Garden Variety Mutations: Using Primary Data to Understand the Central Dogma in Large-Lecture Introductory Biology
Editor: Jenna Hicks
Published online:
Abstract
The ability to interpret and create an argument from data is a crucial skill for budding scientists, yet one that is seldom practiced in introductory courses. During this argumentation module, students in a large lecture class will work in groups to understand how a single mutation can lead to an obvious phenotypic change among tomatoes. Before the module begins, students are provided with background information on mutations and techniques to give them a starting point to explain what they will see in the data. In class, students will use data from the primary literature to understand the relationship between single amino acid mutations and phenotypic variation within the context of a “big question” about garden tomatoes that ripen without turning red. Over two days, small groups will negotiate data, create and evaluate hypotheses, and consolidate their understanding through clicker questions and writing tasks. Together, they will craft an argument for how mutations can lead to phenotypic changes, even if they do not lead to disease like in many common examples. Through this activity, the instructor and students work together to understand an engaging and relevant example of the central dogma. During our implementation of this activity, we observed high engagement with the in-class and out-of-class aspects of the argumentation activities to explain how a single mutation could result in a visible change to the flesh of a tomato.
Primary Image: Garden Variety Mutations. Students work in small groups, interpreting data and evaluating hypotheses that explain how a single nucleotide change can alter tomato plant coloration.
Citation
Woodbury J, Arneson JB, Anderson J, Collins L, Cavagnetto A, Davis W, Offerdahl EG. 2022. Garden Variety Mutations: Using Primary Data to Understand the Central Dogma in Large-Lecture Introductory Biology. CourseSource 9. https://doi.org/10.24918/cs.2022.43Society Learning Goals
Biochemistry and Molecular Biology
- Information storage and flow are dynamic and interactive
- How does the nucleotide sequence of the gene lead to biological function?
Genetics
- Molecular biology of gene function
- How is genetic information expressed so it affects an organism's structure and function?
- Genetic variation
- How do different types of mutations affect genes and the corresponding mRNAs and proteins?
Lesson Learning Goals
Students will:- connect changes in DNA to an organism’s phenotype.
- understand that gene expression varies between different organisms and within the same organism.
- understand that mutations do not always lead to disease.
- practice interpreting data and evaluating hypotheses.
- From Biochemistry and Molecular Biology Learning Framework:
- “How does the nucleotide sequence of the gene lead to biological function?”
- From Genetics Learning Framework:
- “How is genetic information expressed so it affects an organism’s structure and function?
- “How do different types of mutation affect genes and the corresponding mRNAs and proteins?”
Lesson Learning Objectives
Students will be able to:- explain how a single nucleotide mutation can cause molecular changes that may lead to phenotypic differences.
- interpret photographic, northern blot, and protein sequence data from the primary literature.
- use primary data to evaluate hypotheses about the effects of mutations.
- identify how mRNA expression varies over an organism’s life cycle, in different environments, or in different cell types.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
Introduction
Understanding the flow of biological information is critical for life science majors (1). At the heart of this key concept lies the central dogma which is notoriously difficult for students to master (2-4). Students grapple with gene function, how proteins give rise to observable traits, and how changes in the DNA (e.g., mutations) may lead to altered gene expression or phenotypic differences (2-4). Students’ difficulties have been attributed to the “invisible” nature of the biological structures involved in information flow and to the need for students to engage in multilevel reasoning to generate explanations that ultimately connect the sub-molecular mechanisms to physiological outcomes (5-7). Given these challenges, creating activities that foster a deep understanding of the central dogma is crucial to students forming the foundation for in-depth learning of further biological phenomena. Importantly, it has been suggested that creating a “need to know” will increase the likelihood that students will make the causal connections necessary to master the central dogma (8).
Not surprisingly, several CourseSource activities have been developed to support student understanding of information flow in biological systems (e.g., 9-12). Many of these activities use common animal models such as Drosophila or C. elegans (9, 11), apply an inheritance- or population-based approach to understanding information flow (9, 11, 12), and use examples of human health and disease as a context to understand the central dogma (10, 12). These approaches constrain student thinking to animal models, often overlook the mechanisms of information flow, and may lead to a misconception that mutations inevitably lead to a disease state. This lesson was designed with plants as the model organism which is notable because plant-based examples are used less often in introductory biology (13). Using plant-based examples can promote student interest and knowledge in non-animal organisms, which are crucial to understanding life as a whole (14). Instead of an inheritance- or population-based approach, our lesson shifts the focus to the molecular changes linking mutation to phenotypic differences. Further, we intentionally chose an example where mutation does not lead to a disease phenotype, hoping to disconnect the assumption that mutations necessarily lead to cancer or diseases. Finally, our activity is framed by a “big question” that creates a need to know for the students.
Beyond understanding foundational concepts, undergraduate students should also develop competency in the skills and practices commonly used by biologists (15). The ability to craft and evaluate evidence-based arguments is key in biology and requires that biologists have proficient skills in data analysis, pattern recognition, and drawing appropriate conclusions. The importance of integrating argumentation-based pedagogies in the biology classroom has long been recognized in K-12 contexts (16) as studies have shown these pedagogies can improve students’ abilities to craft evidence-based arguments (17-20) and deepen students’ understanding of science concepts (21, 22).
