate, backbone, small molecule) best represents stereochemistry?” |
| After class |
|
||
| Preparation for Serine Protease Problem Solving Class Session | |||
| Pre-class preparation (At least one day prior to class) |
|
10-15 minutes prior to class |
|
| Serine Protease Problem Solving Activity Class Session | |||
| At beginning of class, instructor-led Q&A | Ask if students have questions/comments. | ~5 minutes | |
| Students begin activity in small groups | Hand out the printed group activities OR open the LMS activity. Remind students to share and to use soft hands. The models are breakable. | ~20-30 minutes | |
| Class wide discussion | Go over questions. | ~20 minutes |
Also pose the following questions for the class (we usually draw on the white board):
WEAR → (chymotrypsin) W + EAR → (trypsin) W + EAR → (elastase) W + EA + R Overall answer: W + EA + R (Our students have their one and three letter codes memorized along with the structures of all the amino acids and can draw them at any pH between 0 – 14.)
|
| After class ends – clean-up |
|
10-15 minutes | |

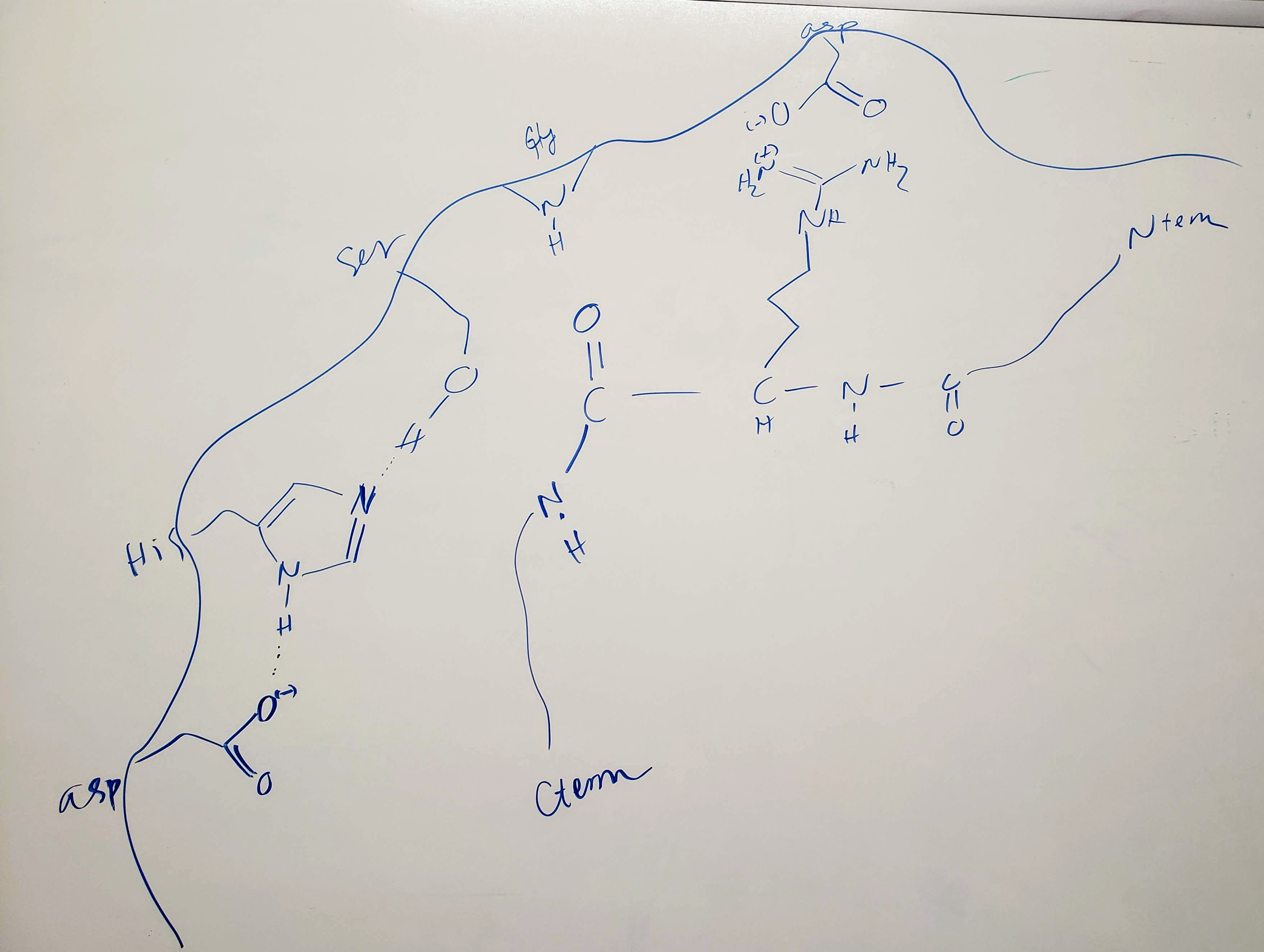
As indicated above, we created an instructor guide video that highlights the features of the models and how we use these models with the students (Supporting File S3: Enzyme Substrate Interaction - Instructor Guide Video on Teaching using Serine Protease models). Watching this video can help prepare instructors for this set of lessons. Additionally, we describe the video timeline in Table 2.
Serine Protease Model Exploration Activity In-Class Session
Prior to the start of class, the model kits are placed on the tables with all of the pieces separate. Class begins with an instructor-led recall discussion (see Table 3. Lesson Plan for the Model Exploration and Problem Solving Activities for script) using a whiteboard drawing of a serine protease active site (see Figure 5). We then orient students to the models (see Table 3. Lesson Plan for the Model Exploration and Problem Solving Activities for script) and follow that by either passing out the hard copy of the activity worksheet or directing students to the online LMS version. Since many students want to play with the models and ignore the activity, we remind the students to work through the questions in order. Although each student has a copy of the in-class activity, they are encouraged to work together. Our students readily interact with one another on this assignment, in part because they must share the model set. As students work though the questions, we wander through the room and answer questions. We frequently help re-orient students with the models; like