Building Trees: Introducing evolutionary concepts by exploring Crassulaceae phylogeny and biogeography
Published online:
Abstract
Teaching evolutionary theory is foundational for all biological sciences and a key aspect of overall science literacy. The conceptual framework for understanding evolution relies on thinking clearly about evolutionary trees (phylogenetics) and how geological history influences biological processes and diversity. Central to a student's comprehension of evolutionary research is an understanding of how scientists infer evolutionary relatedness and how they integrate geographic data. To address these concepts, we developed a series of lessons suitable for a typical introductory biology course in which students learn to infer phylogenies for the plant family, Crassulaceae. In the first part of the lesson, students develop phylogenetic hypotheses based on both morphology and DNA sequence data, use software (MEGA: Molecular Evolutionary Genetics Analysis, FigTree) to infer a phylogeny, and compare trees constructed from the different data sources and statistical models. In the second part of the lesson, students use their phylogenies and additional software (RASP) to reconstruct the biogeographic history of Crassulaceae. The lessons described here help students better understand how geological changes during Earth history can influence evolutionary processes and species diversification. Students should come away from the lesson with an improved understanding of phylogenetic tree construction and interpretation, molecular dating, the geological time scale, and the role of biogeographic factors in macroevolution. The lessons are designed to be used sequentially, and incorporate various evidence-based teaching and learning strategies. Lessons were designed to complement lecture-based instruction for an introductory biology course, but suggestions for expanding the activities or adapting them to new audiences are provided.
Citation
Karimi, N., Parks, B.M., Rouse, D., Martin, K., Dong, X., Rajangam, P.C., Baum, D.A., and Heitz, J.G. 2017. Building Trees: Introducing evolutionary concepts by exploring Crassulaceae phylogeny and biogeography. CourseSource. https://doi.org/10.24918/cs.2017.16Lesson Learning Goals
Students completing lessons described in this article will understand:- How inferring the evolutionary history and relationships among organisms is used in taxonomy.
- How initial evolutionary hypotheses can be developed using phylogenetic principles.
- How morphological characters and analysis of DNA sequences can be used to infer phylogenetic relationships and test prior phylogenetic hypotheses.
- How convergent evolution (e.g., homoplasy) can explain discrepancies between molecular- and morphology-based phylogenetic analyses.
- The geologic time-scale and how geological processes have influenced species diversification.
Lesson Learning Objectives
Students will be able to:- Estimate phylogenetic trees using diverse data types and phylogenetic models.
- Correctly make inferences about evolutionary history and relatedness from the tree diagrams obtained.
- Use selected computer programs for phylogenetic analysis.
- Use bootstrapping to assess the statistical support for a phylogeny.
- Use phylogenetic data to construct, compare, and evaluate the role of geologic processes in shaping the historical and current geographic distributions of a group of organisms.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Our understanding of the process of evolution and our ability to make sense of the biodiversity both depend on an ability to construct and interpret phylogenetic trees, an endeavor that requires some visual thinking (1). Given the many misconceptions about evolution that are common among students, it is not surprising that undergraduate students often struggle with interpreting phylogenetic trees properly (2,3,4). To address this tree-thinking deficiency, a number of resources and student exercises on phylogenetic inference and interpretation have been developed (e.g., 2,5,6,7,8,9). For all their strengths, these tools rarely incorporate notions of absolute time, instead emphasizing degree of evolutionary relationship and, hence, the relative time of different branching events. We propose that it is important to expose students to notions of absolute time to help place macro-evolutionary processes in context, something that is rarely done in introductory biology (10). Furthermore, although teaching evolution often draws on evidence from the fossil record and emphasizes the importance of geologic time (11), to our knowledge no published activity integrates thorough tree-thinking (phylogenetic) activities with historical biogeographical analyses (11). Therefore, we here developed lessons that address familiar tree-thinking misconceptions while also including considering absolute by using a biogeographic case study.
We teach a two-semester introductory biology course for majors that covers cell biology, molecular biology, genetics, and animal physiology in the first semester and evolution, plant biology, and ecology in the second semester. Our course's organizational structure includes lecture and correlated laboratory components. A consistent challenge for us, as reflected in student evaluations, is to maintain tangible connections between the lecture and lab, especially for the topics of evolution and ecology. This challenge is compounded by the difficulty of fostering interest in these topics among undergraduate students who wish to pursue careers in human health.
Initially, we taught core concepts of evolution using pictures of selected animal species and had students develop a phylogeny based on morphological characteristics. This activity was then coupled with a lab in which students used provided sequence data to infer the phylogeny for the same animals. We found, however, that this approach did not result in adequate gains in the understanding of macro-evolutionary processes, including concepts such as the Earth's geological time scale or biogeography. Furthermore, using animals to introduce phylogenetics did not align well with the rest of that semester, which focuses on plant physiology and ecology.
In an effort to improve integration across the three topics taught in the second semester of introductory biology (evolution, plant biology, and then ecology), we designed a semester-long sequence of laboratory units that all relate to a single plant, 'Mother of Thousands' (Kalanchoe daigremontiana, Crassulaceae), or as we came to call them MoTs. Using MoTs for all laboratory activities provides a strand of integration among the three course modules. We have found that the biology and ecology of Crassulaceae species can be used to teach key concepts and help students see how these distinct areas of biology are interrelated.
Here, we describe the evolution laboratory lessons, which precede the plant biology and ecology units. This set of three laboratory activities were designed to reinforce core evolutionary concepts related to phylogenetic inference, the interpretation of phylogenetic trees, and biogeography. These laboratory lessons were intended to supplement and reinforce lecture instruction on evolution. Nonetheless, we have provided sufficient teaching resources in the form of PowerPoint presentations, worksheets and quizzes to support other course contexts.