While argumentation-based activities are increasingly implemented in undergraduate laboratory courses (23-25), they are less common in large-lecture courses due to the logistical challenges associated with high-enrollment classes. Activities that target some of the skills involved in argumentation often take place over multiple weeks and are better suited for smaller classrooms or laboratory sections as they require access to computers or laboratory equipment (e.g., 9, 11). Lessons that have been designed for the large-lecture environment prompt students to predict phenotypic changes (e.g., 10, 12). Our argumentation module extends on this by requiring students to reflect on, revise, or perhaps refute their initial predictions after collaborating with peers to interpret relevant data and consider alternative explanations.
Intended Audience
This lesson was designed for the molecular biology and genetics semester of a large-enrollment (~500 students) undergraduate introductory biology course at a research-intensive, land-grant university. This course serves a variety of majors and pre-professional programs, consisting primarily of undergraduate science students ranging from first-year students to seniors.
Though we designed this lesson to strengthen student understanding of the central dogma and genetic mutation in introductory biology, the lesson could be adapted for use in genetics or molecular and cellular biology courses. This lesson has been implemented in a fixed-seating lecture hall but is well suited for more flexible classroom environments designed for small group work.
Required Learning Time
The module was designed to be implemented across two 50-minute class periods. Students complete three online homework assignments: a pre-class quiz before Day 1 to prepare them for data interpretation, and a written summary task after each class session to demonstrate how they are making sense of the provided data and hypotheses.
Prerequisite Student Knowledge
The lesson was implemented after students received formal instruction on the central dogma, the processes of transcription and translation, amino acid properties, as well as protein folding and function. Students should be familiar enough with the concept of mutation to understand changes to the DNA sequence may (or may not) have downstream effects on transcription, translation, protein folding, and/or protein function. Students should also have some understanding of and practice with interpreting the results of gel electrophoresis. In this course, western blots were discussed during the unit on protein structure, and the background slides (S1. Garden Variety Mutations – Background Slides) prompt students to extend this understanding to northern blots. The pre-class quiz (S2. Garden Variety Mutations – Pre-Quiz) also reinforces their knowledge of gel electrophoresis in preparation for the activity.
Prerequisite Teacher Knowledge
Instructors should understand the processes of gene expression and the nature of mutations. They should also be familiar with amino acid properties and protein structure/function in order to explain how small changes to DNA can lead to phenotypic variation. Background information can be found in an introductory biology textbook (see for example, Chapters 3 & 15 of the open-source textbook Biology 2e).
Instructors should also have a basic understanding of the methods (e.g., leaf discoloration assay, northern blot) used to generate the data students are interpreting. The data sets for this activity were based on the following paper:
Barry, C. S., McQuinn, R. P., Chung, M.-Y., Besuden, A., & Giovannoni, J. J. (2008). Amino Acid Substitutions in Homologs of the STAY-GREEN Protein Are Responsible for the Green-Flesh and Chlorophyll Retainer Mutations of Tomato and Pepper. Plant Physiology, 147(1), 179–187. https://doi.org/10.1104/pp.108.118430.
Scientific Teaching Themes
Active Learning
High-intensity active learning practices (defined as spending more than two-thirds of class time on active learning) have been shown to produce more equitable outcomes among diverse populations of students (27). This module exemplifies a high-intensity design developed specifically for a large-lecture environment. Students are provided materials and a pre-class quiz to prepare for the in-class module, during which they spend the majority of their time working collaboratively to interpret data and craft written responses about what the data mean. Students also individually answer clicker questions and engage in whole-class and small-group discussion to draw connections between the clicker questions and their data interpretation. After the argumentation module, students engage in an individual writing activity to consolidate ideas from group and class discussion. Our decision to include individual writing activities for the argumentation modules was rooted in writing-to-learn research (28); most notably, research into the scientific writing heuristic approach to encourage individual knowledge consolidation outside of the group work throughout the activity (29, 30).
Assessment
We used several formative assessments to diagnose student learning throughout the argumentation modules (see below) and to capture students’ progress in interpreting data to answer the big question. In-class activities provided opportunities for students to engage with and make inferences from the data both as a group and independently. Outside of class, students completed short activities designed to help them prepare for each in-class session and to support their learning by articulating their explanation for the observed phenotypic differences. The instructor could also use responses from these activities to address potential misunderstandings or to help frame whole-class discussion. Ultimately, student understanding of the concepts covered in this lesson was assessed in a summative way on the unit exam.
Student learning was assessed by:
-
A multiple-choice quiz (pre-class on learning management software [LMS]; S2. Garden Variety Mutations – Pre-Quiz) – students individually make basic interpretations of Northern blot results.
-
Data interpretation questions (in-class; Supporting Files S3, S4) – students interpret figures from primary literature in small groups.
-
Clicker questions (in-class; Supporting Files S5, S6) – students individually use the provided data to evaluate competing hypotheses.
-
A data synthesis question (Day 1 homework on LMS; S5. Garden Variety Mutations – Day 1 Slides) – students individually review and submit a written synthesis of the figures from Day 1 of the activity.
-
The summary writing task (Day 2 homework on LMS; S6. Garden Variety Mutations – Day 2 Slides) – students individually complete a written summary to answer the “Big Question” using data from both days.
-
Exam questions (end of unit; S7. Garden Variety Mutations – Exam Questions) – students answer multiple-choice questions assessing (1) conceptual understanding of gene expression and mutation, and (2) interpretation of electrophoresis data.
Inclusive Teaching
We designed this activity to encourage more students to bring different perspectives to scientific data and discussion. Students formed small groups, leveraging different experiences and backgrounds to interpret data and evaluate hypotheses. The variety of formative assessments and whole-class discussions throughout the argumentation module allowed students to express diverse ways of knowing, and by inviting groups to contribute to a whole-class model of the phenomena, the instructor highlighted more voices and gave credibility to more ideas beyond their own. The entire activity moved students away from a strictly traditional lecture style of class and into a group effort where more students are encouraged to contribute.
