Isolation and Functional Analysis of a Pancreatic Enzyme in an Introductory Student Lab
Editor: Neena Grover
Published online:
Abstract
Structure and function are correlated at all levels of biology. This topic is typically addressed early in an undergraduate class in general or molecular biology before students have gained much skill or knowledge in molecular biology. However, an understanding of the chemical bonds involved in forming and maintaining the structure of proteins is critical to understanding how enzymes function and how their activity can be regulated. Here, a laboratory activity is described that is suitable for undergraduate biology students. This activity examines the activity of carboxypeptidase A (CPA), an abundant pancreatic enzyme with a rich history in enzymology and structural biology. The abundance of CPA in pancreatic tissue allows for a series of common biochemical techniques to be easily performed under the constraints of an undergraduate teaching lab, including the separation of proteins by simple precipitation methods, the examination of resulting proteins by SDS-PAGE and Coomassie staining, and the analysis of enzyme function through the determination of constants such as Vmax and Km. These steps illustrate the importance of noncovalent bonds in protein structure and the use of common biochemical instruments in the lab, while providing students with an opportunity to hone analysis skills in their consideration of the resulting data. Finally, this lab may be modified in many ways to make it suitable for upper division classes, CURE approaches to the undergraduate lab, and even to the pre-college classroom.
Primary Image: Carboxypeptidase structure and function. The crystal structure of a human homolog (CPA2; PDB ID 1AYE; image created in PyMol) of carboxypeptidase A is shown. The prodomain is at the top left and the enzymatic domain is below, with the required zinc in the active site shown as a gray sphere. Cleavage of a synthetic substrate by carboxypeptidase A can be seen using a spectrophotometer as a decrease in absorbance.
Citation
Lyons PJ. 2022. Isolation and Functional Analysis of a Pancreatic Enzyme in an Introductory Student Lab. CourseSource 9. https://doi.org/10.24918/cs.2022.39
Society Learning Goals
Biochemistry and Molecular Biology
- Energy is required and transformed in biological systems
- How do enzymes catalyze biological reactions?
- Macromolecular Structure Determines Function and Regulation
- What factors contribute to the size and complexity of biological macromolecules?
- What factors determine structure?
- How are structure and function related?
- What is the role of noncovalent intermolecular interactions?
- How is structure (and hence function) of macromolecules governed by foundational principles of chemistry and physics?
- How are a variety of experimental and computational approaches used to observe and quantitatively measure the structure, dynamics and function of biological macromolecules?
- Discovery requires objective measurement, quantitative analysis and clear communication
- What skills are needed to access, comprehend and communicate science?
Lesson Learning Goals
The following are learning goals that will be accomplished through this series of student labs:
- Students will appreciate how enzymes function to rapidly catalyze chemical reactions.
- Students will observe the impact of salt and pH on the chemical bonds that are involved in protein solubility.
- Students will know the theory behind standard methods used in biochemistry and molecular biology for the analysis of proteins.
Learning goals addressed from the Learning Framework for the CourseSource Biochemistry and Molecular Biology Course, developed by The American Society for Biochemistry and Molecular Biology:
- Energy is required and transformed in biological systems
- How do enzymes catalyze biological reactions?
- Macromolecular Structure Determines Function and Regulation
- What factors contribute to the size and complexity of biological macromolecules?
- What factors determine structure?
- How are structure and function related?
- What is the role of noncovalent intermolecular interactions?
- How is structure (and hence function) of macromolecules governed by foundational principles of chemistry and physics?
- How are a variety of experimental and computational approaches used to observe and quantitatively measure the structure, dynamics, and function of biological macromolecules?
- Discovery requires objective measurement, quantitative analysis, and clear communication
- What skills are needed to access, comprehend, and communicate science?
The learning goals of this activity will be supported by prior learning in the classroom. Most introductory biology and biochemistry textbooks include sections or chapters covering macromolecular structure, chemical bonding, enzymatic function, and perhaps some of the laboratory methods used to collect such information. These will have been considered in the lecture portion of the class or through textbook readings prior to this laboratory experience.
Lesson Learning Objectives
Through this series of labs, students will be able to:
- fractionate a complex protein sample using standard precipitation methods.
- determine the concentration of protein samples using spectrophotometry.
- separate proteins by SDS-PAGE and analyze the sizes of major proteins following protein staining.
- measure the amount of carboxypeptidase enzyme present in each fraction by measuring enzymatic activity.
In addition to these practical lab skills, students will be able to:
- recognize protein domains and utilize relevant mass measurements when discussing proteins.
- describe some examples of specific proteolysis important to most living organisms.
- connect the chemistry of noncovalent bonds to the folding and unfolding of complex protein structures.
- explain the relationship of enzyme and substrate concentrations to a variety of kinetic constants used to describe enzyme function.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
Introduction
Students in introductory biology, molecular biology, and biochemistry courses are introduced early to the core idea that structure and function are intimately related. This concept is central to these subjects, and is listed as such in the Vision and Change document of 2011 (1) and the foundational educational concepts described by the American Society for Biochemistry and Molecular Biology (ASBMB). This structure and function concept is not unique to biology. Each tool or device we utilize in our everyday life (cooking utensil, vehicle, electronic device, etc.) has a particular structure that enables its function. However, in biochemistry and molecular biology the items of interest are mostly invisible to the naked eye, and so we require specialized tools, techniques, and skills to investigate invisible structures and functions. Enzymes, for example, have functions that are closely tied to their structures. The change of a substrate into a product catalyzed by an enzyme can sometimes be observed as a change in reagent color that can be measured with a spectrophotometer. These scientific tools and skills undergird much of what we teach in biochemistry and molecular biology.
A number of experimental protocols and kits have been described that can be used to address the functions of enzymes in biochemistry and molecular biology courses. These include the analysis of dihydrofolate reductase (2), alkaline phosphatase (3), and threonine dehydrogenase (4), amongst others. These protocols are typically multi-week labs in which students are guided through the process of transforming competent bacteria with an expression plasmid, inducing expression of the enzyme encoded by this plasmid, and finally analyzing expression and activity of the expressed enzyme. These labs cover a wide range of topics, some which may not be covered in the lecture class that often focuses on protein structure and function first and gene expression later, if at all.
Lab activities that introduce the basic concepts of protein structure and function to students of intermediate-level biochemistry and molecular biology courses are often useful. Intermediate-level courses commonly introduce the tools and techniques used for the analysis of enzymes and other methods that may not have been encountered earlier in the general biology lab. Quantitative data analysis and interpretation of data through graphing are other important competencies that should be taught at this level and throughout the curriculum (1).