In the first lab activity, students are introduced to phylogenetic tree thinking and work in groups to create a phylogenetic tree based on observations of the morphology of ten different species of Crassulaceae. In the second lab activity, students work with DNA sequence data for twenty species of Crassulaceae to obtain a molecular phylogeny. The morphological and molecular trees are compared and various topics such as monophyly, homoplasy, and parsimony are explored. To help students further grasp tree thinking and the role of phylogenetic inference in research, the set of DNA sequence data results in several different, equally parsimonious trees. In the third lab activity, students explore the geological timescale, learn about some major changes in the positions of the Earth's continents over time, and relate their phylogenetic trees to the processes of species divergence, by examining the timing of land mass movements and species' current geographical distributions. Students must critically analyze the data and create hypotheses to explain the patterns they observe: since the divergence of Crassulaceae clades are estimated to have occurred well after the breakup of land masses, divergence more likely resulted from long-distance dispersal and not vicariance (geographic separation). There are numerous options for extending the activities described here within introductory biology courses or in upper-level courses. The activity as a whole, while heavily scripted, accurately models the kinds of research questions and methods used in the field of phylogenetic systematics.
INTENDED AUDIENCE
The intended audience includes freshman and sophomore students who are planning to major in the biological sciences. In the course, students had either completed, or placed out of, the first semester of college-level introductory biology, which covers molecular and cellular biology, genetics, and animal physiology.
REQUIRED LEARNING TIME
As presented, the lesson activities operate within three-hour lab sessions that meet weekly, with additional activities to be completed by the students outside of lab. Supplementary materials provided (PowerPoint mini-lectures, pre-lab worksheets, and quizzes) can be completed either independently or collaboratively, in class or outside, per the instructor's preference.
PRE-REQUISITE STUDENT KNOWLEDGE
Students should be instructed on or already know the basic concepts of evolution (variations arise and are inherited in subsequent generations), and tree thinking (phylogenetic trees depict relatedness), as commonly presented in any introductory biology textbook.
PRE-REQUISITE TEACHER KNOWLEDGE
This series of lessons assumes the instructor has some background in evolution, as well as constructing and interpreting phylogenetic trees. Reading the cited literature is strongly recommended. Instructors must also be comfortable learning how to use the various software programs used in the lessons presented here, including MEGA (12), RASP (13) and FigTree (http://tree.bio.ed.ac.uk/software/figtree/). Tutorials for each program are available online.
SCIENTIFIC TEACHING THEMES
ACTIVE LEARNING
Students participate in a variety of ways that promote active learning, including group activities, discussions, and inquiry-based learning. Students develop and analyze their own data to construct phylogenetic trees and compare their results with those of other students. Students are asked to develop hypotheses to explain differences among the morphological and molecular phylogenetic trees that they construct. Students also need to brainstorm and test different biogeographic hypotheses to best explain current species distributions and the inferred evolutionary relationships. To accomplish these tasks, students work in small groups and then evaluate their ideas and findings, in comparison with the work of other class participants.
ASSESSMENT
We assess student learning formatively and summatively through group and individual worksheets (S3, S12, S18), and individual quizzing (S13). Students evaluate their own progress by comparison of their work to their peers in small-group and full-class discussions.
INCLUSIVE TEACHING
Students work collaboratively in instructor-assigned groups to complete the majority of the work. Some out-of-class assignments are completed independently, but these activities may be modified for in-class and/or group completion. All computer-based activities use open-access software programs available for free download and are completed in-class on institutional desktop computers, thus avoiding disadvantaging students who may not otherwise have access. These lessons focus on incorporating visual, auditory, and kinesthetic learning experiences, as well as offering activities that are both social and solitary.
LESSON PLAN
Please see Tables 1 and 2 for Teaching Timelines of the lesson (also found as supporting file S1). The lessons described below were designed for student groups of two to four individuals. Our laboratory rooms have six stations, each of which can accommodate four students, and is equipped with a computer and monitor. We organize groups either randomly or according to instructor preference based upon prior student work behaviors noted earlier in the semester. In smaller classes, instructors may prefer students to work independently until the Think-Pair-Share and group discussion portions of the activities are reached. All lesson documents are provided as Supporting Materials.