showing them how to find the N-terminus of each peptide substrate, asking them where the catalytic versus binding residues are located, and/or showing how to find the best placement of the substrate in the active site. Often, we notice that the female students are reluctant to work with the models, whereas the male students are not, so we give gentle reminders to share and be equitable with the models. In the last ten to fifteen minutes, we lead a class-wide discussion of the activity, models, commonly missed concepts, and reflection (see Table 3. Lesson Plan for the Model Exploration and Problem Solving Activities for script). The activity worksheets are collected or submitted online, and the entire activity is graded on correctness (See Supporting File S6: Enzyme Substrate - Model Exploration Activity Answer Key and Rubric). Prior to leaving the classroom, students help place the models back on the cart.
Preparation for Serine Protease Problem Solving Class Session
A day prior to the second in-class session we check over the models and prepare a cart for ease of transport (See Table 3. Lesson Plan for the Model Exploration and Problem Solving Activities). The in-class activity worksheet (Supporting File S5: Enzyme Substrate -Problem Solving Activity) is either printed so that each student has a copy or created in our LMS (we use Canvas, so this activity is set up as a “Quiz”).
Serine Protease Problem Solving Activity In-Class Session
Prior to the start of class, the model kits are placed on the tables with all of the pieces separate. At the beginning of class, the instructor covers any questions posed by the students. Typically, there are few, if any, questions at this time. This shorter introduction to class enables a longer end-of-class discussion. We then hand out the printed activities or direct students to open the LMS activity. Again, we move through the room and answer questions as students work through the activity. At the end of class, or throughout, we cover a series of questions as a class (See Table 3. Lesson Plan for the Model Exploration and Problem Solving Activities for script). We have whiteboards along every wall in our classroom, so one student from each group will be at the whiteboard answering a question. Groups rotate through students for each question. At the end of the class the activity worksheets are collected or submitted online. For this activity we grade the entire activity on correctness (See Supporting File S8: Problem Solving Activity Answer Key and Rubric - the answer key and rubric to the Problem Solving activity). Before leaving the classroom, students help store the models.
