Sanger Sequencing by Hand: Using Paper Clips to Demonstrate Chain Termination
Editor: Rou-Jia Sung
Published online:
Abstract
Sanger sequencing is commonly taught with a hands-on approach. Sanger sequencing involves chain termination by dideoxynucleotides, because they are missing the oxygen on the 3’ carbon atom, which is required for the addition of subsequent nucleotides. This is a first-generation sequencing method, but it is still relevant to genetics today. It is also simpler than next-generation sequencing. Therefore, Sanger sequencing is a helpful introduction to sequencing techniques for students. The concept of chain termination can be visualized in a variety of ways using different objects, such as candy or cut pieces of paper, to represent deoxynucleotides and dideoxynucleotides. However, we believe the chemical explanation for chain termination is often lost in these activities. In our activity, we designed an affordable and manageable way to highlight the functional groups that are important in the polymerization of nucleotides. By using paper clips, we provide a new method for showing chain termination. Most significantly, our paper clips show a simplified version of the chemical reaction of bond formation and prevent addition of another nucleotide once the dideoxynucleotide is attached. This helps students to understand why dideoxynucleotides cause chain termination more directly than some other activities currently available.
Primary Image: “Sanger Sequencing by Hand” activity. A completed set of DNA fragments which have been terminated by fluorescently labeled dideoxynucleotides.
Citation
Smith CE, Ho Pao C. 2023. Sanger Sequencing by Hand: Using Paper Clips to Demonstrate Chain Termination. CourseSource 10. https://doi.org/10.24918/cs.2023.7Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
Introduction
Sanger sequencing is a type of first-generation sequencing technology developed by Frederick Sanger and colleagues in 1977 (1). The book Genetics: Analysis & Principles published by McGraw Hill (2) and a review article on sequencing techniques in the peer-reviewed journal Genome Research (3) provide a review of the Sanger sequencing method currently in use. Dideoxynucleotides (ddNTPs) are engineered nucleotides that cause chain termination because they are missing the hydroxyl (—OH) group at the 3’ site. In the Sanger sequencing reaction, deoxynucleotides (dNTPs) and ddNTPS of all four DNA bases are combined with DNA polymerase and the amplified DNA to be sequenced. A corresponding primer is annealed to the template DNA, and nucleotides are added sequentially according to the template DNA. When a ddNTP is added to the growing DNA strand, the strand is terminated because there is no 3’ oxygen with which a new nucleotide can bind. In this reaction, nucleotides will be added according to the normal base pairing rules following the template strand. Yet, whether dNTPs or ddNTPs are added will be random, resulting in termination at every possible point in the sequence, corresponding to every possible DNA fragment size (2). In automated Sanger sequencing, the ddNTPs are fluorescently labeled, with a different color corresponding to each nitrogenous base. Once the fragments are run on a gel, the DNA sequence is electronically determined based on fluorescence detection (3). The synthesized complementary sequence is read from the bottom to the top of the gel since distance run corresponds to size, with the smaller fragments traveling further (2).
Sanger sequencing is an easy way to introduce sequencing methods to students. Next-generation sequencing has replaced Sanger sequencing in many cases (4, 5). However, next-generation sequencing is more complicated, and Sanger sequencing allows students to grasp the concept of sequencing while they learn valuable historical developments of the genetics field. Research has shown that students have difficulties understanding how physics and chemistry are connected to biology (6). Our lesson addresses this by showing the functional groups and utilizing a hands-on activity that reinforces the basic genetics and specifically sequencing concepts.
We designed this lesson in three steps for students to learn, interpret, and apply their knowledge.