Lesson Plan
Course Context
This argumentation activity (Table 1) was integrated into a large-lecture introductory biology course that covers molecular and cellular biology and genetics. This course is typically delivered in an interactive lecture format with periodic clicker questions and whole-class discussions led by the instructor. The instructor delivers the course in an amphitheater-style classroom with a daily attendance of ~450 students. We implemented this activity during the Fall 2019 and Spring 2020 semesters (prior to emergency remote teaching), reflecting on students’ discussions and performance to refine the materials each semester using a design-based research approach (31).
Table 1. Lesson Plan Timeline. The Lesson Plan for the Module allows time for small-group and whole-class discussions centered on interpreting data and evaluating hypotheses from the 3 Data Sets.
| Activity | Description | Estimated Time | Notes |
|---|---|---|---|
| Preparation for Class, Day 1 | |||
| Instructor Activities, Pre-Class Day 1 |
1. Post slides and pre-quiz. 2. Print Data Sets 1 and 2. 3. Organize materials for deployment. |
2-4 hours, depending on size of class |
See Supporting Files S1–S3. Collate printouts for quick distribution based on number of seats/classroom arrangement. |
| Student Activities, Pre-Class Day 1 |
1. Review background slides & overview. 2. Complete pre-quiz. |
1-2 hours | |
| In-Class Day 1 | |||
| Group Formation | Students self-assemble into small groups and elect a “scribe.” | <2 minutes | Project instructions before class to reduce time needed. |
| Introduce the Argumentation Module | Review background information from slides and introduce the “Big Question.” | 5 minutes | If students struggled with the pre-quiz, briefly discuss how to interpret gel electrophoresis. |
| Data Interpretation 1 | Display/ distribute Data Set 1. Students work in groups to answer questions. | 10 minutes | See Supporting Files S5 and S3. |
| Clicker Question 1 |
Present CQ 1 and poll class. Elicit student reasoning in whole class discussion. |
2-3 minutes for CQ + 7-8 minutes to discuss | Clicker Questions are found in Supporting File S5. Garden Variety Mutations – Day 1 Slides. |
| Data Interpretation 2 | Display/ distribute Data Set 2. Students work in groups to answer questions. | 10 minutes | Data Set 2 is found in Supporting File S3. Garden Variety Mutations – Day 1 Handouts. |
| Clicker Question 2 |
Present CQ2 and poll class. Elicit student reasoning in whole class discussion. |
10 minutes | Clicker Questions are found in Supporting File S5. Garden Variety Mutations – Day 1 Slides. |
| Day 1 Wrap-Up | Summarize the day’s activities and introduce the homework for Day 2. | 3 minutes | |
| Pre-Class Day 2 | |||
| Instructor Activities, Pre-Class Day 2 |
1. Post Pre-Class Homework. 2. Prepare Data Set 3 handouts. 3. Review student pre-class answers to sample student reasoning. |
2-4 hours |
Homework prompt found in S5. Garden Variety Mutations – Day 1 Slides. Data Set 3 is found in Supporting File S4. Garden Variety Mutations – Day 2 Handouts. |
| Student Activities, Pre-Class Day 2 |
1. Review Data Sets 1 & 2. 2. Answer writing prompt. |
15-30 minutes | |
| In-Class Day 2 | |||
| Introduction |
1. Ask students to re-form their groups from Day 1. 2. Display Data Sets 1 and 2 and lead brief discussion on interpretation of each. |
5 minutes | See Supporting File S6. Garden Variety Mutations – Day 2 Slides. |
| Clicker Question 3 | Present CQ3 and poll class. | 5 minutes | This question is meant to gauge student understanding of Day 1 data and remind students of the ruled-out hypothesis. |
| Data Set 3 |
Display/ distribute Data Set 3. Students work in groups to answer questions. |
10 minutes | Data Set 3 is found in Supporting File S4. Garden Variety Mutations – Day 2 Handouts. |
| Clicker Question 4 |
Present CQ4 and poll class. Elicit student reasoning in whole class discussion. |
2-3 minutes for CQ + 7-8 minutes to discuss | CQ 4 can be found in Supporting File S6. Garden Variety Mutations – Day 2 Slides. |
| Answering the Big Question | Display the Big Question and start small group discussions. | 5 minutes | Allow time for small groups to share their individual answers to the Big Question with each other. |
| Clicker Question 5 | Present CQ5 and poll class. | 2-3 minutes for CQ | CQ 5 can be found in Supporting File S6. Garden Variety Mutations – Day 2 Slides. |
| Answering the Big Question as a class | Lead the class in discussing answers to the Big Question. | 5-10 minutes | See Instructor Notes in S6. Garden Variety Mutations – Day 2 Slides for suggestions in whole class discussion. |
| Wrap-up | Summarize the Big Question discussion and introduce summary writing assignment. | 2 minutes | |
| Post-Argumentation Activity | |||
| Instructor Activities, Post-Class Day 2 | Post the Individual Student Summary Writing activity as a homework assignment. | 15 minutes | Homework prompt is found in Supporting File S6. Garden Variety Mutations – Day 2 Slides. |
| Individual Student Summary Writing | Students use Data Sets 1-3 to answer the Big Question. | 15-30 minutes | |
Pre-class Day 1
The pre-class slides provide students with an overview of the activity, a review of the central dogma and potential effects of mutations, and information about the northern blot technique (S1. Garden Variety Mutations – Background Slides). Before coming to class, students should review the slides before taking a short pre-class quiz that walks them through an example interpretation of gel electrophoresis data to help prepare them for the activity (S2. Garden Variety Mutations – Pre-Quiz). As students in our course typically are allowed multiple attempts on pre-class quizzes, we allowed students two attempts on this quiz. However, the instructor can opt to limit students to one attempt.