Laboratory activities that might fit into such an intermediate-level class have been described, often using a purified and commercially-available enzyme. These include the analysis of phytase (5), alkaline phosphatase (6), tyrosinase (7), and papain (8). These experiments demonstrate important aspects of enzymology and use enzyme material that can be purchased easily. Other experiments have been reported that analyze enzymes derived from foods or other readily-available materials. For example, the activity of turnip peroxidase is commonly analyzed in student labs (9). This enzyme is readily extracted from turnips and other vegetables, and the oxidation of guaiacol is accompanied by a color change from clear to brown. Unfortunately, guaiacol is toxic and therefore extra care must be taken in handling and disposal. Other labs that utilize readily available materials to teach protein structure and enzyme function include the purification and analysis of lysozyme from egg whites (10) and invertase from baker’s yeast (11), the study of polyphenol oxidases within avocados (12), and the analysis of lactase from LactAid tablets (13).
The laboratory activity described herein provides educators with another option for the teaching of protein structure and function using an enzyme obtained directly from its natural source. It provides students with an illustration of protein structure and conditions that can modify this structure as they study enzyme function. The methods utilized in this lab, including precipitation, spectrophotometry, and gel electrophoresis, are standard protein isolation and characterization methods that are central to biochemistry and that enable quantitative and qualitative analysis of data. The enzyme studied is well-known and a standard example of a hydrolytic enzyme. Although neither the methods nor the subject matter of this activity are particularly unique, the current laboratory activity benefits from the use of a natural and abundant enzyme, carboxypeptidase A (CPA), derived from the cow pancreas. This abundance results in a laboratory activity with relative ease of setup and with potential for student learning through variations on the theme of enzyme function.
Scientific Background: The Pancreas and its Secreted Enzymes
The pancreas is an important secretory organ. In fact, greater than 90% of proteins made by the pancreas may be secreted (14). The pancreas secretes a number of small proteins and peptides, such as insulin and glucagon, in an endocrine manner, to regulate blood glucose levels. Banting and Best are most well-known for their extraction of insulin from cow and pig pancreases for the treatment of diabetes (15). In addition to the endocrine function of the pancreas, acinar cells of the pancreas produce large amounts of a few (less than 20) digestive enzyme precursors, including amylase, lipase, trypsin, chymotrypsin, and carboxypeptidase (16, 17). These are packaged as inactive enzymes (zymogens) into large secretory vesicles called zymogen granules for storage prior to exocrine release into the pancreatic ductal system. As much as 30% of acinar cell protein is found in the contents of the zymogen granules, and most of this protein content is made up of amylase, trypsin, and carboxypeptidase precursors (17). Upon reaching the duodenum, these enzymes are activated by proteolytic cleavage, usually leading to the removal of a prodomain so that the enzymes can function in digestion.
Most organisms contain three pancreatic metallocarboxypeptidase genes, which produce very similar proteins with different substrate specificities. All precursor forms are about 45 kDa in size, decreasing to about 35 kDa upon activation by proteolytic removal of the prodomain (Figure 1A). Carboxypeptidase A1 (CPA1), upon activation, cleaves aliphatic or aromatic C-terminal amino acids. Carboxypeptidase A2 (CPA2), upon activation, preferentially cleaves larger aromatic C-terminal amino acids such as phenylalanine and tryptophan (Figure 1B). Carboxypeptidase B (CPB) cleaves the basic amino acids, lysine and arginine, from the C-terminus of a substrate protein (18). The story is somewhat simplified in the cow, as there is only one carboxypeptidase A gene (CPA), rather than the two that exist in most other vertebrates. The catalytic mechanism used by carboxypeptidases is a general acid-base approach to hydrolysis, with intermediates stabilized by the zinc cofactor (see Figure 1B). The binding of substrate is facilitated by structural changes of the enzyme, thus a good example of the “induced fit” model of substrate binding first proposed by Koshland (19).
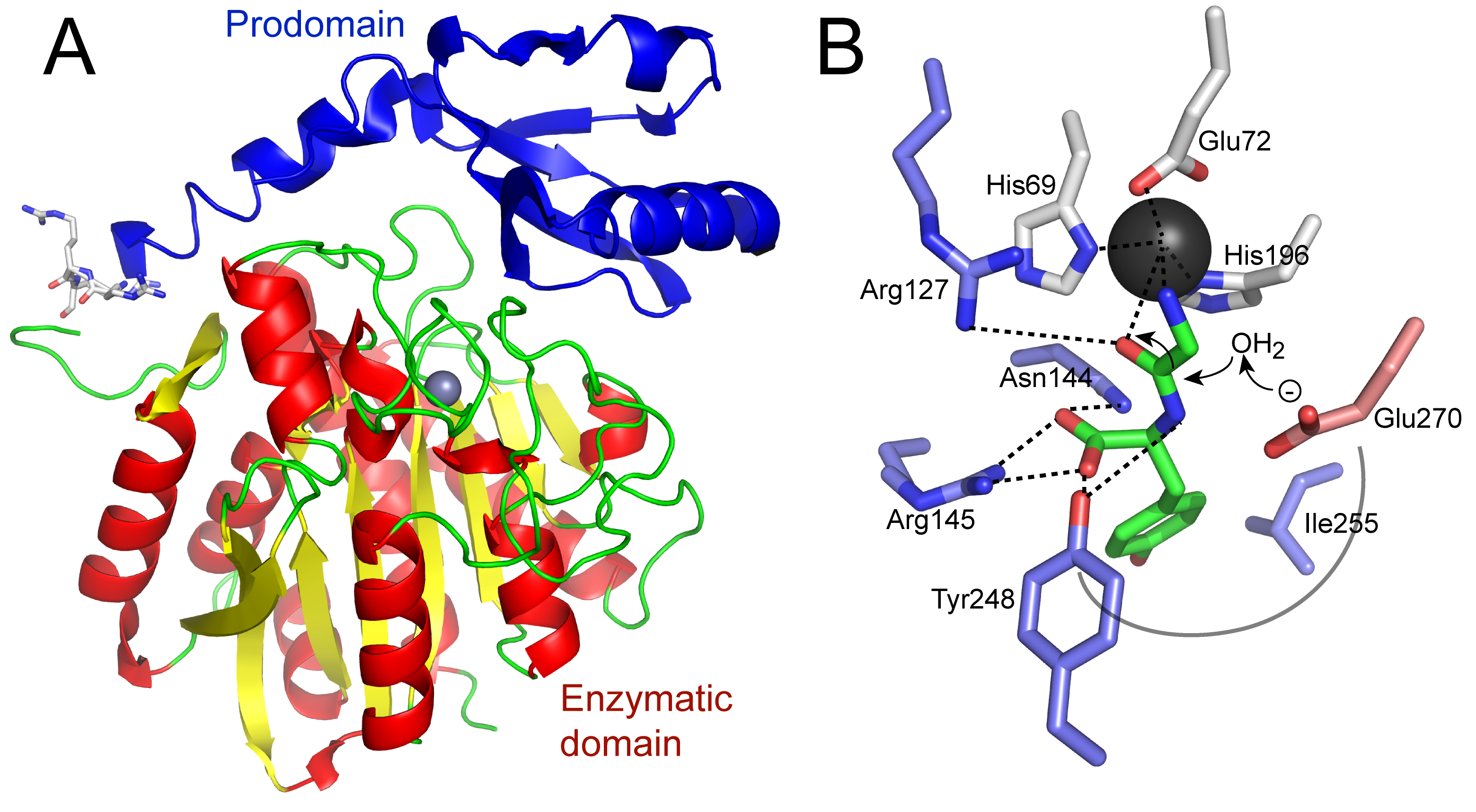
A number of features make pancreatic carboxypeptidase A an ideal subject for study in the undergraduate biochemistry or molecular biology laboratory. 1) It is available through the donation of a bovine (or other) pancreas from a local slaughterhouse. 2) As described above, there is an abundance of CPA in pancreatic tissue, and it is quite stable at -20 °C. We have stored the tissue for 10 years at this temperature without appreciable loss of enzyme activity. 3) The assay for enzyme activity is easy to perform. Even a novice student will detect activity. 4) The biochemistry of the carboxypeptidases nicely illustrates 1°, 2°, and 3° protein structure. We teach students that the 1° structure of a protein is written N-terminal to C-terminal. This becomes very important for the study of a carboxypeptidase enzyme, as the C-terminus is the end where the carboxypeptidase cuts. The carboxypeptidase requires activation by removal of a prodomain, illustrating domains and 3° structure (see Figure 1A). There is a wide array of three-dimensional structures available in the Protein Data Bank for the carboxypeptidases, which can be used to further illustrate the 1°, 2°, and 3° structure of proteins. 5) The biochemistry of the carboxypeptidases illustrates the exquisite specificity of many enzymes (see Figure 1B). CPA only cuts at the C-terminus, and then only cuts aliphatic/aromatic amino acids; CPB only cuts basic amino acids. This presents an opportunity to review the properties of amino acid side chains as well. 6) There is a history to this enzyme as one of the first enzymes to be studied in detail and to have its structure solved by X-ray crystallography in 1965 (20), following the crystal structure of the first enzyme (hen egg white lysozyme) in 1962 (21, 22). Bovine CPA was one of only nine structures to be deposited in the initial Protein Data Bank (PDB) in 1973. For a biochemistry course, the catalytic mechanisms of the carboxypeptidases are very well characterized and could be further explored (see Figure 1B).