Learn
Several instructors and universities have developed hands-on learning methods to teach Sanger Sequencing. Many currently available activities involve objects such as candy (7) or pieces of paper (8) that cannot be physically connected and do not show the chemical structure of the nucleotide. Additionally, some commercially available kits may have connectable pieces but still do not show the functional groups that are driving chain synthesis and termination (e.g., 3D Molecular Designs Biotechnology Kit). Our activity involves paper clips that can be connected to synthesize a complementary DNA strand by hand. The paper clips have paper attached that show the 5’ phosphate and the 3’ hydroxyl group on the nucleotide. However, it is not possible to add more paper clips once a paper clip with only hydrogen (ddNTP) instead of a hydroxyl group is added. Physical obstruction has also been demonstrated using unifix cubes and tape (9). However, in that activity, the cubes were not labeled with the nucleotide base and did not show the chemical bonding between nucleotides. Thus, our goal was to provide an updated activity that directly incorporates the chemical explanation for chain termination.
Interpret
We also wanted students to be able to understand how their fragments would be visualized on a gel. Others have included a visualization of the gel in their activities (e.g., 7, 8), but we created a handout that was more clearly labeled. The nucleotide numbers along the side of the gel helps students determine which direction to read the gel, as done in a helpful YouTube video that explains Sanger sequencing. Students also color in a gel handout with the corresponding fluorescent dye for each nucleotide—a helpful visual not provided in other activities. In these ways, students are guided to analyze their results on several levels.
Apply
The last step in our activity provides a practical application concerning mutations (10). Students will better understand the relevance of Sanger sequencing by finding a mutation in a sequenced piece of DNA. We believe this application is valuable and is missing from the currently available activities. Helping students make connections and understand the relevance of technology in the real world helps them to learn better and appreciate biology concepts on a broader scale (11).
Pre-class preparation
Instructors should print out two copies of the assessment quiz (Supporting File S1; answer key is available in Supporting File S2) and a student activity pack (Supporting File S3) for each student.
Instructors should prepare the “nucleotides” (paper clips) by wrapping and taping small pieces of white paper around the inner loop of jumbo paper clips. These papers may be labeled by hand or printed.
For each pair of students, the instructor should prepare the proper number of dNTP and ddNTP paper clips following options 1 or 2 below. This ratio of paper clips allows for a greater chance of producing all of the needed fragments.
-
dNTP paperclips:
-
6 “A”
-
6 “T”
-
5 “G”
-
4 “C”
-
-
ddNTP paperclips:
-
2 “T”
-
2 “C”
-
1 “A”
-
1 “G”
-
Option 1: Handwritten labels
The base letter should be written in the middle of each paperclip. At the top of the paper clip, write the letter “P” for the 5’ phosphate and write “OH” at the bottom of the paper clip for the 3’ hydroxyl group (Figure 1).
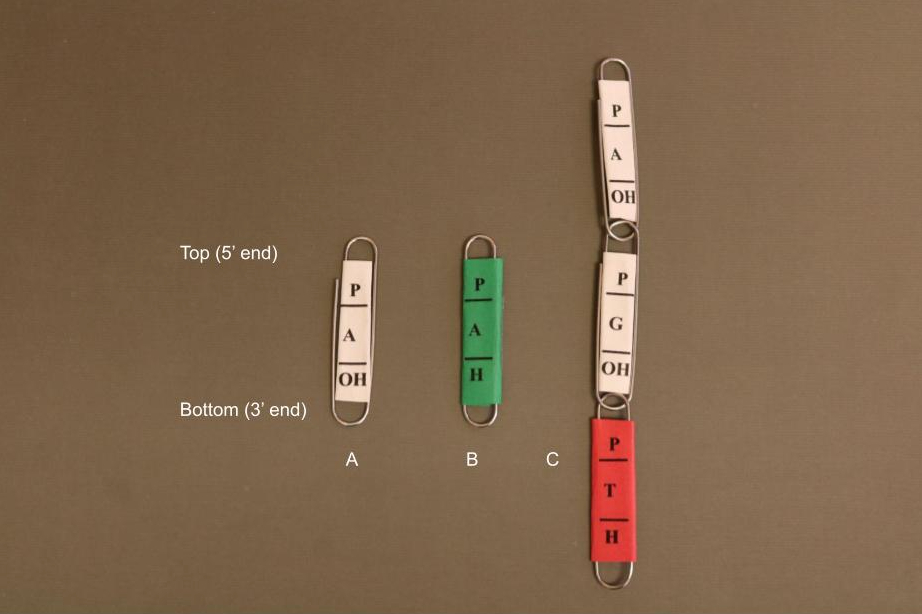
Instructors should also make ddNTP paper clips. They should have the letter “H” instead of “OH.” For ddNTPs, wrap the paper around the outside of the paper clip so it cannot hook to another paper clip (i.e., no additional nucleotides can be added to the chain). Each base corresponds to a different color paper: blue for C, red for T, green for A, and yellow for G. These are the fluorescent dyes usually used for these bases in Sanger sequencing (2).