In Class Day 1
Introduction
As students arrive, the instructor should direct them to form small groups and display a slide with this instruction (S5. Garden Variety Mutations – Day 1 Slides). Once students are seated in their groups, the instructor should introduce the activity using the Big Question (S5. Garden Variety Mutations – Day 1 Slides):
How does DNA mutation lead to differences in the phenotype of [Instructor]’s tomatoes?
To frame this activity, the instructor should explain to students that they will reinforce their understanding of foundational biology concepts by acting as biologists engaging in scientific community and discourse to interpret data and evaluate hypotheses.
Data Set 1
While distributing Data Set 1 handouts (S3. Garden Variety Mutations – Day 1 Handouts) to all students, the instructor should broadcast the Figure 1 slide, which shows that the mutant plant has retained some green pigment after two weeks in the dark (S5. Garden Variety Mutations – Day 1 Slides), and emphasize the importance of each small group coming to a consensus when answering the associated questions. Displaying the slide as the data sets are passed out will provide students a little time to individually reflect on the data, so that once the physical copy is in front of them, groups can be ready to begin discussing their interpretation of Data Set 1 and answering the questions listed on the handout:
What do the results of these two experiments mean?
How would you explain what is happening in a tomato plant with the A → T mutation in the gf gene?
The instructor should give the class about 10 minutes to work through these problems but allow groups more time if needed for discussion.
Next, the instructor should present Clicker Question 1 (S5. Garden Variety Mutations – Day 1 Slides):
How would you explain what is happening in a tomato plant with the A → T mutation in the gf gene?
-
The mutation prevents translation of the gf mRNA into protein
-
More chlorophyll was produced in the mutant tomato plants than in wildtype plants
-
The GF protein produced did not work as well in mutant as compared to wildtype plants
-
The mutation prevented transcription of the gf gene in mutant plants
The four options represent the most common explanations students generated in a previous semester when asked to interpret Data Set 1 in an open-response question. At this point, the data provided cannot rule out any of these hypotheses, so the clicker question serves as a means to capture initial thoughts about the impact of mutation on GF expression. After students answer the clicker question, the instructor should display the distribution of student responses and ask for volunteers to explain the reasoning behind their selections. The follow-up discussion should address all four options, and the instructor should affirm for the class that, despite whatever trend may emerge in student responses, there is not a single correct answer. While the hypotheses provided were the most common, they are by no means the only possible explanations for the phenotypic differences between wildtype and mutant tomatoes. Thus, the instructor should prompt students to share other ideas their groups had and write those down on the overhead or slide to give credit to students who have come up with another possible explanation.
Data Set 2
While distributing Data Set 2, the instructor should emphasize the need for more information to rule out some of the potential explanations. This northern blot data reveals the mutant gf gene is expressed during the same stages of development as the wildtype GF. Since introductory-level students may be less familiar with northern blots, the instructor could provide a brief explanation of the technique to review the information from the pre-class activity before groups begin discussion. Ten minutes are an appropriate starting point to work through the new set of data and answer the following questions (S3. Garden Variety Mutations – Day 1 Handouts):
How does the expression of the mutant gene compare to that of the wildtype gf gene?
How, if at all, do the results in Figure 2 alter your prediction about the effect of the A → T mutation from Figure 1?
The instructor should verify that small groups are coming to consensus while discussing their answers and allow more time for discussion if needed. While groups are working and discussing, the instructor should move through the classroom, listening to the reasoning groups are coming up with, clearing up confusion, and answering questions.
Next, the instructor should present Clicker Question 2 (S5. Garden Variety Mutations – Day 1 Slides):
Based on Data Set 2, which explanation can you rule out?
-
The mutation prevents translation of the gf mRNA into protein
-
More chlorophyll was produced in the mutant tomato plants than in wildtype plants
-
The GF protein produced did not work as well in mutant as compared to wildtype plants
-
The mutation prevented transcription of the gf gene in mutant plants
During a follow-up whole-class discussion, student volunteers should explain why they ruled out a particular hypothesis and why they could not rule out the others. The instructor should be sure to note when students bring up evidence from one of the data sets.
Day 1 Wrap-up
Following whole-class discussion, the instructor should summarize the ideas posed by students about Data Sets 1 & 2, and then introduce the homework assignment to be completed before Day 2 (S5. Garden Variety Mutations – Day 1 Slides).
Pre-class Day 2
Students should complete the pre-class activity for Day 2, one open-ended question requiring a written response (S5. Garden Variety Mutations – Day 1 Slides):
What is your current explanation for how DNA mutation leads to differences in the phenotype of the tomatoes?
Before class, the instructor could skim responses for general trends and potential misunderstandings or misinterpretations and use this information to shape the introductory discussion on Day 2.
In Class Day 2
Day 2 Introduction
Again, students should be directed to form groups upon entering the classroom. Though we asked students to work in the same group for both days, this may not be necessary. When groups are settled, the instructor should remind the class about the Big Question and display the first two data sets. The instructor could share or ask students to share the conclusions drawn from the first two data sets and remind the class of information from the activity background. They could also incorporate trends seen in the pre-class activity for Day 2 into this initial discussion. Next, the instructor should present Clicker Question 3 (S6. Garden Variety Mutations – Day 2 Slides).