In this two-week laboratory activity, students complete four major activities. First, they fractionate crude pancreatic lysates using salting out and isoelectric point precipitation procedures. While these are relatively crude methods of protein enrichment and may not be often used now due to the availability of column and affinity chromatography, they nicely illustrate concepts of amino acid polarity and hydrophobicity and the effects of these on protein solubility. The steps necessary for these precipitation procedures are few and simple. Following this fractionation, students measure protein concentration using a standard Bradford assay. This will give them some information regarding the outcome of the precipitation methods. The following week, students run an SDS-PAGE gel and stain it with Coomassie Blue. Nearly all students are able to see distinct protein bands and make predictions regarding identity based on molecular weight, since even in the crude lysate there are only a few highly expressed proteins. Finally, while the SDS-PAGE is running, students measure the enzyme activity of each fraction, with the goal of identifying one fraction enriched in CPA activity. Using data supplied to them, students apply their knowledge of enzyme function to make and analyze a Lineweaver-Burk plot.
Intended Audience
This laboratory activity is best suited for an introductory cell and molecular biology or biochemistry course for majors in the biological sciences. It is used by the author in a cell and molecular biology course for biology majors, most in their second year, at a primarily undergraduate institution. The course typically has 40-50 students divided between two lab sections. A version of this activity has also been offered to a select group of motivated students enrolled in freshman-year general biology.
Required Learning Time
This lab activity requires two three-hour sessions to complete.
Prerequisite Student Knowledge
Students should be familiar with protein structure. As presented, students should also have been introduced to basic enzyme kinetics, so that they can apply this knowledge to this lab. In addition to these content areas, students should have some prior experience in the use of a spectrophotometer and micropipette, as incorrect cuvette placement or micropipette usage will negatively impact all data collection in this lab. Finally, some prior experience performing basic laboratory calculations (for example, determining concentrations and making dilutions) is necessary before beginning this laboratory. Rudimentary skills in these areas will help to prevent frustration and aid in understanding the big picture.
Prerequisite Teacher Knowledge
The teacher should understand key concepts in protein structure and basic enzymology. These topics are typically addressed within the first several chapters of any biochemistry or cell biology textbook. The teacher should also be knowledgeable in the use of basic molecular biology and biochemistry tools including the spectrophotometer, gel electrophoresis apparatus, and micropipettes.
Scientific Teaching Themes
Active Learning
The laboratory activity is entirely hands-on, allowing students to use equipment and perform techniques that they have previously learned about. They are able to observe experimental results first hand and are required to actively interpret the data that they collect. For example, students can actually see proteins precipitating, they must determine the approximate molecular weights of proteins observed on the stained gel and compare these with sizes predicted for key proteins, and they must manipulate and graph enzyme activity data so that they can arrive at a conclusion. All of these activities are typically done in groups of two, allowing for peer discussion and teaching.
Assessment
Students complete a five-question short-answer quiz at the beginning of each lab session to ensure understanding of basic concepts described in the lab handout assigned for prior reading. Following each lab session, students are required to write a short summary of the lab activities that they completed. Following the lab sequence, students assemble their lab methods summaries into a methods section for a lab report. To this, they add a one-page introduction in which they report their brief exploration of the scientific literature on the topic, a written description of their observations (results), and a short discussion in which they compare their results with other scientific literature describing enzyme purification and/or activity. Key to this lab report is the inclusion of a figure showing their stained gel, a figure showing graphs of enzyme activity and substrate binding (Michaelis-Menten and Lineweaver-Burk plots), and a table showing possible enrichment of CPA in fractions. Detailed instructions and examples for each of these components are provided, with the goal to model clear presentation of data. These instructions are abbreviated, placed into table format, and used as a rubric for grading (S3. Pancreatic Enzyme – Report Rubric).