Teaching Discussion
The previously documented research about misconceptions of enzyme-substrate interaction expressly points to a need for better teaching tools, particularly those that are three-dimensional (5-7). We developed two such lessons with 3D physical models of serine proteases. These lessons have several benefits. First, by having all three serine protease models, students are able to physically manipulate and see their similarities and differences. This enables a physical understanding of the role of size, stereochemistry, and electrostatic forces in enzyme-substrate interactions. In doing so, students also engage in four visual literacy skills (3), and while we cannot directly measure the development of mental models, we perceive this is happening based on discussions with students and from their responses to the activity questions. Physically manipulating the three enzymes also helps students with the misconception that a “specificity pocket is not a binding site.” Additionally, the models make it easy for the instructor to address other misconceptions, such as “the ‘key’ image represents the enzyme” and “the enzyme will bind most tightly to the active site.” For example, asking students to hold up a substrate, then to hold up an enzyme, and then to point to an active site helps to address the “active site is on the substrate” misconception. Furthermore, asking students to hold up the molecules with a peptide bond addresses the “enzyme is a protein and therefore a protein cannot be a substrate” misconception. Additionally, these activities facilitate robust discussions that can help students build better shared mental models of enzyme-substrate interactions. These discussions help students teach each other, confront misconceptions, and gain further mastery of the material in a low-stakes environment. The formative activities also give the students further practice to master the material. In addition to the small group collaborations and varied active learning strategies, these lessons incorporate relevant examples and promote development of spatial reasoning that facilitate an inclusive teaching environment.
Improvements and Adaptions
These lessons were employed by sections of Biochemistry I with one instructor and up to 33 students in a section. Due to the amount of interaction between the instructor and student groups scaling this up to larger section sizes would likely require additional teaching help, from either teaching assistants or instructional colleagues. Additionally, these activities could be adapted to a laboratory setting. Here the instructor could expand the activities to include enzyme inhibition.
In general, students needed more time with the in-class activity when the physical models were present, compared to the students who used the activity without the models. Models must be introduced in a systematic way, and students need time to explore both the meaning and the use of models (20). Our observations of students interacting with the physical models affirm this finding. There is more to process and connect when transitioning from 2D representations to the 3D physical models. As such, the beginning of class time is spent orienting students to the models around the meaning of colors, orientation of the catalytic site, what the surface plate represents, which pieces represent the enzyme versus the substrate, and the intended purpose of providing the models to the students. This initial orientation to the models helps creates a shared mental model facilitating discussion that enriches student learning.
Anecdotally, we observed some student difficulties during the lessons. Some students struggle to identify the catalytic triad and the binding pocket. They seem to have trouble reconciling that these are unique, geographical locations in the active site of serine proteases. Additionally, some students refer to the surface plate and backbone as ‘separate molecules’ and talk about the amino acid residues shown on the backbone as molecules - instead of as part of the protein. Further refinement of the activity and/or corollary class wide discussions with the instructor modeling what they ‘see’ when looking at this physical model set could improve student understanding. Also, many students will treat the model set as a puzzle-solving experience and ignore the activity, so it’s important to remind students early that there is an activity with questions that are meant to be followed in order.
In order to better help students prepare for the lesson, the pre-class activity could be altered so that students create backbone and surface renderings of these enzymes using a virtual modeling program, like UCSF Chimera or PyMOL. There is some evidence that combining physical and virtual models leads to better learning gains (14). The pre-class activity could also have students label the catalytic triad and binding pocket. Additionally, conceptual physical models could be used for students to gain a general understanding of the geometric, electronic, and stereochemical aspects of enzyme-substrate interactions prior to introducing the serine protease kit (for example Enzymes in Action and the Substrate Specificity kits from 3D Molecular Designs). These types of conceptual models could be introduced in first- and second-year biology courses, like introductory and cellular biology, to better prepare students to use the serine protease set in biochemistry. These alterations to the curriculum may decrease the perceived cognitive load of these models for the students and lead to deeper learning.