How would you explain what is happening in a tomato plant with the A → T mutation in the gf gene?
-
The mutation prevents translation of the gf mRNA into protein
-
More chlorophyll was produced in the mutant tomato plants than in wildtype plants
-
The GF protein produced did not work as well in mutant as compared to wildtype plants
-
The mutation prevented transcription of the gf gene in mutant plants
Once students have entered their answers, the instructor should affirm that only one explanation can be ruled out at this point and emphasize the need for more data to evaluate the remaining hypotheses. The instructor should then introduce the third piece of data (S4. Garden Variety Mutations – Day 2 Handout).
Data Set 3
Data Set 3 depicts a portion of the primary structure of the wildtype and mutant proteins, demonstrating the mutation resulted in one amino acid residue being substituted by a different residue. After Data Set 3 is displayed and distributed, students should discuss the following questions with their team (S4. Garden Variety Mutations – Day 2 Handout):
How, if at all, did the A → T nucleotide change affect the protein’s primary structure?
How, if at all, did the A → T nucleotide change affect the function of the wildtype protein?
The instructor should monitor student groups to determine how students are working with the data, what explanations student groups have generated for Data Set 3, and the degree to which there is any confusion about the data.
Next, the instructor should start Clicker Question 4 (S6. Garden Variety Mutations – Day 2 Slides):
Based on Data Set 3, which explanation can you rule out?
-
The mutation prevents translation of the gf mRNA into protein
-
More chlorophyll was produced in the mutant tomato plants than in wildtype plants
-
The GF protein produced did not work as well in mutant as compared to wildtype plants
-
The mutation prevented transcription of the gf gene in mutant plants
After the class has answered this clicker question individually, the instructor could start the discussion by asking students to explain what information they gained from Data Set 3. Using that information, volunteers can justify why they ruled out their selected explanation and why they could not rule out the other hypotheses. Some students may need prompting to think deeper about the structural and functional consequences of substituting amino acids in the primary sequence. While facilitating the discussion, the instructor should highlight when student volunteers offer details about amino acid properties, protein folding, or the impact of structure on function. If necessary, the instructor can pose guiding questions to elicit those details and connections.
Answering the Big Question
As the final group component of the activity, the instructor should display the Big Question on the slides (S6. Garden Variety Mutations – Day 2 Slides). The instructor should direct students to start a discussion of the Big Question in their groups:
How does DNA mutation lead to differences in the phenotype of [Instructor]’s tomatoes?
After a few minutes of group discussion, the instructor should start Clicker Question 5 (S6. Garden Variety Mutations – Day 2 Slides):
How would you explain what is happening in a tomato plant with the A → T mutation in the gf gene?
-
The mutation prevents translation of the gf mRNA into protein
-
More chlorophyll was produced in the mutant tomato plants than in wildtype plants
-
The GF protein produced did not work as well in mutant as compared to wildtype plants
-
The mutation prevented transcription of the gf gene in mutant plants
Following the clicker question, the instructor should begin a whole class discussion, eliciting multiple groups’ ideas and prompting students to offer additional details to support or to counter provided explanations. Students should be encouraged to use the three data sets as evidence to support their explanations and to discuss the limitations of the provided data. The instructor could also prompt students to think about what additional data they might collect to further evaluate the remaining hypotheses. At the end of class, the instructor should summarize the results of the whole-class discussion and introduce the individual homework to be completed outside of class.
After Day 2
Students should work independently to answer the Big Question as a homework assignment (S6. Garden Variety Mutations – Day 2 Slides):
“How does the single nucleotide mutation in the GF gene affect the phenotype of [Instructor]’s tomato plants?”
Teaching Discussion
Observations
This argumentation module provided students with an opportunity to engage each other in meaningful discussion in class and to articulate their own reasoning through writing assignments. Typically, students in this course would have multiple attempts to complete five multiple-choice reading questions before coming to class, where they would participate by answering a few clicker questions sprinkled throughout the lecture. Despite the significant departure from typical class expectations, the argumentation module was still only worth a small number of points (homework and in-class participation for the two-day module comprised ~1.5% of the course grade). Based on this and the scheduling of the module right before the first exam, we were initially concerned students might not engage in pre-class activities or in-class argumentation.