Option 2: Printed labels
Alternatively, the dNTP and ddNTP labels (Supporting File S4) can be printed on appropriately colored paper. The proper number of labels can be cut out for each group of two students as indicated above.
Preparing nucleotide sets
Mix the dNTPs and the ddNTPs together. Put all of the nucleotides for base A (dNTPs and ddNTPs) in one envelope, all of the nucleotides for base C in a different envelope, and so on. Fill four envelopes for each group of two students.
Instructors can also prepare a completed reference example (Figure 2) for part three of the activity. This may be useful if the class size is small and it is less likely that all fragment sizes will be made.

Supplies for each group of two students:
-
2 student activity packs (Supporting File S3)
-
4 envelopes of nucleotides (dNTPs and ddNTPs)
-
Colored pencils: yellow, blue, green, red
Lesson Plan
Pre assessment
The assessment questions (Supporting File S1) should be distributed to the class, and answered question sheets turned in before starting the activity. Students should write “pre” on the top of the sheet.
Optional additions
At the beginning of the class period, instructors may choose to present an overview of Sanger sequencing (e.g., the chapter 20.3 section of the PowerPoint slides from [2]).
Instructors may wish to provide a review of nucleotide structure and the polymerization of nucleotides to provide the chemical context for sequencing. Such information can be found in most textbooks that cover basic genetics (including this chapter provided by LibreTexts).
Finally, if time allows, a discussion on the ethical implications of sequencing can be added. For example, whole genomes can now be sequenced, which raises the question of whether sequences can be owned (12). There has also been concern over the ethical treatment of marginalized groups. For example, of specific concern has been the sequencing of a deceased indigenous man’s genome from a hair sample when it would be difficult to receive consent from all those who may be concerned (13). Also, the issue of race has been a major controversial topic in genetics research. Using race as a category for genetics research has led to confusion due to race being a socially constructed category and many races having mixed heritage (14). Although such research is intended to uncover important answers to diseases (14), it has also increased controversy and created a new means for stereotyping (15).
Instructors can discuss the ethics of sequencing technology in a lecture, or assign a review article (15) to be read before class. Then as another option, students can have a group discussion about the assigned article.
Part 1: Random addition of ddNTPs
To begin the activity, instructors ask students to form pairs. The instructors provide each pair with two student activity packs (Supporting File S3), paperclips in envelopes, colored pencils (see supplies list above) and walk the students through the instructions listed below.
Students will pretend they are the DNA polymerase and add the complementary nucleotides after the primer for the DNA to be sequenced using the provided paperclips:
-
“Randomly select and connect nucleotides (paper clips) out of envelopes of the four bases, which include a mix of dNTPs and ddNTPs. Notice there is no way to add another nucleotide once a ddNTP is added.
-
“Each student makes three DNA fragments this way.”
Part 2: Combining Sanger Sequencing fragments
Students follow these instructions next:
- “Combine fragments in each group with the fragments from the entire class.
-
“Line the fragments up from longest (on top) to shortest (on bottom). Exclude any identical fragments or unterminated fragments.”
The instructor should then check the results to ensure they are correct before moving onto the third part of the activity.
Part 3: Interpreting fragments
Students continue working with their partner to complete the following:
-
“Color in the bands with the fluorescent dye color that corresponds to the proper nucleotide in the gel.
-
“If there are any fragments missing, refer to the instructor's reference set to finish coloring in the bands.”