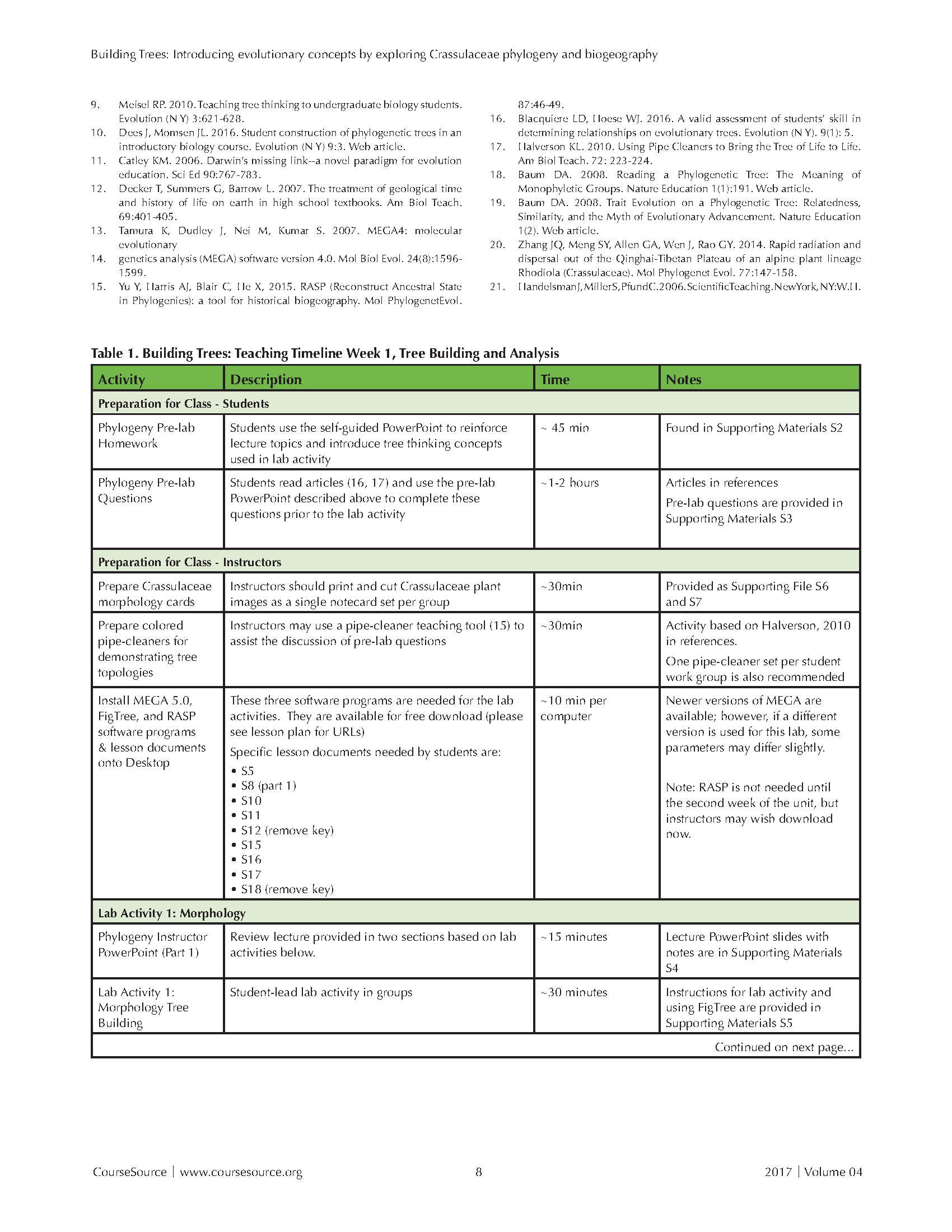
Table 1. Building Trees-Teaching Timeline Page 1
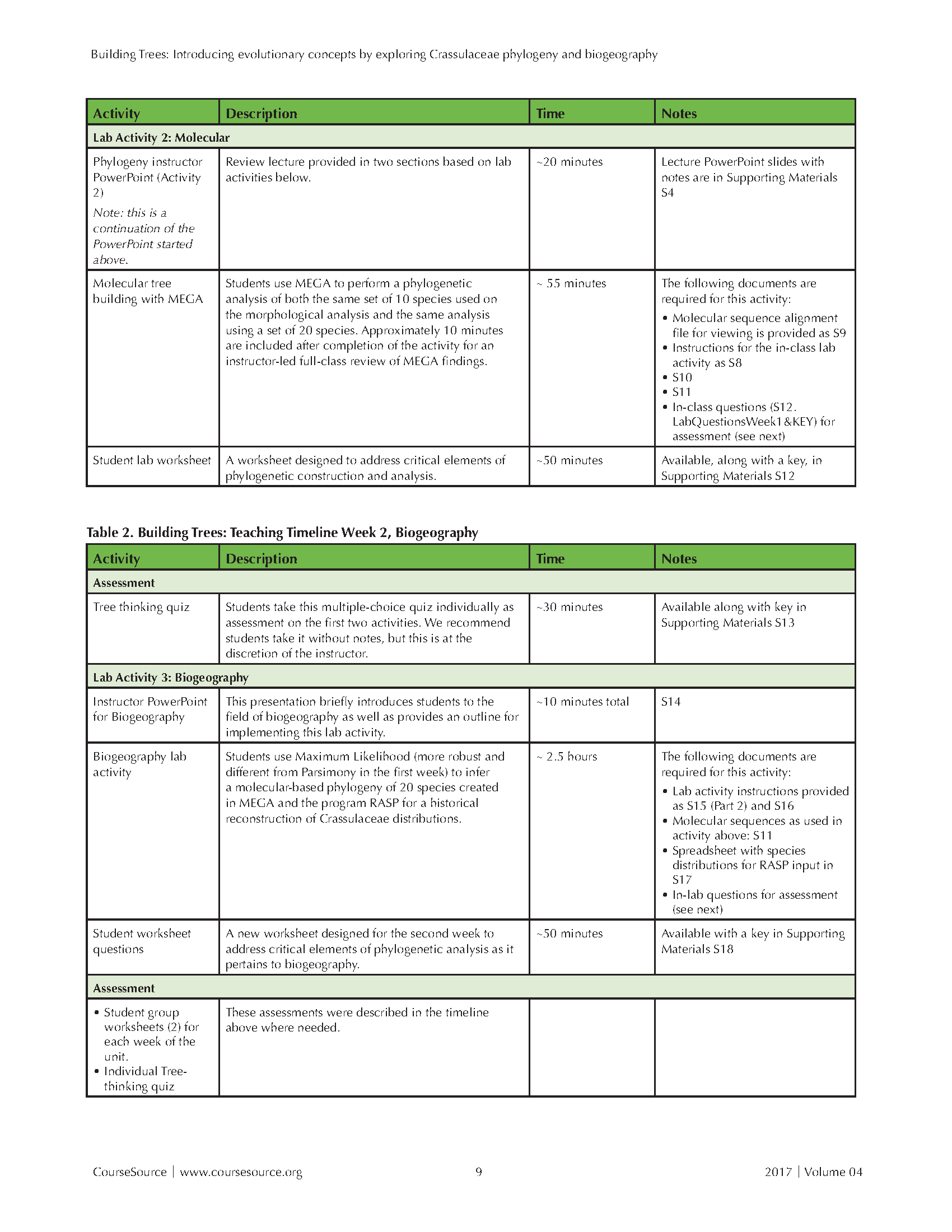
Table 1. Building Trees-Teaching Timeline Page 2
LAB 1: Activities 1 (Morphology-based trees) and 2 (Molecular-based trees)
BEFORE LAB 1
Prior to the first lab activity, assign as homework the self-guided presentation (S2) and pre-lab questions (S3). This assignment walks students through the basic concepts of tree thinking. We suggest beginning each in-class activity with a discussion of key concepts, assumptions, and conclusions (from the pre-lab resources above). We have found that visual representation of phylogenetic trees is most helpful to convey concepts, but it is also important to repeatedly quiz students to make sure they are correctly interpreting the trees (14). If they have trouble with tree thinking, which is likely, we recommend instructors use pipe cleaners as a teaching tool (15); students can create phylogenetic trees out of pipe cleaners similar to the ones shown in the student pre-lab assignment (S3). Using pipe cleaners as a physical teaching aid is especially useful to illustrate that rotation of branches around nodes does not alter the relationships or other kinds of evolutionary information.
ACTIVITY 1: MORPHOLOGY-BASED PHYLOGENETIC TREES
Prior to starting this activity, instructors present the phylogeny instructor PowerPoint lecture on phylogenetic tree construction and analysis (S4). This lecture is different from the presentation (S2) students use as part of their individual pre-lab preparation, which is constructed to be a self-explanatory/learning guide. In the lecture, we encourage instructors to note the distinction between taxonomy and phylogeny, to use pipe cleaners (15) to illustrate how rotation of branches around nodes does not alter the evolutionary information conveyed, and to review the key concepts from the pre-lab including the use of parenthetical format to depict nested sets of relationships [e.g. - ((A,(B,C)),(D,E,F));] and evolutionary histories. Note: There is a break in the PowerPoint presentation between morphological trees and molecular phylogenetic trees; we let students begin Lab Activity 1 at this point, before continuing with the molecular section of the presentation.
Instructors can introduce students to FigTree software, available both as Mac and Windows executables (http://tree.bio.ed.ac.uk/software/figtree/), by using the S5 provided. Following the introduction to FigTree, instructors should give students the Crassulaceae image notecards to try and build a morphologically-based phylogenetic tree. Students then infer a phylogeny using images of ten Crassulaceae species (S6 and S7), based on morphological characters of their choice. Students receive unlabeled images of plants on notecards because scientific names provide some information on the likely topology of the currently accepted phylogenetic tree. Students then share their trees with the class and explain what visible characteristics they used to inform their groupings of organisms. After all groups have completed discussing their trees with the class, they receive the species names for each photo and then modify their trees to include the scientific names.