Inclusive Teaching
Throughout the lab activity, students are reassured that the goal is not to determine the “right answer” but rather to identify a clear and reasonable interpretation of the data. This reassures many students when their data is not what they may have expected. In our experience, almost every student is able to get some reasonable data in this series of experiments due to the robust nature and abundance of the enzyme being studied. While the enzyme activity data is quantitative and must be graphed and carefully analyzed, the gel and observations throughout the two-week series are visual and therefore more qualitative, demonstrating that both qualitative and quantitative analyses are important in this discipline. Quantitative analysis and graphing might be challenging to some students with less preparation in this area. In some cases, the quantitative analysis could be reduced or simplified; for example, the purification table could be eliminated (this is considered further in the Teaching Discussion). However, we attempt to bring every student along through extensive peer teaching with the use of undergraduate teaching assistants in addition to the availability of the course instructor. The diversity of data collection and analysis provided by this laboratory exercise illustrates the advice given by E. O. Wilson to the young scientist: “For every scientist, whether researcher, technologist, or teacher, of whatever competence in mathematics, there exists a discipline in science for which that level of mathematical competence is enough to achieve excellence.” (23) p. 41.
Lesson Plan
This two-week lab sequence corresponds to in-class lecture topics on protein structure and enzyme function. Therefore, students should be familiar with general protein structure, the role of enzymes in biological systems, and the impact of pH on enzymes, proteins, and the chemical bonds found within. Students will come to lab having read the introductory material provided to them in the lab handout (S1. Pancreatic Enzyme - Student Lab Handout) and so should be familiar with the carboxypeptidase enzyme to be investigated, its role in biological systems, and the theory behind the methods to be performed. Students have previously learned the use of micropipettes and spectrophotometers.
In the first three-hour lab session, students complete two major tasks and prepare materials for the next three-hour session. Specifically, they fractionate a pancreatic lysate using basic precipitation methods and then use a Bradford assay to determine the total protein concentration of each fraction. This is followed by the preparation of samples for SDS-PAGE, which will occur in the following week along with the analysis of enzyme activity.
Pre-Lab Preparation (See Table 1 for details)
Prior to lab, a number of reagents need to be obtained (S2. Pancreatic Enzyme – Reagent List). Most importantly, a source of pancreas needs to be determined. A bovine pancreas can be obtained from a local slaughterhouse, keeping it cool with ice packs in a cooler until processing into smaller pieces and freezing at -20 °C in individual freezer bags. The enzyme in this frozen pancreas tissue will remain viable for more than ten years, likely much longer.
Just before lab Session 1, fifty grams of pancreas tissue is blended in a standard kitchen blender with 150 mL of Tris-buffered saline (50 mM Tris, 150 mM NaCl, pH 7.3). This material is then filtered through several layers of cheesecloth or one layer of crafting t-shirt to remove debris and fatty material. We have found that even after this filtering and a subsequent centrifugation step, there still remains some lipid material floating on the surface. This does not seem to hinder subsequent steps. The addition of 0.1% NP-40 detergent to the lysis buffer eliminates some of this floating lipid, but also reduces the accuracy of subsequent Bradford assay protein concentration measurements.
In addition to the preparation of the pancreatic lysate, bovine serum albumin (for Bradford assay) and Laemmli sample buffer (for SDS-PAGE) require preparation (see Table 1). Preparing individual student group aliquots of most materials prevents congestion at common lab tables.
Table 1. Prelab Preparation. These notes assume preparation for 24 groups of two students each.
| Activity | Description | Estimated Time | Notes |
|---|---|---|---|
| Preparation prior to the first lab session | |||
| Obtain pancreas | A local meat processing facility may be happy to provide pancreas tissue. | Location dependent |
Tissue can be frozen at -20 °C in 50 g portions for an indefinite time period. |
| Prepare lysis buffer | Prepare 300 mL of Lysis buffer, a simple Tris-buffered saline: 50 mM Tris, 150 mM NaCl, pH 7.3. Adjust pH with pH meter. | 30 minutes | If a pH meter is unavailable, pH paper will be sufficient. |
| Prepare pancreatic lysate | Slice about 50 g of pancreas tissue into small pieces and place in a standard kitchen blender with 150 mL of lysis buffer. Blend on high until homogenous. Filter through several layers of cheesecloth or a layer of cotton crafting t-shirt to remove remaining pieces. | 15 minutes | Some lipid may still remain floating on the top. You might try other filtration methods to get rid of this, but it does not impact subsequent steps much. |
| Prepare BSA solution and Laemmli sample buffer | Dilute bovine serum albumin (BSA) concentrate to 0.1 mg/mL in distilled water. Mix 20 μL of 1M DTT into every 300 μL of 2x Laemmli sample buffer. | 10 minutes | BSA is usually purchased as a concentrated solution. DTT must be added to Laemmli sample buffer to ensure reduction of protein disulfide bonds. |
| Aliquot and distribute materials to student workstations |
Each workstation receives:
|
1-2 hours | Preparing aliquots of reagents for each group prevents traffic jams at common tables. |
Lab Session 1 (See Table 2 for details and timeline)
Student groups are provided two milliliters of clarified lysate. This is centrifuged at high speed (15,000 rpm) to pellet any remaining particulate matter and 1 mL is transferred to a new tube for the start of the precipitation procedures (the remainder is saved for later analysis of crude lysate). First, 2M acetic acid is added in 10 µL volumes, testing the pH with pH paper each time until reaching pH 5.0. Students may see some solids precipitating in this step, although minimal. After centrifuging and transferring the supernatant to a new tube, the supernatant is again precipitated, this time by the addition of 30% (0.3 g) ammonium sulfate. Again, the sample is centrifuged, supernatant transferred to a new tube, and another 0.3 g of ammonium sulfate is added, for a total of 60% ammonium sulfate. Extended vigorous shaking may be required to dissolve this last portion of ammonium sulfate. If a small amount of ammonium sulfate refuses to dissolve, the addition of a small amount of lysis buffer is recommended. While this changes the volume, any undissolved ammonium sulfate will be pelleted with the precipitate and will affect the ability to resolve these samples by SDS-PAGE. The sample is centrifuged as before and the supernatant is transferred to a new tube. It is important at this point to emphasize that any remaining liquid still present in tubes containing a precipitate must be discarded, as the low pH and high concentration of ammonium sulfate impacts the subsequent SDS-PAGE. One milliliter of lysis buffer along with 20 µL of 2M NaOH is added to dissolve the precipitates so that they can be used in subsequent steps.