SUPPORTING MATERIALS
- Supporting File S1: Enzyme Substrate Interaction – Pre-class Activity Serine Proteases. An activity for students to complete after watching/learning about serine proteases and before attending class.
- Supporting File S2: Enzyme Substrate Interaction – Pre-class Activity Serine Proteases Answer Key. The answer key for the pre-class activity.
- Supporting File S3: Enzyme Substrate Interaction – Instructor Guide Video on Teaching using Serine Protease models. A video for instructors demonstrating how to use the serine protease model set.
- Supporting File S4: Enzyme Substrate Interaction – Model Exploration Activity. The first activity for students that accompanies the model kit.
- Supporting File S5: Enzyme Substrate Interaction – Problem Solving Activity. The second activity for students that accompanies the model kit.
- Supporting File S6: Enzyme Substrate Interaction – Model Exploration Activity Answer Key and Rubric. The answer key and rubric to the Model Exploration activity.
- Supporting File S7: Enzyme Substrate Interaction – Problem Solving Activity Answer Key and Rubric. The answer key and rubric to the Problem Solving activity.
- Supporting File S8: Enzyme Substrate Interaction – Sample Exam Questions.
Acknowledgments
This work was partially supported by the National Science Foundation under award number 1711402 to the University of Minnesota, Rochester and award numbers 1323414 and 1725940 to Milwaukee School of Engineering. We thank Jeffery Ewertz, Thor Wirth, Naima Yusuf for helping design the serine protease physical models. We thank Michael Warden for final model design and construction of the models.
This study was approved by the IRB at the University of Minnesota under STUDY00002273: Seeing and Learning Biochemistry and STUDY0000026: Modeling for the Enhancement of Learning Chemistry, and per IRB guidelines all identifying information was removed for data analysis and dissemination.
References
- American Society for Biochemistry and Molecular Biology Learning Framework. https://www.coursesource.org/courses/biochemistry-and-molecular-biology. Last accessed 05 February 2021.
- Loertscher, J. 2011.Threshold concepts in Biochemistry. Biochemistry and Molecular Biology Education, 39 (1): 56–57. https://doi.org/10.1002/bmb.20478.
- Schönborn KJ, Anderson TR. 2010. Bridging the educational research-teaching practice gap: Foundations for assessing and developing biochemistry students’ visual literacy. Biochemistry and Molecular Biology Education, 38 (5): 347–354. https://doi.org/10.1002/bmb.20436.
- Schönborn KJ, Anderson T. 2009. A model of factors determining students’ ability to interpret external representations in Biochemistry. International Journal of Science Education, 31 (2): 193–232. https://doi.org/10.1080/09500690701670535.
- Linenberger KJ, Bretz SL. 2012. Development of the Enzyme-Substrate Interactions Concept Inventory. Biochemistry and Molecule Biology Education, 40 (4): 229–33.
- Linenberger KJ, Bretz SL. 2014. Biochemistry students’ ideas about shape and charge in enzyme-substrate interactions. Biochemistry and Molecular Biology Education, 42 (3): 203–212. https://doi.org/10.1002/bmb.20776.
- Linenberger KJ, Bretz SL. 2015. Biochemistry students’ ideas about how an enzyme interacts with a substrate. Biochemistry and Molecular Biology Education, 43 (4): 213–222. https://doi.org/10.1002/bmb.20868.
- Cooper AK, Oliver-Hoyo MT. 2017. Creating 3D physical models to probe student understanding of macromolecular structure. Biochemistry and Molecular Biology Education, 45 (6): 491–500. https://doi.org/10.1002/bmb.21076.
- Babilonia-Rosa MA, Kuo HK, Oliver-Hoyo MT. 2018. Using 3D printed physical models to monitor knowledge integration in Biochemistry. Chemistry Education Research and Practice, 19 (4): 1199–1215. https://doi.org/10.1039/C8RP00075A.
- Srivastava A. 2016. Building mental models by dissecting physical models. Biochemistry and Molecular Biology Education, 44 (1): 7–11. https://doi.org/10.1002/bmb.20921.