Using NotePad (Microsoft Windows application) or any text editor, students write their hypothesized relationships in parenthetical "set" format in a .txt text file. The text file can then be input into FigTree (http://tree.bio.ed.ac.uk/software/figtree/), which allows students to display a tree graphically. Instructions on how to use FigTree are found in Supporting Materials as S5. Student groups should export and save JPEG images of their hypothesized morphological tree, including annotations that describe the differences/character changes for the tips of the tree, for submission at the end of the lesson. Different student groups typically create very different trees based on differing decisions as to which morphological characters to emphasize. Comparing different phylogenetic trees provides an important segue into the next activity, which uses molecular markers to infer phylogenetic trees for the same set of species.
Morphology-based Phylogenetic Tree: Discussion
Having students compare their trees to those generated by other groups, and discuss similarities and differences and why they might exist, provides the instructor with an opportunity to lead a class discussion on factors (convergent evolution, errors in scoring, etc.) that might lead to the inference of incorrect relationships using a small number of morphological characters. It could be highlighted that morphology-based trees less likely to yield the correct phylogenetic tree, when there is abundant homoplasy, which is to say a situation where similar character states in two taxa trace to different evolutionary origins. Furthermore, incorrect phylogenies can occur if the traits selected evolved very rapidly relative to the history of branching such that they retain little if any information regarding evolutionary history. Also, with few characters, it is possible for characters with misleading patterns to obscure the subset of characters that contain good phylogenetic information. This is likely to be a factor in the current exercise, which is based on images, meaning that potentially important morphological characteristics may be absent (e.g., flowers & reproductive structures).
Activity 2: DNA Sequence-based (Molecular) Phylogeny
Instructors should return to and conclude the PowerPoint lecture (S4). The presentation will now explain bootstrapping and the concept of pseudo-replicates, which are needed to complete the activity. When introducing bootstrapping, emphasize that it is a statistical procedure that provides relative support/confidence of different branches of a tree and does so without needing to obtain multiple replicate data sets. This is possible under the assumption that the sampled are randomly sampled from the same historical process. Under such an assumption (which is not unreasonable for a large sequence alignment), by resampling the observed characters we can estimate how well supported different phylogenetic topologies would be in the case that we had obtained a different data set.