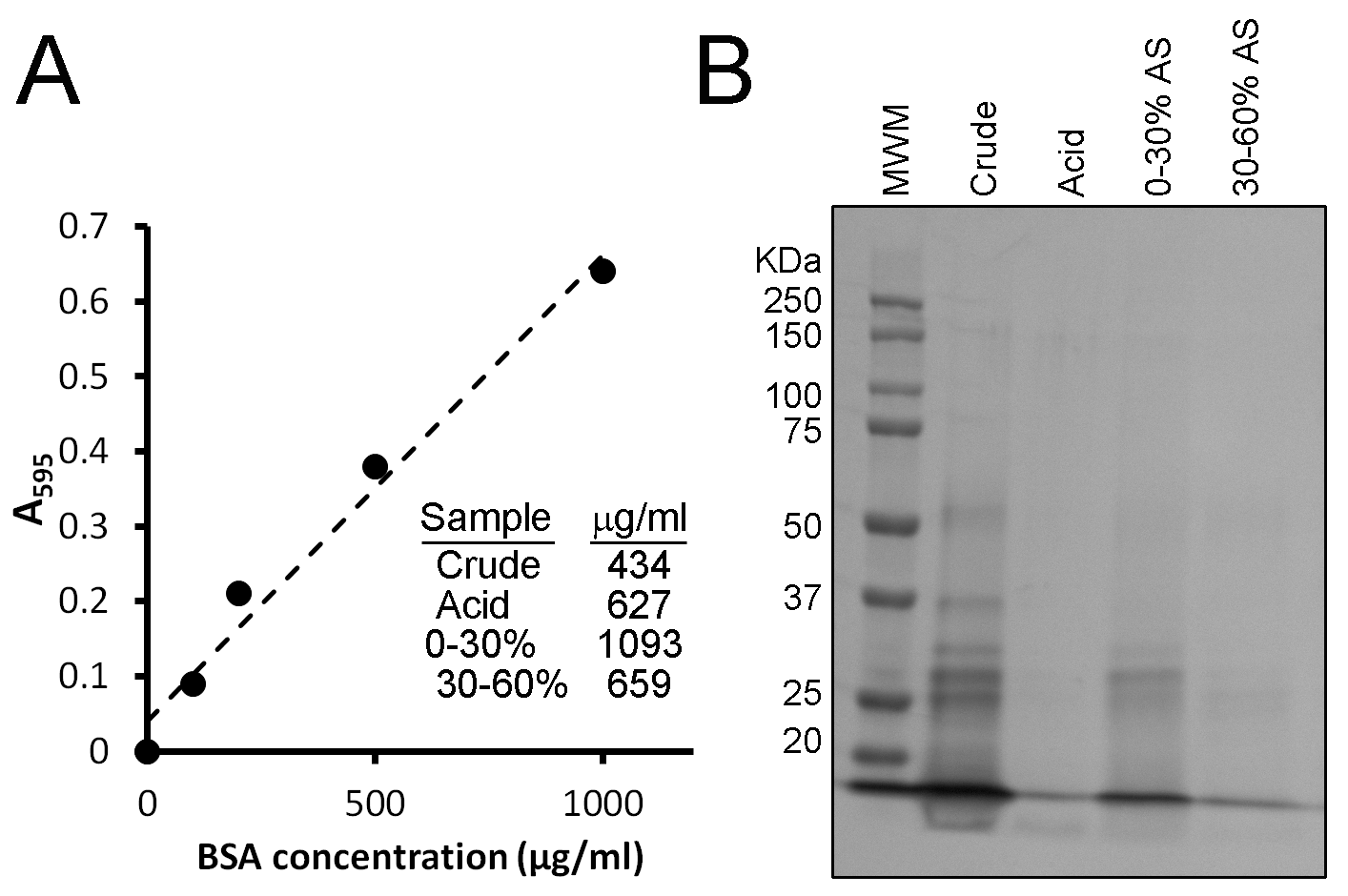
Upon completion of lysate fractionation, the second major activity for Session 1 is to determine the concentration of protein in these fractions. This is done using a Bradford assay. The Bradford reagent makes use of the Coomassie Brilliant Blue G-250 dye, which at acidic pH binds to basic amino acids within proteins and turns from a brown color to a blue color. This color change can be measured by an increase in absorbance at 595 nm (A595), where a greater absorbance means a greater concentration of proteins. A standard curve is first prepared using known concentrations of bovine serum albumin, where the concentrations are plotted against the determined A595 for a range of concentrations (Figure 2A). The A595 is then determined for the lysate fractions and the concentration of protein within these fractions determined by using the information in the standard curve (see Figure 2A for representative data).
The final step for this first lab session is to prepare samples for SDS-PAGE to expedite the following week’s activity and to denature all proteins to ensure that minimal protein degradation occurs in the intervening week. Students mix their fractions with an equal amount of SDS-PAGE sample buffer, heat them at 95 °C for 3 minutes, and place in a box for storage at -20 °C. In addition to these SDS-PAGE samples, all protein fractions must also be stored at -20 °C for later analysis of enzyme activity. Students occasionally forget and discard these fractions. Students also occasionally add SDS-PAGE sample buffer to their entire protein fractions, with the result of denaturing everything. In these cases, students are asked to measure the enzyme activity of another group’s samples. This, unfortunately, does not allow them to make any correlations between enzyme activity and SDS-PAGE results.
Table 2. Timeline and notes for Lab Session 1.
| Activity | Description | Estimated Time | Notes |
|---|---|---|---|
| Lab Session 1 | |||
| Quiz | A short 5-question short answer quiz to test basic knowledge from lab handout. | 10-15 minutes | |
| Lysate centrifugation | To eliminate any remaining particulate matter, centrifuge 2 mL of pancreatic lysate for 5 minutes at 15,000 rpm. | 10 minutes |
|
| Lysate fractionation |
|
60 minutes |
|
| Analysis of protein concentration |
|
60 minutes | |
| Preparation for SDS-PAGE |
|
10 minutes | Also store remainder of protein fractions to which no sample buffer has been added at -20 °C until the next lab session as well. |
Lab Session 2 (See Table 3 for details and timeline)
In this session, students perform SDS-PAGE (we use precast gels from Bio-Rad) and measure the activity of carboxypeptidase A in each protein fraction from the previous week. Pre-lab preparation is minimal, consisting only of diluting SDS-PAGE running buffer to 1x and dissolving the carboxypeptidase substrate in previously-made Tris-buffered saline to a 0.5 mM concentration.