Part 4: Application
Next, students work alone to answer the following question corresponding to the completed gel image in the student activity pack (Supporting File S3):
“Sanger sequencing generated fragments are represented on the gel below. Researchers are trying to discover if a patient has a mutation that commonly causes cancer in a specific fragment of a gene. The normal sequence is 5’—TCGGTCGACAATCTG—3’. Write out the mutated gene sequence and circle the mutation.”
Accommodation
If unable to print part 4 in color, the instructor can print a black and white sheet (Supporting File S5) and fill in the gel with colored pencils or markers. If any students are colorblind, the instructor should ensure that the pencils or markers are colorblind-friendly.
Post assessment
Once the students have completed the activity, a new copy of the assessment questions (Supporting File S1) can be distributed to the class to determine how much their knowledge has improved after the activity. Students should write “post” on the top of the sheet. The instructor can use the answer key (Supporting File S2) to grade the pre and post assessments. The instructor might also choose to have students explain their answers aloud.
Instructors may wish to provide an additional form of assessment. In the supplemental material of an education journal article, there is a 16-item survey that can aid the instructor’s understanding of student engagement with the activity (16).
Teaching Discussion
Our undergraduate genetics students had little difficulty completing the activity and expressed their enjoyment in participating. Additionally, everyone scored higher on the post assessment.
The ratio of dNTPs to ddNTPs appeared appropriate. Even with a class size of 6 (3 sets of pairs), every possible fragment size was made and the reference set was not needed.
Scientific Teaching Themes
Active Learning
Our activity incorporates multiple strategies to allow for different learning styles and active engagement (17). Students are paired for most of the activity. Having students in small groups encourages participation from each (17). Students are not able to rely on their partner completing the activity packet because part 4 requires students to work independently to ensure that everyone understands the concepts. Students also work together as a class to combine their fragments to try to make a complete set of all possible fragment sizes. After they take the post assessment quiz, students are encouraged to explain their answers. These changes in dynamics allow students to actively participate on different levels including learning, explaining, and applying the information. The activity is also very hands-on. Students physically connect paper clips and color in a gel to complete the activity. These simple tasks translate to understanding more complex information in a way that is straightforward and approachable, as has been previously demonstrated using models for understanding genetics concepts (18).
Assessment
Our main assessment was based around the assessment questions (Supporting File S1) given to the students before and after the activity to determine whether the activity helped them grasp the major concepts. The assessment was also based on the proper completion of the activity pack (Supporting File S3).
Inclusive Teaching
All students actively participate in our activity with a partner and the whole class. This allows all students to be included. Quieter students or students who need more help will not be lost in a large group setting since they are originally grouped in pairs (17). Then, bringing everyone together encourages large group teamwork once they are more familiar with the concepts. Part 4 of the activity is designed to be completed individually, since students should understand the activity enough to confidently work alone by this last step. In these ways, our activity encourages all students to participate and contribute to completing the activity on different levels.
Supporting Materials
-
S1. Sanger Sequencing by Hand – Assessment Questions
-
S2. Sanger Sequencing by Hand – Answer Key for Assessment Questions
-
S3. Sanger Sequencing by Hand – Student Activity Pack
-
S4. Sanger Sequencing by Hand – dNTP and ddNTP Sheet
-
S5. Sanger Sequencing by Hand – Black and White Student Activity Pack Part 4
Acknowledgments
We would like to thank the students in Trinity International University’s fall 2021 genetics course who participated in this activity and the Smith family who helped to test the activity before it was used in a classroom setting.
References
- Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 74:5463–5467. doi:10.1073/pnas.74.12.5463.
- Brooker RJ. 2021. Genetics: Analysis and principles, 7th ed. McGraw-Hill Education, New York, NY.
- Metzker ML. 2005. Emerging technologies in DNA sequencing. Genome Res 15:1767–1776. doi:10.1101/gr.3770505.
- Park ST, Kim J. 2016. Trends in next-generation sequencing and a new era for whole genome sequencing. Int Neurourol J 20:S76–83. doi:10.5213/inj.1632742.371.