We suggest instructors demonstrate MEGA v5.0 (http://www.megasoftware.net) to the class by using one of the pre-packaged datasets provided with that software package. After the demonstration, students can follow the instructions (S8) to estimate a molecular phylogeny based on parsimony analysis of molecular sequences. We ask them to use MEGA to infer the evolutionary relationships among Crassulaceae species. The specific DNA sequences are from the nuclear ribosomal ITS (Internal Transcribed Spacer) region for each of the species. There are two datasets to use: One is limited to the ten species used for morphological comparison (for direct comparison of these two data sources, i.e. plant images versus DNA sequences) and the other is a full set of twenty species. A sequence alignment file is also provided for students to view (S9).
Students use MEGA to find the most parsimonious trees for both the ten- and twenty-taxon datasets (S10 and S11), and then evaluate robustness by conducting a bootstrap analysis for the large dataset. Note: the MEGA alignment files (S10 and S11) cannot be opened outside of this program. While students are working, instructors should circulate to provide assistance and show other tree-presenting features within MEGA (e.g. - unchecking "Topology only" shows the tree with branch lengths calculated proportional to the number of changes mapped on by parsimony). Students save their trees as .tiff or .pdf, so that they can compare them to the tree they developed based on morphological characteristics.
During the activity described above, students answer in-class questions within their groups (S12). NOTE: Because they used eyeball analysis of morphology, but parsimony analysis of DNA, there are most likely differences between the trees. We recommended in "Options to Extend the Lesson" that students also try to use parsimony on the morphological trees.
The ITS dataset results in multiple equally parsimonious trees, which should lead to a discussion about uncertainty in the estimation of evolutionary histories. Although researchers would clearly like to have unambiguous results, finding a single tree with bootstrap support for all branches rarely occurs. We have found that the uncertainty that student's experience in this exercise offers teachable moments on the nature of science. However, it should be noted that some clades are shared by all equally parsimonious trees and are associated with high bootstrap support. This shows that, although much uncertainty remains, molecular phylogenetics can yield rigorous inferences about the evolutionary history.
Molecular Phylogeny: Discussion
Instructors now organize a full-group discussion to share students' findings, starting with a group of students displaying their own generated bootstrap trees and explaining what they found. We often start with a discussion of just the tree topology, asking what are the clades and what do they mean? Instructors can either show a tree on the screen, or start with a group of students displaying their results. A major question to address is, what clades do students have confidence in, and why? It might be helpful to ask them what they would expect to find if they had sequences from a different gene. Would they expect the same tree? Which clades on the ITS tree are more or less likely to be shared with a new gene tree? It is best to not provide the students with answers directly; asking questions incorporates active learning more effectively. We encourage instructors to highlight the following key concepts during this discussion:
- Phylogenetic trees students constructed in MEGA 5.0 were based on parsimony, as described in the PowerPoint lecture.
- Construction of Maximum Parsimony trees is based only on the scored data and the idea that the tree that can explain the observed variation with the fewest evolutionary changes (i.e., the "simplest" explanation) is most likely to be true. A phylogeny is a good hypothesis of relationships until we have other information to make us question our assumptions. Even if we assume that parsimony is appropriate, there is some uncertainty in relationships: construction of phylogenetic trees is like any other scientific estimate: the best inference with the data at hand but not an absolute.
- Conclusions derived from the use of parsimony are best estimates; hence the use of statistical methods (bootstrapping) to provide confidence in our tree constructions.
- Using multiple datasets can improve our conclusions, especially if the same clades are consistently recovered.
- Bootstrapping yields a guide to statistical confidence in the clades on a given tree. For example, with bootstrap values under 50, we are better off collapsing those branches of the tree since there are other conflicting clades that are plausibly true given these data.
- A bootstrap number of 52, for example, means that for 52 out of 100 times (or 52%), pseudoreplicate data sets analyzed by parsimony yielded that particular clade.
As an assessment, the phylogeny lab quiz (S13) covers concepts introduced in the two Nature Education essays (16,17), the pre-lab and lab activities, and the instructor's PowerPoint lecture. We suggest that the quiz be conducted without use of notes and be given at the end of week one or at the beginning of week 2 lab, just before the biogeography activity described below.
LAB 2: Activity 3 (Biogeography)
BEFORE ACTIVITY 3
Instructors should begin the lab with a review in a student-driven, large-group discussion, focusing on what was learned in the previous lab. Main topics to cover include:
- Differences between a phylogeny and taxonomy.
- How the two kinds of phylogenetic trees were developed in the last lab (morphological and molecular; intuitive and computational) and why these might have yielded different phylogenies.
- What parsimony is, and how it is utilized to generate most parsimonious trees.
- What a bootstrap analysis is and how such a statistical approach might be used to obtain estimates of how confident we should be in each of a tree's branches.
A missing and critical question that the phylogenetic trees in isolation do not address is how these plants spread over the Earth through geological time. The goal for the second activity is to use the estimated phylogeny and current distribution to infer: (1) How the current distribution of the species arose via dispersal across the planet? (2) Over what timescale did the migrations of ancestral populations happen? In other words, we introduce the field of biogeography.
As a starting point, it is important to reinforce that one of the most important drivers of speciation is physical separation of a subset of a population of interbreeding individuals (allopatric speciation) from other members of the ancestral population. This might happen when a population undergoes a rare dispersal event to a distant landmass or when a single landmass breaks up by continental drift. Instructors can use the second PowerPoint lecture to introduce this lab and the field of biogeography (S14).