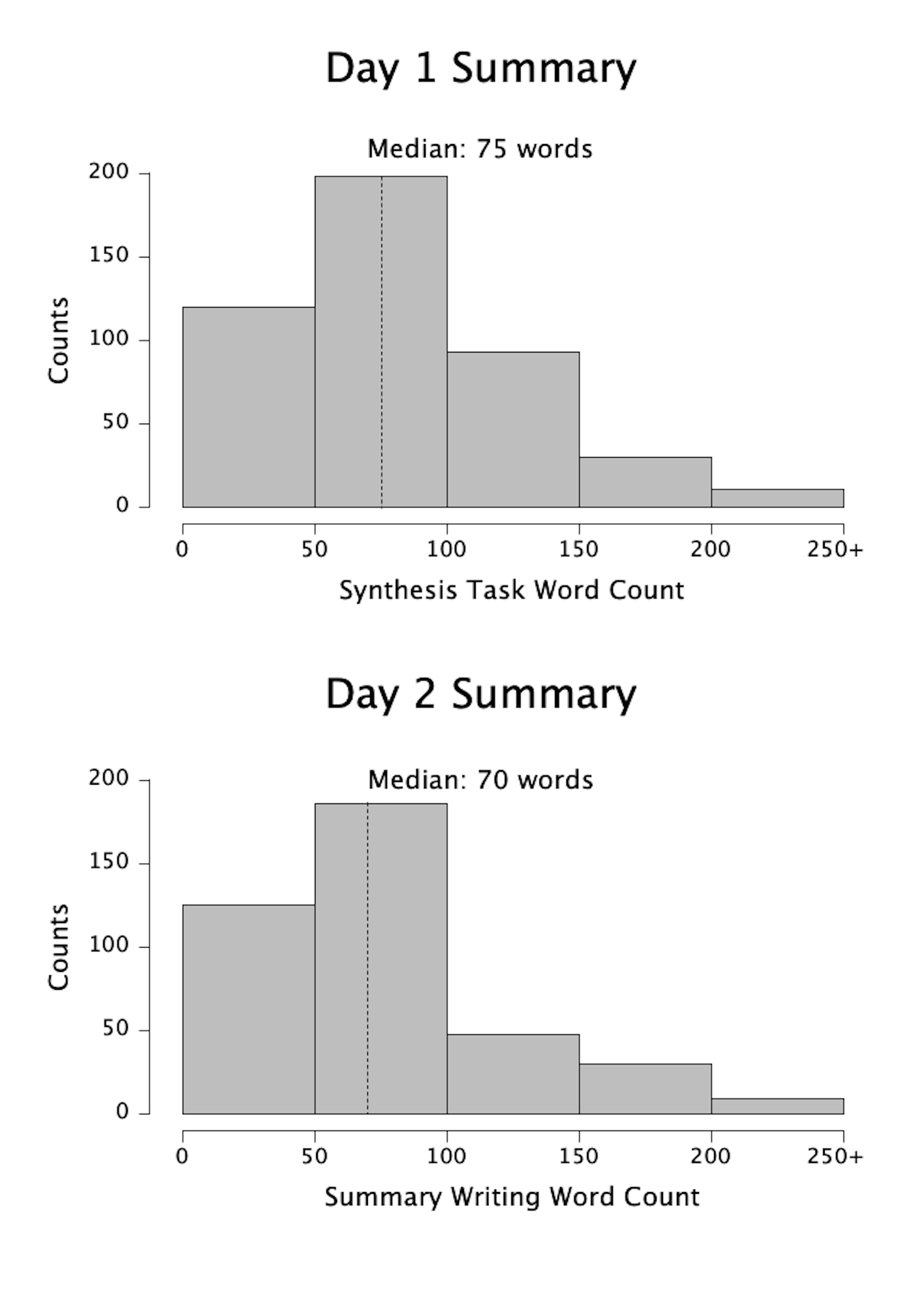
However, we were pleased to see that students completed their homework and participated in clicker questions to a similar extent during the argumentation module as they had during typical class days (Table 2). After the clicker questions, students engaged in lively discussions within their small groups, and several volunteers offered their groups’ ideas during whole-class discussion. On the daily writing assignments, most students submitted at least a paragraph (median word counts ~70; Figure 1) in which they leveraged the provided data and hypotheses to answer the Big Question. Many students evaluated multiple hypotheses in their responses and even used the argumentation module resources to generate their own alternative, testable explanations.
Table 2. Student participation in the clickers and homework fell within the typical ranges of participation for the course.
| Typical Class Sessions | During the Argumentation Module | |
|---|---|---|
| Clicker Participation | 77-93% |
Day 1: 84% Day 2: 92% |
| Homework Completion | 87-97% |
Pre-Quiz for Day 1: 79% Pre-Quiz for Day 2: 91% Summary Activity: 79% |
Suggestions for possible improvements or adaptations
When implemented, the Day 2 activities and discussions did not require the entire session, leaving the instructor 10-15 minutes to use for general exam review. While this suited the needs for our course, we recognize some instructors may want to extend the module, especially if students are less accustomed to engaging in discussion. On Day 1, for instance, groups could be asked to draw models that make explicit the connections between each of the hypotheses and the mutant tomatoes’ phenotype following Clicker Question 1. These models could potentially help students connect the hypotheses to the data sets and the molecular processes involved.
Additional time could also be used to facilitate student discussion on Day 2. Instead of beginning with a whole-class discussion, the instructor could first ask students to discuss how they answered the Day 1 writing assignment in their small groups. This could help students refresh their memory of the previous day’s data and consider more diverse ideas before the instructor brings the class back together with a review clicker and discussion.
Alternatively, an additional task could be assigned at the end of the module. Since there are multiple explanations that have not been ruled out by the provided data, groups could work together to brainstorm what other data they could collect or devise an experiment to further test hypotheses. For an upper-level course, students could make use of their knowledge of molecular techniques to assist in this experimental design.
Reflections
In a previous implementation of this module, student responses revealed they were mainly trying to discern minute differences in signal intensities within the northern blot and neglecting the broader picture; the gene is expressed during the same stages of ripening in both plants. This prompted us to make three important changes to the module. Specifically, we 1) were more thoughtful in introducing the experimental method, explicitly describing why loading controls are used, 2) altered the final hypothesis to have them evaluate if transcription was prevented rather than affected, and 3) made sure to discuss discrepancies in interpreting the blot during follow-up, highlighting why it can be difficult to visually determine differences in signal intensity. We considered but ultimately rejected adding quantitative measurements to the data set as it would likely detract students from trying to make sense of and debating their interpretations of the actual blot if they could simply glean from a bar chart that there was no difference.