- Hagemann IS. 2015. Overview of technical aspects and chemistries of next-generation sequencing, p 3–19. In Kulkarni S, Pfeifer JD (ed), Clinical genomics. Academic Press, Boston, MA.
- Queloz AC, Klymkowsky MW, Stern E, Hafen E, Köhler K. 2017. Diagnostic of students' misconceptions using the Biological Concepts Instrument (BCI): A method for conducting an educational needs assessment. PLoS One 12:e0176906. doi:10.1371/journal.pone.0176906.
- Conley JE, Meisel AJ, Smith JJ. 2016. Using M&M's to model Sanger's dideoxy DNA sequencing method. Am Biol Teach 78:516–522. doi:10.1525/abt.2016.78.6.516.
- O'Leary-Driscoll S. 2016. DNA sequencing activity. Illinois Mathematics and Science Academy. Retrieved from https://digitalcommons.imsa.edu/bioinfo_sequencing/8 (accessed 22 January 2022).
- Young J. 2018. Sanger sequencing – A hands-on simulation. Genetics Society of America Peer-Reviewed Education Portal (GSA PREP); 2018. 003. doi:10.1534/gsaprep.2018.003.
- Gomes A, Korf B. 2018. Genetic testing techniques, p 47–64. In Robin N, Farmer MB (ed), Pediatric cancer genetics. Elsevier, St. Louis, MO. doi:10.1016/B978-0-323-48555-5.00005-3.
- Redfield RJ. 2012. “Why do we have to learn this stuff?”—a new genetics for 21st century students. PLoS Biology 10:e1001356. doi:10.1371/journal.pbio.1001356.
- Robertson JA. 2003. The $1000 genome: Ethical and legal issues in whole genome sequencing of individuals. Am J Bioeth 3:35–42. doi:10.1162/152651603322874762.
- Callaway E. 2011. Aboriginal genome analysis comes to grips with ethics. Nature 477:522–523. doi:10.1038/477522a.
- Yudell M, Roberts D, DeSalle R, Tishkoff S. 2016. Taking race out of human genetics: Engaging a century-long debate about the role of race in science. Science 351:564–565. doi:10.1126/science.aac4951.
- Foster MW. 2006. Ethical issues in medical-sequencing research: Implications of genotype-phenotype studies for individuals and populations. Hum Mol Genet 15:R45–R49. doi:10.1093/hmg/ddl049.
- Wiggins BL, Eddy SL, Wener-Fligner L, Freisem K, Grunspan DZ, Theobald EJ, Timbrook J, Crowe AJ. 2017. ASPECT: A survey to assess student perspective of engagement in an active-learning classroom. CBE Life Sci Educ 16:ar32. doi:10.1187/cbe.16-08-0244.
- Tanner KD. 2013. Structure matters: Twenty-one teaching strategies to promote student engagement and cultivate classroom equity. CBE Life Sci Educ 12:322–331. doi:10.1187/cbe.13-06-0115.
- Read CY, Ricciardi CE, Gruhl A, Williams L, Vandiver KM. 2016. Building genetic competence through partnerships and interactive models. J Nurs Educ 55:300–303. doi:10.3928/01484834-20160414-12.
Article Files
Login to access supporting documents
Smith-Ho Pao-Sanger Sequencing by Hand Using Paper Clips to Demonstrate Chain Termination.pdf(PDF | 305 KB)
S1. Sanger Sequencing by Hand - Assessment Questions.docx(DOCX | 14 KB)
S2. Sanger Sequencing by Hand - Answer Key for Assessment Questions.docx(DOCX | 14 KB)
S3. Sanger Sequencing by Hand - Student Activity Pack.docx(DOCX | 25 KB)
S4. Sanger Sequencing by Hand - dNTP and ddNTP Sheet.docx(DOCX | 104 KB)
S5. Sanger Sequencing by Hand - Black and White Student Activity Pack Part 4.docx(DOCX | 22 KB)
- License terms

Comments
Comments
There are no comments on this resource.