Lab Activity 3a - Maximum Likelihood in MEGA & RASP
In this activity, students use MEGA to build a phylogeny and RASP (Reconstruct Ancestral States in Phylogenies, http://mnh.scu.edu.cn/soft/blog/RASP), to produce a historical reconstruction of species' geographical distributions based on that phylogeny. This activity requires that students conduct a more sophisticated phylogenetic analysis in MEGA than in Lab Activity 1; specifically they build a maximum likelihood bootstrap tree in MEGA, which requires more time to complete than the parsimony analysis. To build the maximum likelihood tree, students use different MEGA instructions (S15) than previously used. MEGA instructions can be distributed to students per instructor's preference (e.g. projected at the front of the room, printed one per group, or have each student bring their own copy). Lab Activity 3a compares the parsimony-based tree inferred in week one with a maximum likelihood tree. Maximum Likelihood generally yields a more accurate tree than parsimony and is needed to estimate the number of mutations per site occurring on each branch. Insofar as we are willing to assume that the rate at which mutations in ITS sequences arise and are fixed in evolving Crassulaceae lineages is more or less constant over time, then these branch lengths should be proportional to time (in millions of years). Using this principle, students can consider that a greater number of differences between a pair of sequences implies a greater time since they diverged from each other and, thus, permits students to equate longer branches with longer time.
We do not expect the students to be able to develop a sophisticated understanding of the differences between parsimony analysis (covered and explained in the first week) and maximum likelihood. However, if these approaches are included in lecture, the instructor here is at liberty to expound on maximum likelihood and why it might be considered to be a more accurate method. We use both methods in this activity as a way for students to get used to working with MEGA, but also because teaching phylogenetic inference by parsimony helps scaffold student learning. As demonstrated in the student pre-lab activity (S2), teaching parsimony helps students grasp the principles that all molecular sequence data to use for phylogenetic inference and also helps students visualize character state changes on the branches of a phylogenetic tree.
After students generate their maximum likelihood tree in MEGA, they use this phylogeny and the present day locations of species in RASP (S16) to generate an ancestral-state reconstruction as needed for historical biogeography. Students input their optimal maximum likelihood tree into RASP to generate an inference of ancestral states based on the present day location of species. The resulting phylogeny displays the probability of alternative historical geographical distributions for common ancestors. Students use the Bayesian binary Markov chain Monte Carlo technique, which makes the assumption that all living species are the same total distance from the root node, while allowing for chance variation in the number of substitutions on different branches.
Students are provided with RASP Instructions (found in Supporting Files as S16) and information on the current geographic locations of the species (S17). They then try an integrate knowledge of geological history to generate historical species distributions based on the topology of the phylogeny. NOTE: To load properly into RASP, the Crassulaceae Origins spreadsheet MUST be saved with ".csv" extension in the actual filename. For example, save the file specifically as Crassulaceae_Origins.csv as provided.
During the RASP activity, students answer questions on the worksheet (S18) for submission at the end of class. Once all groups have constructed and saved their RASP tree, students should walk around the room and note similarities and differences between trees. In contrast to the Maximum Parsimony tree, there typically are few to no differences between the RASP trees. NOTE: You may wish to provide a map of Africa to each group or display a map of Africa at the front of the room for students with limited geography knowledge.
Lab Activity 3a - Discussion
We recommend this discussion occur at end of Lab Activity 3a after students build their trees in MEGA and RASP. Back in a large-group discussion, ask students to interpret the meaning of their new trees.
- What do the pie charts mean? When the pie has multiple piece colors, they represent the relative probability of the geographic origins of the common ancestor. Uncertainty in one node needs to be factored into the next deepest node (i.e. - the previous ancestor).
- Are the graphs generated by each group similar? Why or Why not? Remember, they are all working with the same .csv origins file and they used this same geographic data in conjunction with a phylogeny generated from the same data set (by the same program). Therefore, the trees should be very similar or identical between groups.
- What is missing from this tree? Age or time. We can see proportional time, but there are no actual years associated with Earth's timeline.
Instructors can now ask the class how they might incorporate geological time into this tree specifically in the case that there are no known fossils for the group under study. Answering this question may be difficult for students, but they might be led to the insight that speciation often results from the physical (geographic) separation of a single population into regions that are no longer in contact (e.g., - a river that separates an interbreeding population or perhaps even a new sea or ocean). If we knew of dated geological events that corresponded to at least one node on the tree, we would be able to extrapolate to obtain actual ages for all other nodes.
To initiate this thought process, the instructor should first try to help students connect small separations such as caused by new rivers to much larger separations such as continent splitting events. The instructor should have the students discuss and explore how a new sea or ocean might be generated on a geological timescale (plate tectonics). Therefore, one approach we can take is to map the geological history of Earth, as reconstructed in great detail by geologists, to pinpoint periods when now-isolated landforms (such as Madagascar and continental Africa) were once connected. Next, we can contrast the geologic timeline of land movements with the predicted phylogeny and species distributions for various Crassulaceae taxa to identify nodes that represent a continental splitting event.
Lab Activity 3b - Paleobiology Navigator
Students now use the interactive web-based map and database "Paleobiology Navigator" (https://paleobiodb.org/navigator/), to show how the locations of Earth's continents changed over time. Using the Paleobiology Navigator, students can answer questions focusing on Crassulaceae historical biogeography on their worksheets (S18). The worksheet leads students through the various Earth eras to see how the continents change over time and explore whether continental movements can explain any branches of the Crassulaceae tree.