SDS-PAGE samples made in the previous week (25 µL each) as well as a protein molecular weight marker (5 µL) are loaded onto a gel and run using standard techniques. One sample, however, is omitted. This is the sample prepared with the supernatant containing 60% ammonium sulfate. This amount of salt prevents the proper migration of samples through the gel. When the dye front reaches the bottom of the gel, the run is terminated. The gels are washed three times with 50 mL of water in plastic containers. After the water is poured off, 50 mL of Biosafe Coomassie Stain is added and the gels are left to stain overnight. The following day, gels are scanned and files uploaded to the learning management system for student analysis. Typical student results are shown in Figure 2B. While the identities of the bands are unknown, predictions can be made based on known protein sizes. In addition, this approach can be compared to that of Western blotting, which would enable the protein of interest to be more precisely identified.
While the gel is running, students measure the activity of carboxypeptidase A in each fraction using a synthetic substrate, FA-Phe-Phe-OH (Bachem), which absorbs light at 340 nm. Five microliters of each fraction are added to 1 mL of 0.5 mM substrate, immediately mixed, and placed in the spectrophotometer. Upon cleavage of the terminal amino acid from this substrate, the absorbance at 340 nm decreases. A greater rate of decrease indicates the presence of a greater amount of enzyme. Students record the A340 every 30 seconds for 2.5 minutes. This data is then graphed and the slopes of each line determined. Typical student data is shown in Figure 3A.
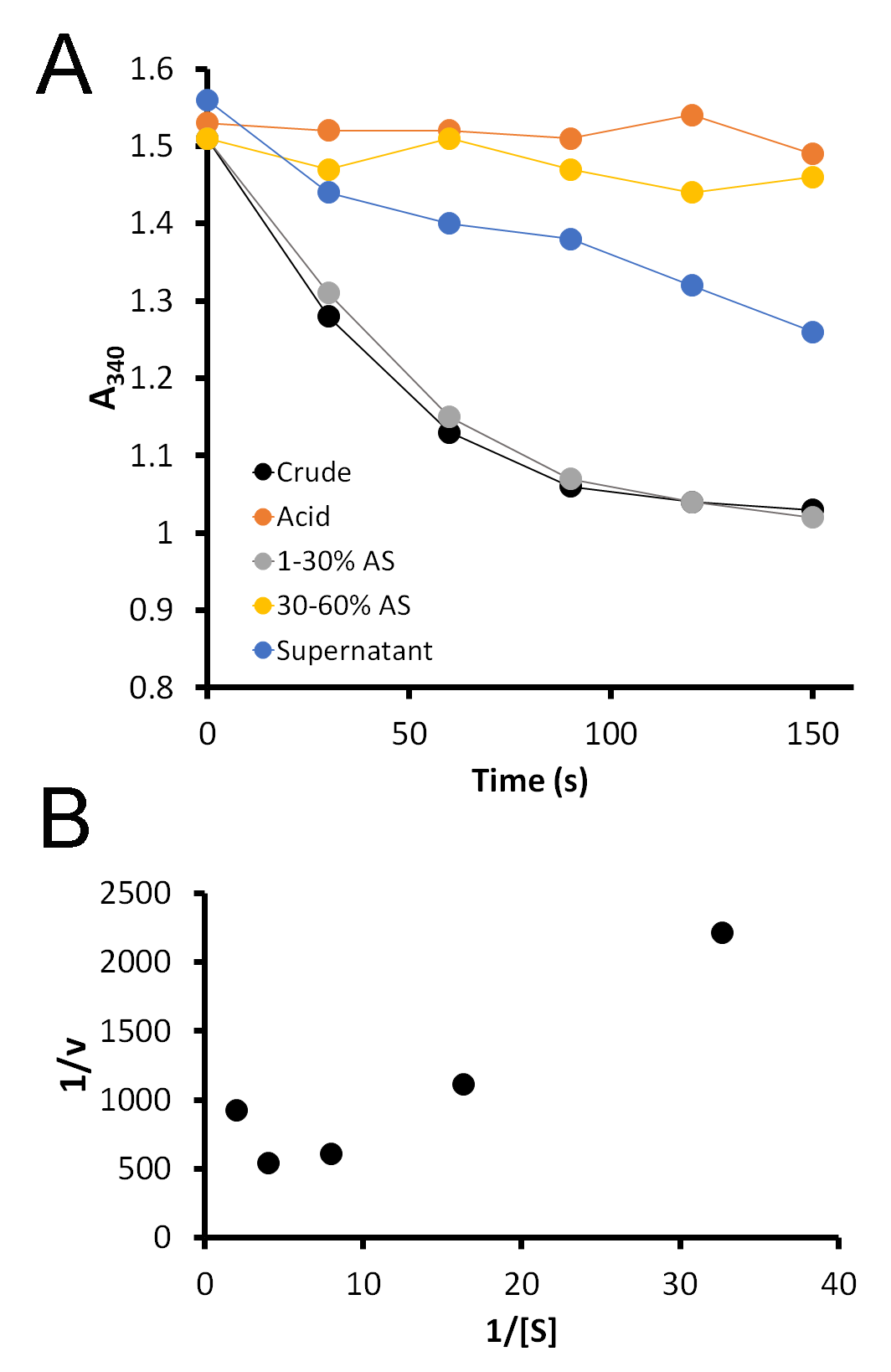
In our class, we learn about Km and Vmax. Although we do not have time in lab to measure the activity of carboxypeptidase A with a range of substrate concentrations in order to determine Km and Vmax, this system presents a perfect opportunity to do so. I take the middle ground and provide them with data that I have collected previously (S4. Pancreatic Enzyme – Sample Enzyme Kinetics Data). This data can be collected by incubating a range of substrate concentrations (the sample data uses concentrations from 0.5 to 0.03 mM FA-FF) with a small amount of enzyme and collecting A340 data over time. Students determine the rate of reaction (ΔA340/time) at each substrate concentration, and then the inverse of the rate is plotted against the inverse of the substrate concentration for a Lineweaver-Burk plot. This gives students the opportunity to analyze such data and determine, from the equation y = mx + b, Vmax (Vmax = 1/b) and Km (Km = m*Vmax). Typical results are shown in Figure 3B. Notably, this Lineweaver-Burk graph does not appear to be a straight line as we would expect, as CPA is known to exhibit substrate inhibition at high concentrations (24). This may confuse students at first, until they are directed to consider the substrate concentrations. Thus, students are required to use some higher order thinking skills to analyze the data and determine how it deviates from the textbook examples.
Upon completion of this two-week lab series, students prepare a lab report. They are provided with clear guidance regarding what data should be included in this lab report to model typical approaches to scientific reporting. One of the items requested of students is a purification table, modeled after examples often presented in protein purification studies (25). A typical table is shown in Table 4. This table gives the students time to reconsider what they did during these labs, in terms of lysate fractionation and analysis of protein concentrations and enzyme activities within.