The other figures were less challenging for students to interpret. We recommend, however, encouraging students to draw on their prior knowledge to consider the biological implications of the results during small group and whole class discussions. For Figure 3, for instance, many students merely identified the substituted amino acid or classified the type of mutation that took place. Additional prompting was necessary to help students think more deeply about the consequences of such a mutation and how those changes in protein structure may lead to the observed phenotypic changes. Students were largely able to rule out the appropriate hypotheses after each data set interpretation, and they predominantly selected Hypothesis C as the most likely explanation by the end of the module. Some students, however, still felt Hypothesis B was the more likely explanation. This was the ideal result for us because it set the discussion up for limitations of data and potential alternative explanations.
We intentionally designed this activity to be simpler in terms of both content and data, so as to provide students an opportunity to practice argumentation skills in a review situation, before asking them to do so while diving into new concepts and more challenging data (32). We still recommend that students be provided opportunities to practice extracting information from common biological representations (e.g., graphs, schematics, gels) before engaging in argumentation to help scaffold their skill development.
This argumentation module was one of two such modules designed to augment the interactive lecture course (see [32] for the other module). While we felt students got a lot out of this module, we believe that students’ argumentation skills will grow even more with practice and use in different content areas. It would be exciting to see an introductory course where the majority of time was structured around close inspection of data, hypothesis testing, and scientific argumentation.
Supporting Materials
-
S1. Garden Variety Mutations – Background Slides
-
S2. Garden Variety Mutations – Pre-Quiz
-
S3. Garden Variety Mutations – Day 1 Handouts
-
S4. Garden Variety Mutations – Day 2 Handouts
-
S5. Garden Variety Mutations – Day 1 Slides
-
S6. Garden Variety Mutations – Day 2 Slides
-
S7. Garden Variety Mutations – Exam Questions
Acknowledgements
This material is based upon work supported by the National Science Foundation under Grant No. 1822490. This work was reviewed by the WSU Office of Research Assurances and determined to satisfy the criteria for Exempt Research at 45 CFR 46.101 (b)(1) and 45 CFR 46.101(b)(2).
References
- Brownell SE, Freeman S, Wenderoth MP, Crowe AJ. 2014. BioCore Guide: A tool for interpreting the core concepts of Vision and Change for biology majors. CBE Life Sci Educ 13(2):200–211. doi:10.1187/cbe.13-12-0233.
- Todd A, Romine WL, Correa-Menendez J. 2019. Modeling the transition from a phenotypic to genotypic conceptualization of genetics in a university-level introductory biology context. Res Sci Educ 49(2):569–589. doi:10.1007/s11165-017-9626-2.
- Wright LK, Newman DL. 2013. Using PCR to target misconceptions about gene expression. J Microbio Bio Educ 14(1):93–100. doi:10.1128/jmbe.v14i1.539.
- Wright LK, Fisk JN, Newman DL. 2014. DNA → RNA: What do students think the arrow means? CBE Life Sci Educ 13(2):338–348. doi:10.1187/cbe.cbe-13-09-0188.
- Marbach-Ad G, Stavy R. 2000. Students’ cellular and molecular explanations of genetic phenomena. J Biol Educ 34:200–205. doi:10.1080/00219266.2000.9655718.
- Duncan RG, Reiser BJ. 2007. Reasoning across ontologically distinct levels: Students’ understandings of molecular genetics. J Res Sci Teach 44:938–959. doi:10.1002/tea.20186.
- Duncan RG. 2007. The role of domain-specific knowledge in generative reasoning about complicated multileveled phenomena. Cogn Instr 25:271–336. doi:10.1080/07370000701632355.
- Southard K, Wince T, Meddleton S, Bolger MS. 2016. Features of knowledge building in biology: Understanding undergraduate students’ ideas about molecular mechanisms. CBE Life Sci Educ 15(1):ar7. doi:10.1187/cbe.15-05-0114.
- Ross J. 2016. Predicting and classifying effects of insertion and deletion mutations on protein coding regions. CourseSource 3. doi:10.24918/cs.2016.18.
- Pelletreau KN, Andrews T, Armstrong N, Bedell MA, Dastoor F, Dean N, Erster S, Fata-Hartley C, Guild N, Greig H, Hall D, Knight JK, Koslowsky D, Lemons P, Martin J, McCourt J, Merrill J, Moscarella R, Nehm R, Northington R, Olsen B, Prevost L, Stolzfus J, Urban-Lurain M, Smith MK. 2016. A clicker-based case study that untangles student thinking about the processes in the central dogma. CourseSource 3. doi:10.24918/cs.2016.15.
- Dean DM, Deitcher DL, Loehlin DW, Banta LM. 2020. Mapping a mutation to its gene: The “fly lab” as a modern research experience. CourseSource 7. doi:10.24918/cs.2020.51.
- DeVito SR. 2021. Targeting misconceptions in the central dogma by examining viral infection. CourseSource 8. doi:10.24918/cs.2021.31.
- Brownlee K, Parsley KM, Sabel JL. 2021. An analysis of plant awareness disparity within introductory biology textbook images. J Biol Educ doi:10.1080/00219266.2021.1920301.
- Parsley KM. 2020. Plant awareness disparity: A case for renaming plant blindness. Plants, People, Planet 2(6):598–601. doi:10.1080/00219266.2021.1920301.