Lab Activity 3b - Paleobiology Discussion
In comparing their RASP outputs with the continental shifts, students should begin to notice that Madagascar and mainland Africa were once part of the same landmass, but then separated in the early Cretaceous, ~130 million years ago. Student trees show that Kalanchoe diverged from a common ancestor shared with the rest of Crassulaceae, and that there is a distinct geographical divergence between Madagascar and mainland African taxa. Comparing the geologic timeline of land movements with the information obtained from RASP, students can begin to propose hypothetical dates to the nodes on their RASP outputs by assuming that a divergence event corresponds to the separation of major landmasses. NOTE: Associating clade divergence directly with the separation of landmasses is an assumption that happens to be false in this case! Students will discover this discrepancy when they learn that the time of the divergence from the common ancestor of Crassulaceae has now been dated (using fossils) to ~100-60 million years ago (e.g., 18) Students must then develop hypotheses to address the discrepancies between the phylogeny, the estimated dates of species divergence, and the timing of Earth's geological history. These discrepancies push students to independently consider various patterns and processes in biogeography. Specific important points they should be encouraged to discuss and think about:
- The worksheet questions expose an inconsistency in the data analyses. How can students resolve this inconsistency?
- Comparing data from the Paleobiology Navigator with the age of Crassulaceae reveal that the continental split of Madagascar from mainland Africa occurred ~130 million years ago, while the Crassulaceae are only ~100-60 million years old. How could the genus of Kalanchoe diverged from other Crassulaceae at ~130 million years ago if the entire family is at most 100 million years old?
- Certain Kalanchoe trace to mainland Africa (as deduced from the RASP output) after the dated split of Madagascar. What processes could have produced this pattern? One explanation is that an over-water migration event occurred after the landmasses had separated. Students should come to this conclusion on their own.
- What critical component is missing from all of these analyses? Missing components that we hope students will recognize are fossils (direct physical evidence) of Crassulaceae that can be attached to a particular branch of the tree, and are accurately dated and placed geographically. How would fossil evidence either support or conflict with the results?
TEACHING DISCUSSION
The hands-on nature of this lesson requires physical/visual manipulation of phylogenetic trees in a variety of formats, including: (1) Pipe cleaner models that allow students to understand how rotation at nodes can occur without changing phylogenetic relations (optional). (2) Inferring shared ancestry and relatedness using morphological characters students select. (3) Interpretation of molecular phylogenetic trees developed using MEGA. (4) RASP analysis to try and develop a biogeographic narrative to explain the evolutionary relationships. These activities facilitate understanding of key evolutionary concepts (such as common ancestry, descent with modification, deep time, speciation and dispersal/migration), which are often very difficult for students to grasp through conventional lecture-based approaches. Students successfully completing these lessons will be well-prepared to participate in undergraduate research opportunities in phylogenetic biology.
Previously, these lessons were taught using mammals as the study organisms. While using animals had some benefits, as they are generally more familiar to students, we have found that using a family of plants provides integration with the other topics covered in the rest of the semester (ecology, plant physiology). Furthermore, by removing student's preconceived notions about animal relatedness, the lessons force students to analyze phylogenetic trees more objectively. We found that there was no significant difference in student's understanding of the concepts or in exam grades when we switched from mammals to Crassulaceae as our organisms of study. However, student comments on written evaluations at the end of the semester often emphasized that these phylogenetic labs were reinforcing concepts learned in lecture. Students also stated how they enjoyed learning new tools that are relevant to scientists in the field, particularly the programs for phylogenetic tree inference and biogeography.
OPTIONS TO EXTEND THE LESSON
Although there are numerous opportunities for instructors to extend the lessons presented in this article, expanding the datasets and types of phylogenetic analyses would be the easiest to implement. The activities described in this article were developed using DNA sequences for only twenty species of Crassulaceae, but we have sequences and images for almost fifty species. Such a large dataset is not appropriate for a large enrollment introductory biology course, though it might be practical for smaller courses, courses with more time flexibility, or higher-level courses. We would be happy to supply these extra sequences upon request. Alternatively, instructors can utilize morphological and sequence data from organisms of their choice.
Instructors could expand upon the assessment methods used, by developing new quizzes aimed at assessing student comprehension of concepts or summative evaluations relating to laboratory materials could be included in final exams. Additionally, in courses that require independent research projects, student groups could be taught to download sequence alignments (for example from treebase.org or phylota.net) and conduct analogous analyses using MEGA and RASP. Likewise, the activities could be expanded to utilize some of the many other statistical methods for biogeographic inference (either within RASP or another program), with students being asked to compare the outputs from different methods. Directly incorporating fossils in the biogeography lesson would also be a nice extension. Instructors may also wish to spend more time on activities addressing underlining concepts that students often struggle with, including the statistical approaches for different phylogenetic reconstruction methods and measures of accuracy (i.e. parsimony, maximum likelihood, bootstrapping).