Table 3. Timeline and notes for Lab Session 2.
| Activity | Description | Estimated Time | Notes |
|---|---|---|---|
| Lab Session 2 | |||
| Prelab preparation |
|
15 minutes |
|
| Quiz | A short 5-question short answer quiz to test basic knowledge from lab handout. | 10-15 minutes | |
| SDS-PAGE |
|
60-80 minutes |
|
| SDS-PAGE staining |
|
20 minutes |
|
| Enzyme activity measurement |
|
About 15 minutes per student group. More needed if sharing spectrophotometers. This is completed while gels are running. | It is important that students start reading immediately after adding enzyme. If they take too much time, the reaction will be done before they even start reading, especially for those samples with more enzyme. |
| Graphing Lineweaver-Burk plot | This can be done by students at home, but we encourage students to do it in lab while help is immediately available. | 30 minutes | |
Table 4. Purification Table. This table of typical student data is modeled after a similar one found in Marinkovic D.V. et al. (1977), Purification of carboxypeptidase B from human pancreas. Biochem J. 163:253-260. Students are given the Marinkovic et al. table as an example.
| Protein Fraction | Volume (mL) | CPA activity (units) | Mass of Protein (mg) | Specific Activity (unit/mg) | % Recovery | Fold Purification |
|---|---|---|---|---|---|---|
| Crude | 1 | 0.767 | 2.434 | 0.315 | 100.0 | 1.00 |
| Acid | 1 | 0.030 | 0.627 | 0.048 | 4.0 | 0.15 |
| 0-30% AS | 1 | 0.667 | 1.093 | 0.610 | 87.0 | 1.94 |
| 30-60% AS | 1 | 0.133 | 0.659 | 0.201 | 17.3 | 0.64 |
| Supernatant | 1 | 0.179 | 0.755 | 0.236 | 23.3 | 0.75 |
Teaching Discussion
This two-week laboratory activity enabled students to meet a number of learning goals in large part due to the simplicity of the approach and the abundance of enzyme. Students were able to observe the function of an enzyme and to determine how manipulation of the enzyme’s environment resulted in changes in its solubility. The labs did not require extensive background information unrelated to the learning goals of protein structure and function. The techniques required the effective use of micropipettors and spectrophotometers but could be completed without advanced experience and knowledge – the abundance of enzyme meant that almost any student could get usable data, both in the form of bands on a stained SDS-PAGE gel and enzyme activity. The materials used were relatively inexpensive, assuming the availability of micropipettors and spectrophotometers.
Students responded favorably to the activity. While not all students obtained accurate data, many commented on how nicely the labs connected with material covered in the lecture portion of the cell and molecular biology course. Student comments included “Interesting labs were done to show real applications of research in cell biology,” “The analytical biology application allowed me to see biology in a broader sense,” and “Labs were a great extension over topics discussed in lecture.” It is gratifying to see students’ responses to seeing the invisible world of biology!
Analysis of student lab practical exam responses following this activity suggested that learning outcomes were reached, although with room for improvement. Topics included on the exam included concentrations, molecular weights, qualitative analysis of SDS-PAGE results, analysis and preparation of a standard curve graph, and the proper use of a spectrophotometer in the collection of enzyme assay data. The average score on this exam over three years and one hundred students was 78%. No direct comparison can be made with learning prior to the implementation of this lab activity as a similar lab practical exam was not given.
This lab activity could be modified or extended in many ways. For example, in a more advanced setting or one where more lab sessions could be devoted to this activity, one could have students collect their own data to determine Vmax and Km. Additionally, students could use different substrates, therefore measuring the activity of different enzymes. Primarily, cow pancreas has two carboxypeptidase enzymes; carboxypeptidase A acts upon FA-Phe-Phe-OH, while carboxypeptidase B acts upon FA-Ala-Lys-OH (also available from Bachem). Other similar substrates available from Bachem include FA-Phe-Ala-OH, FA-Glu-Glu-OH, and FA-Ala-Arg-OH, which might be acted upon to differing extents by these enzymes. A variety of inhibitors are known for these enzymes, for example EDTA, EGTA, o-phenanthroline, 2-benzylsuccinic acid, and potato carboxypeptidase inhibitor (26-28), which could be made available, providing the ability to analyze different mechanisms of inhibition. Depending on availability, one could imagine obtaining pancreases from different organisms and comparing Km for carboxypeptidase A or B from these different organisms. Other tissues could also be assessed for carboxypeptidase-like activity. Perhaps, if equipment for column chromatography is available, the initial precipitation steps could be replaced by anion or cation exchange chromatography (10, 25, 29). This would also illustrate chemical bonding and the use of these features in protein analysis. There are many ways that students could take the skills learned in this two-week series and apply them in an experimental design of their own.
This activity could be modified for a lower division class. One possible simplification could be to eliminate the first precipitation steps, simply examining the carboxypeptidase enzyme activity present in crude pancreatic lysates. This has been done in a local junior high classroom, where a demonstration of enzyme activity was performed. The setup was simple, requiring the instructor to bring just FA-Phe-Phe-OH substrate, crude lysate, and a spectrophotometer. Students were introduced to proteins and their component amino acids through a brief lecture. This was followed by an engaging activity in which students constructed a portion of insulin with “pop beads” of various colors and shapes, one type of bead per amino acid. The topic of proteolysis was then described in reference to the maturation of insulin, and this was illustrated by the rapid decrease in absorbance of the FA-Phe-Phe-OH substrate in the presence of enzyme (as compared with a control where no enzyme was added).
Supporting Materials
-
S1. Pancreatic Enzyme - Student Lab Handout
-
S2. Pancreatic Enzyme - Reagent List
-
S3. Pancreatic Enzyme - Report Rubric
-
S4. Pancreatic Enzyme - Sample Enzyme Kinetics Data
Acknowledgments
Many thanks to the students and teaching assistants who have been involved in these labs, providing much-appreciated feedback on their improvement.
References
- American Association for the Advancement of Science (AAAS). 2011. Vision and change in undergraduate biology education: A call to action. AAAS, Washington, DC. http://visionandchange.org/finalreport
- Lau J, Gilbert M. 2012. Teaching biochemistry and molecular biology using dihydrofolate reductase as an expression system. The FASEB Journal 26:620.5. doi:10.1096/fasebj.26.1_supplement.620.5.
- Witherow DS. 2016. A ten-week biochemistry lab project studying wild-type and mutant bacterial alkaline phosphatase. Biochem Mol Biol Educ 44:555-564. doi:10.1002/bmb.20982.