- Forbes-Lorman RM, Harris MA, Chang WS, Dent EW, Nordheim EV, Franzen MA. 2016. Physical models have gender-specific effects on student understanding of protein structure-function relationships. Biochemistry and Molecular Biology Education, 44 (4): 326–335. https://doi.org/10.1002/bmb.20956.
- Newman DL, Stefkovich M, Clasen C, Franzen MA, Wright LK. 2018. Physical models can provide superior learning opportunities beyond the benefits of active engagements. Biochemistry and Molecular Biology Education, 46 (5): 435–444. https://doi.org/10.1002/bmb.21159.
- Terrell CR, Franzen MA, Herman T, Malapati S, Newman DL, Wright LK. 2019. Physical models support active learning as effective thinking tools. In ACS Symposium Series; Bussey, T. J., Linenberger Cortes, K., Austin, R. C., Eds.; American Chemical Society: Washington, DC; Vol. 1337, pp 43–62. https://doi.org/10.1021/bk-2019-1337.ch003.
- Harris MA, Peck RF, Colton S, Morris J, Chaibub Neto E, Kallio JA. 2009. Combination of hand-held models and computer imaging programs helps students answer oral questions about molecular structure and function: A controlled investigation of student learning. CBE—Life Sciences Education, 8 (1): 29–43. https://doi.org/10.1187/cbe.08-07-0039.
- Roberts JR, Hagedorn E, Dillenburg P, Patrick M, Herman T. 2005. Physical models enhance molecular three-dimensional literacy in an introductory biochemistry course. Biochemistry and Molecular Biology Education, 33 (2): 105–110. https://doi.org/10.1002/bmb.2005.494033022426.
- Geldenhuys WJ, Hayes M, Van der Schyf CJ, Allen DD, Malan SF. 2007. Receptor surface models in the classroom: Introducing molecular modeling to students in a 3-D world. Journal of Chemical Education, 84 (6): 979. https://doi.org/10.1021/ed084p979.
- Tanner K. 2013. Structure matters: Twenty-one teaching strategies to promote student engagement and cultivate classroom equity. CBE – Life Sciences Education. 12: 322-331. https://doi.org/10.1187/cbe.13-06-0115.
- Voyer D, Voyer S, Bryden MP. 1995. Magnitude of sex differences in spatial abilities: A meta-analysis and consideration of critical variables. Psychological Bulletin, 117(2): 250. https://doi.org/10.1037/0033-2909.117.2.250.
- Linn MC, Petersen AC. 1985. Emergence and characterization of sex differences in spatial ability: A meta-analysis. Child Development, 56(6): 1479–1498. https://doi.org/10.2307/1130467.
- Harrison AG, Treagust DF. 1998. Modeling in science lessons: Are there better ways to learn with models? School Science and Mathematics, 98 (8): 420–429.
Article Files
Login to access supporting documents
Terrell-Its a SubstrateIts a ProteinNo-Its an Enzyme.pdf(PDF | 1 MB)
S1.Enzyme Substrate Interaction-Pre-class Activity Serine Proteases.docx(DOCX | 27 KB)
S2.Enzyme Substrate Interaction-Pre-class Activity Serine Proteases Answer Key.docx(DOCX | 142 KB)
S3.Enzyme Substrate Interaction-Instructor Guide Video on Teaching using Serine Protease models.docx(DOCX | 19 KB)
S4.Enzyme Substrate Interaction-Model Exploration Activity.docx(DOCX | 1 MB)
S5.Enzyme Substrate Interaction-Problem Solving Activity.docx(DOCX | 4 MB)
S6.Enzyme Substrate Interaction-Model Exploration Activity Answer Key and Rubric.docx(DOCX | 1 MB)
S7.Enzyme Substrate Interaction-Problem Solving Activity Answer Key And Rubric.docx(DOCX | 4 MB)
S8.Enzyme Substrate Interaction-Sample Exam Questions.docx(DOCX | 2 MB)
- License terms


Comments
Comments
There are no comments on this resource.