- Clemmons AW, Timbrook J, Herron JC, Crowe AJ. 2020. Bioskills Guide: Development and national validation of a tool for interpreting the Vision and Change core competencies. CBE Life Sci Educ 19(4):ar53. doi:10.1187/cbe.19-11-0259.
- Cavagnetto, AR. 2010. Argument to foster scientific literacy a review of argument interventions in K–12 science contexts. Review of Educational Research 80(3):336–371. doi:10.3102/0034654310376953.
- Andriessen J, Baker M, Suthers DD. 2003. Arguing to learn: Confronting cognitions in computer-supported collaborative learning environments. Springer, Dordrecht, Germany. doi:10.1007/978-94-017-0781-7.
- Osborne J, Erduran S, Simon S. 2004. Enhancing the quality of argumentation in school science. J Res Sci Teach 41(10):994–1020. doi:10.1002/tea.20035.
- Garcia-Mila M, Gilabert S, Erduran S, Felton M. 2013. The effect of argumentative task goal on the quality of argumentative discourse. Sci Educ 97(4):497–523. doi:10.1002/sce.21057.
- McNeill KL, Krajcik J. 2008. Scientific explanations: Characterizing and evaluating the effects of teachers’ instructional practices on student learning. J Res Sci Teach 45(1):53–78. doi:10.1002/tea.20201.
- Poock JR, Burke KA, Greenbowe TJ, Hand BM. 2007. Using the Science Writing Heuristic in the general chemistry laboratory to improve students’ academic performance. J Chem Educ 84(8):1371–1379. doi:10.1021/ed084p1371.
- Rudd J, Greenbowe TJ, Hand BM. 2007. Using the Science Writing Heuristic to improve students’ understanding of general equilibrium. J Chem Educ 84(12):2007–2011. doi:10.1021/ed078p1680.
- Yaman F. 2018. Effects of the Science Writing Heuristic approach on the quality of prospective science teachers’ argumentative writing and their understanding of scientific argumentation. Int J Sci Math Educ 16(3):421–442. doi:10.1007/s10763-016-9788-9.
- Hosbein KN, Alvarez-Bell R, Callis-Duehl KL, Sampson V, Wolf SF, Walker, JP. 2020. Development of the investigation design, explanation, and argument assessment for general chemistry I laboratory. J Chem Educ 98(2):293–306. doi:10.1021/acs.jchemed.0c01075.
- Stephenson NS, Sadler-McKnight NP. 2016. Developing critical thinking skills using the Science Writing Heuristic in the chemistry laboratory. Chem Ed Res Pract 17(1):72–79. doi:10.1039/C5RP00102A.
- Barry CS, McQuinn RP, Chung M-Y, Besuden A, Giovannoni JJ. 2008. Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Phys 147(1):179–187. doi:10.1104/pp.108.118430.
- Theobald EJ, Hill MJ, Tran E, Agrawal S, Arroyo EN, Behling S, Chambwe N, Cintrón DL, Cooper JD, Dunster G, Grummer JA, Hennessey K, Hsiao J, Iranon N, Jones L, Jordt H, Keller M, Lacey ME, Littlefield CE, Lowe A, Newman S, Okolo V, Olroyd S, Peecook BR, Pickett SB, Slager DL, Caviedes-Solis IW, Stanchak KE, Sundaravardan V, Valdebenito C, Williams CR, Zinsli K, Freeman S. 2020. Active learning narrows achievement gaps for underrepresented students in undergraduate science, technology, engineering, and math. PNAS 117(12):6476–6483. doi:10.1073/pnas.1916903117.
- Wallace CS, Hand B, Prain V. 2004. Writing and learning in the science classroom. Springer, Dordrecht, Germany. doi:10.1007/978-1-4020-2018-6.
- Gunel M, Hand B, Prain V. 2007. Writing for learning in science: A secondary analysis of six studies. Int J Sci Math Educ 5(4):615–637. doi:10.1007/s10763-007-9082-y.
- Burke KA, Hand B, Poock J, Greenbow T. 2005. Using the Science Writing Heuristic: Training chemistry teaching assistants. J Coll Sci Teach 35(1):36–41.
- Barab S, Squire K. 2004. Design-based research: Putting a stake in the ground. J Learn Sci 13(1):1–14. doi:10.1207/s15327809jls1301_1.
- Arneson JB, Woodbury J, Anderson J, Collins L, Cavagnetto A, Davis WB, Offerdahl EG. 2022. Splicing it together: Using primary data to explore RNA splicing and gene expression in large-lecture introductory biology. CourseSource 9. doi:10.24918/cs.2022.11.
Article Files
Login to access supporting documents
Woodbury-Arneson-Anderson-Collins-Cavagnetto-Davis-Offerdahl-Garden Variety Mutations Using Primary Data to Understand the Central Dogma in Large-Lecture Introductory Biology.pdf(PDF | 254 KB)
S1. Garden Variety Mutations - Background Slides.pptx(PPTX | 158 KB)
S2. Garden Variety Mutations - Pre-Quiz.docx(DOCX | 47 KB)
S3. Garden Variety Mutations - Day 1 Handouts.docx(DOCX | 2 MB)
S4. Garden Variety Mutations - Day 2 Handouts.docx(DOCX | 82 KB)
S5. Garden Variety Mutations - Day 1 Slides.pptx(PPTX | 6 MB)
S6. Garden Variety Mutations - Day 2 Slides.pptx(PPTX | 6 MB)
S7. Garden Variety Mutations - Exam Questions.docx(DOCX | 68 KB)
- License terms

Comments
Comments
There are no comments on this resource.