- Craig PA. 1999. A project-oriented biochemistry laboratory course. J Chem Educ 76:1130. doi:10.1021/ed076p1130.
- Boyce A, Casey A, Walsh G. 2004. A phytase enzyme-based biochemistry practical particularly suited to students undertaking courses in biotechnology and environmental science. Biochem Mol Biol Educ 32:336-340. doi:10.1002/bmb.2004.494032050392.
- Barton JS. 2011. A comprehensive enzyme kinetic exercise for biochemistry. J Chem Educ 88:1336-1339. doi:10.1021/ed100816r.
- Nairn R, Cresswell W, Nairn J. 2015. Mushroom tyrosinase: A model system to combine experimental investigation of enzyme-catalyzed reactions, data handling using R, and enzyme-inhibitor structural studies. Biochem Mol Biol Educ 43:370-376. doi:10.1002/bmb.20887.
- Cornely K, Crespo E, Earley M, Kloter R, Levesque A, Pickering M. 1999. Kinetics of papain: An introductory biochemistry laboratory experiment. J Chem Educ 76:644. doi:10.1021/ed076p644.
- Weinheimer T, White D. 2003. Using peroxidase to demonstrate enzyme kinetics. The American Biology Teacher 65:116-121. doi:10.1662/0002-7685(2003)065[0116:UPTDEK]2.0.CO;2.
- Deal ST, Hurst MO. 1997. Utilizing isolation, purification, and characterization of enzymes as project-oriented labs for undergraduate biochemistry. J Chem Educ 74:241. doi:10.1021/ed074p241.
- Melius P. 1971. Isolation of yeast invertase by sephadex gel chromatography. A biochemistry laboratory experiment. J Chem Educ 48:765-766. doi:10.1021/ed048p765.
- Peres S, Oonk K, Riley K. 2019. The avocado lab: An inquiry-driven exploration of an enzymatic browning reaction. CourseSource 6. doi:10.24918/cs.2019.38.
- Pope SR, Tolleson TD, Williams RJ, Underhill RD, Deal ST. 1998. Working with enzymes - Where is lactose digested? An enzyme assay for nutritional biochemistry laboratories. J Chem Educ 75:761. doi:10.1021/ed075p761.
- Scheele GA, Palade GE, Tartakoff AM. 1978. Cell fractionation studies on the guinea pig pancreas. Redistribution of exocrine proteins during tissue homogenization. J Cell Biol 78:110-130. doi:10.1083/jcb.78.1.110.
- Bliss M. 2019. The discovery of insulin: The twenty-fifth anniversary edition. University of Toronto Press, Toronto, ON. doi:10.3138/9781442621480.
- Chen X, Walker AK, Strahler JR, Simon ES, Tomanicek-Volk SL, Nelson BB, Hurley MC, Ernst SA, Williams JA, Andrews PC. 2006. Organellar proteomics: Analysis of pancreatic zymogen granule membranes. Mol Cell Proteomics 5:306-312. doi:10.1074/mcp.M500172-MCP200.
- Rindler MJ, Xu CF, Gumper I, Smith NN, Neubert TA. 2007. Proteomic analysis of pancreatic zymogen granules: Identification of new granule proteins. J Proteome Res 6:2978-2992. doi:10.1021/pr0607029.
- Garcia-Guerrero MC, Garcia-Pardo J, Berenguer E, Fernandez-Alvarez R, Barfi GB, Lyons PJ, Aviles FX, Huber R, Lorenzo J, Reverter D. 2018. Crystal structure and mechanism of human carboxypeptidase O: Insights into its specific activity for acidic residues. PNAS 115:E3932-E3939. doi:10.1073/pnas.1803685115.
- Koshland DE. 1958. Application of a theory of enzyme specificity to protein synthesis. PNAS 44:98-104. doi:10.1073/pnas.44.2.98.
- Hartsuck JA, Ludwig ML, Muirhead H, Steitz TA, Lipscomb WN. 1965. Carboxypeptidase A, II. The three-dimensional electron density map at 6 Å resolution. PNAS 53:396-403. doi:10.1073/pnas.53.2.396.
- Blake CC, Koenig DF, Mair GA, North AC, Phillips DC, Sarma VR. 1965. Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Angstrom resolution. Nature 206:757-761. doi:10.1038/206757a0.
- Jaskolski M, Dauter Z, Wlodawer A. 2014. A brief history of macromolecular crystallography, illustrated by a family tree and its Nobel fruits. FEBS J 281:3985-4009. doi:10.1111/febs.12796.
- Wilson EO. 2013. Letters to a young scientist, 1st ed. Liveright Publishing Corporation, New York, NY.
- Hartsuck JA, Lipscomb WN. 1971. 1 Carboxypeptidase A, p 1-56. In Boyer P (ed), The Enzymes, vol 3. Academic Press, San Diego, CA. doi:10.1016/S1874-6047(08)60392-3.
- Marinkovic DV, Marinkovic JN, Erdos EG, Robinson CJ. 1977. Purification of carboxypeptidase B from human pancreas. Biochem J 163:253-260. doi:10.1042/bj1630253.
- Byers LD, Wolfenden R. 1972. A potent reversible inhibitor of carboxypeptidase A. J Biol Chem 247:606-608. doi:10.1016/S0021-9258(19)45746-3.
- Ryan CA, Hass GM, Kuhn RW. 1974. Purification and properties of a carboxypeptidase inhibitor from potatoes. J Biol Chem 249:5495-5499. doi:10.1016/S0021-9258(20)79755-3.
- Auld DH. 2012. Carboxypeptidase A, p 1289-1301. In Barrett AJ, Rawlings ND, Woessner J (ed), Handbook of Proteolytic Enzymes, 3rd ed. Academic Press, San Diego, CA.
- Rosenberg IM. 2005. Protein analysis and purification: Benchtop techniques, 2nd ed. Birkhauser, Boston, MA.
Article Files
Login to access supporting documents
Lyons-Isolation and Functional Analysis of a Pancreatic Enzyme.pdf(PDF | 674 KB)
S1. Pancreatic Enzyme - Student Lab Handout_final.docx(DOCX | 99 KB)
S2. Pancreatic Enzyme - Reagent List_final.docx(DOCX | 24 KB)
S3. Pancreatic Enzyme - Report Rubric_final.xlsx(XLSX | 11 KB)
S4. Pancreatic Enzyme - Sample Enzyme Kinetics Data.xlsx(XLSX | 11 KB)
- License terms

Comments
Comments
There are no comments on this resource.