SUPPORTING MATERIALS
- S1. Building Trees: Teaching timeline table for all lab activities
- S2. Building Trees: A student self-guided pre-lab PowerPoint activity to introduce tree thinking
- S3. Building Trees: Assessment worksheet on the self-guided pre-lab
- S4. Building Trees: Instructor presentation for reviewing concepts used in lab activity
- S5. Building Trees: Instructions for visualizing phylogenetic trees in parenthetical format in FigTree
- S6. Building Trees: Image file used to make the image notecards for the development of morphological trees during the lab exercises in the first activity
- S7. Building Trees: Species names added to the Crassulaceae images
- S8. Building Trees: Instructions for the lab activity, inputting sequences into MEGA and performing phylogenetic analyses
- S9. Building Trees: DNA sequence alignment of the ITS region for twenty Crassulaceae species
- S10. Building Trees: DNA Sequence file of ten Crassulaceae species to be uploaded into MEGA
- S11. Building Trees: DNA Sequence file of twenty Crassulaceae species to be uploaded into MEGA
- S12. Building Trees: The student worksheet, with key, to be used for the first activity
- S13. Building Trees: A quiz with key to be used as assessment after both phylogenetic activities
- S14. Building Trees: A brief PowerPoint lecture for instructors to introduce the field of biogeography and outline the subsections of the biogeography activity
- S15. Building Trees: Instructions for performing a bootstrapping phylogenetic analysis in MEGA used in the third activity as input into RASP
- S16. Building Trees: Student instructions for using RASP, a free biogeography analysis software used in the third activity
- S17. Building Trees: A spreadsheet with the current geographic distributions of twenty Crassulaceae species to be used as input into RASP
- S18. Building Trees: In-lab questions for student groups for the third activity on biogeography
ACKNOWLEDGMENTS
Authors would like to thank organizers and participants of the Northstar Summer Institute on Scientific Teaching, the University of Wisconsin - Madison Introductory Biology 152 Course Coordinators and Teaching Assistants.
References
- Baum DA, Smith SD. 2012. Tree Thinking: An Introduction to Phylogenetic Biology. Greenwood (CO): Roberts & Company.
- Baum DA, Smith SD, Donovan SS. 2005. The tree-thinking challenge. Science, 310(5750):979-980.
- Gregory TR. 2008. Understanding evolutionary trees. Evolution (N Y)
- 1(2):121.
- Meir E, Perry J, Herron JC, Kingsolver J. 2007. College students' misconceptions about evolutionary trees. Am Biol Teach. 69:71-76.
- Julius ML, Schoenfuss HL. 2006. Phylogenetic reconstruction as a broadly applicable teaching tool in the biology classroom. J Coll Sci Teach. 35:40-45.
- Singer F, Hagen JB, Sheehy RR. 2001. The comparative method, hypothesis testing and phylogenetic analysis - An introductory laboratory. Am Biol Teach. 63:518-523.
- Lents NH, Cifaentes OE, Carpi, A. 2010. Teaching the process of molecular phylogeny and systematics... CBE Life Sci Educ. 9:513-523.
- Meisel RP. 2010. Teaching tree thinking to undergraduate biology students. Evolution (N Y) 3:621-628.
- Dees J, Momsen JL. 2016. Student construction of phylogenetic trees in an introductory biology course. Evolution (N Y) 9:3. Web article.
- Catley KM. 2006. Darwin's missing link--a novel paradigm for evolution education. Sci Ed 90:767-783.
- Decker T, Summers G, Barrow L. 2007. The treatment of geological time and history of life on earth in high school textbooks. Am Biol Teach. 69:401-405.
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary
- genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 24(8):1596-1599.
- Yu Y, Harris AJ, Blair C, He X, 2015. RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Mol PhylogenetEvol. 87:46-49.
- Blacquiere LD, Hoese WJ. 2016. A valid assessment of students' skill in determining relationships on evolutionary trees. Evolution (N Y). 9(1): 5.
- Halverson KL. 2010. Using Pipe Cleaners to Bring the Tree of Life to Life. Am Biol Teach. 72: 223-224.
- Baum DA. 2008. Reading a Phylogenetic Tree: The Meaning of Monophyletic Groups. Nature Education 1(1):191. Web article.
- Baum DA. 2008. Trait Evolution on a Phylogenetic Tree: Relatedness, Similarity, and the Myth of Evolutionary Advancement. Nature Education 1(2). Web article.
- Zhang JQ, Meng SY, Allen GA, Wen J, Rao GY. 2014. Rapid radiation and dispersal out of the Qinghai-Tibetan Plateau of an alpine plant lineage Rhodiola (Crassulaceae). Mol Phylogenet Evol. 77:147-158.
- Handelsman J, Miller S, Pfund C. 2006. Scientific Teaching. New York, NY:W.H. Freeman.
Article Files
Login to access supporting documents
Building Trees: Introducing evolutionary concepts by exploring Crassulaceae phylogeny and biogeography(PDF | 180 KB)
S1. Building Trees-Teaching Timeline.pdf(PDF | 72 KB)
S2. Building Trees - Student Pre-lab Trees.pptx(PPTX | 2 MB)
S3. Building Trees Pre-lab QuestionsXKEY.docx(DOCX | 118 KB)
S4.Building Trees - Instructor Review.pptx(PPTX | 465 KB)
S5. Building Trees - FigTree Instructions.docx(DOCX | 29 KB)
S6. Building Trees - Crassulaceae Images.docx(DOCX | 3 MB)
S7. Building Trees - Crassulaceae Images KEY.docx(DOCX | 3 MB)
S8. Building Trees - MEGA_Instructions_Part1.docx(DOCX | 197 KB)
S9. Building Trees - SequencesAlignment.pdf(PDF | 3 MB)
S10 and S11.zip(ZIP | 6 KB)
S12. Building Trees -LabQuestionsPart1XKEY.docx(DOCX | 62 KB)
S13. Building Trees -TreeThinkingQuizXKey.docx(DOCX | 339 KB)
S14. Building Trees - Biogeography_Instructions.pptx(PPTX | 862 KB)
S15. Building Trees - MEGA_Instructions_Part2.docx(DOCX | 131 KB)
S16. Building Trees - RASP Instructions.docx(DOCX | 29 KB)
S17. Building Trees - Origins_RASP.xlsx(XLSX | 36 KB)
S18. Building Trees - Biogeography QuestionsXKey.docx(DOCX | 1 MB)
- License terms

Comments
Comments
There are no comments on this resource.