Tackling "Big Data" with Biology Undergrads: A Simple RNA-seq Data Analysis Tutorial Using Galaxy
Published online:
Abstract
Analyzing high-throughput DNA sequence data is a fundamental skill in modern biology. However, real and perceived barriers such as massive file sizes, substantial computational requirements, and lack of instructor background knowledge can discourage faculty from incorporating high-throughput sequence data into their courses. We developed a straightforward and detailed tutorial that guides students through the analysis of RNA sequencing (RNA-seq) data using Galaxy, a public web-based bioinformatics platform. The tutorial stretches over three laboratory periods (~8 hours) and is appropriate for undergraduate molecular biology and genetics courses. Sequence files are imported into a student's Galaxy user account directly from the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA), eliminating the need for on-site file storage. Using Galaxy's graphical user interface and a defined set of analysis tools, students perform sequence quality assessment and trimming, map individual sequence reads to a genome, generate a counts table, and carry out differential gene expression analysis. All of these steps are performed "in the cloud," using offsite computational infrastructure. The provided tutorial utilizes RNA-seq data from a published study focused on nematode infection of Arabidopsis thaliana. Based on their analysis of the data, students are challenged to develop new hypotheses about how plants respond to nematode parasitism. However, the workflow is flexible and can accommodate alternative data sets from NCBI SRA or the instructor. Overall, this resource provides a simple introduction to the analysis of "big data" in the undergraduate classroom, with limited prior background and infrastructure required for successful implementation.
Citation
Escobar, M.A., Morgan, W., Makarevitch, I., and Robertson, S.D. 2019. Tackling “Big Data” with Biology Undergrads: A Simple RNA-seq Data Analysis Tutorial Using Galaxy.CourseSource. https://doi.org/10.24918/cs.2019.13Society Learning Goals
Bioinformatics
- RNA - Information Transfer [TRANSCRIPTOMICS]
- Where are data about the transcriptome found (e.g., expression, epigenomics and structure) and how are they stored and accessed?
- How can bioinformatics tools be employed to examine transfer of genetic information?
Lesson Learning Goals
- Students will know how high-throughput DNA sequence data are stored and retrieved.
- Students will understand how RNA sequencing (RNA-seq) data are analyzed using a computational workflow, including the function(s) of each step in the workflow.
- Students will understand how analyzing RNA-seq data can generate biologically useful insights.
Lesson Learning Objectives
- Students will locate and download high-throughput sequence data and genome annotation files from publically available data repositories.
- Students will use Galaxy to create an automated computational workflow that performs sequence quality assessment, trimming, and mapping of RNA-seq data.
- Students will analyze and interpret the outputs of RNA-seq analysis programs.
- Students will identify a group of genes that is differentially expressed between treatment and control samples, and interpret the biological significance of this list of differentially expressed genes.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
High-throughput DNA sequencing (also called next-generation sequencing) was first described just over a decade ago, with the development of 454 pyrosequencing technology in 2005 (1). Since that first report, dozens of high-throughput DNA sequencing chemistries have been developed, with Illumina sequencing (2) emerging as the current dominant platform (3). As sequence quality has increased and costs have dramatically decreased, high-throughput DNA sequencing has revolutionized almost every discipline within the biological sciences, from plant breeding to microbial ecology to cancer research (4-6).
RNA sequencing (RNA-seq) is a method that uses high-throughput DNA sequencing to analyze global patterns of gene expression. RNA isolated from biological samples is converted to complementary DNA (cDNA) and subsequently sequenced. Tens of millions of individual sequence reads are typically generated from each sample, producing a snapshot of the full transcriptome (7). RNA-seq data has proven to be extremely valuable in genome annotation and the characterization of alternative splicing events. In addition, the comparison of multiple different RNA samples can identify global changes in gene expression, leading to novel biological hypotheses (7-9). However, analyses of RNA-seq data can be challenging, given that each sequence file is typically several gigabytes in size, and substantial computational infrastructure is necessary to process such large files. In addition, scientists trained prior to the high throughput sequencing "era" may lack the background and experience necessary to comfortably incorporate RNA-seq approaches into their teaching and research (10).
Bioinformatics in general, and analyses of large DNA sequence datasets in particular, are increasingly recognized as essential components of modern undergraduate biology curricula (11-12). Further, the ability to manipulate and analyze "big data" is rapidly becoming a prerequisite for students interested in pursuing graduate studies and careers in biology (13). This Lesson can help instructors to introduce "big (sequence) data" analysis to an undergraduate classroom. The tutorial and instructor resources provide both a conceptual overview of RNA-seq technology and step-by-step student instructions for the analysis of RNA-seq data. The specific RNA-seq data set utilized in the Lesson is derived from published research that examined how gene expression in Arabidopsis thaliana roots is altered by infection with the parasitic cyst nematode Heterodera schachtii (14). All data analysis steps are performed using web-based resources, most notably Galaxy, a well-established, user-friendly platform for bioinformatics analyses (15). The RNA-seq data is uploaded directly into a student's (free) Galaxy account from the public National Center for Biotechnology Information Sequence Read Archive (NCBI SRA). This eliminates the need for on-site file storage and enables the entire analysis of differential gene expression to be performed "in the cloud" via Galaxy's graphical user interface. These features make the activity adaptable to a variety of classroom environments and levels of instruction. As such, this resource provides a relatively simple complement to previously described RNA-seq instructional exercises (10,16,17) that require greater computational infrastructure (e.g. access to a Linux cluster) and/or a greater level of computational background (e.g. familiarity with R statistical software and a command-line interface).
Intended Audience
This tutorial has been implemented in the classroom twice, at two different universities. It was first developed for a "Modern Molecular Biology and Genomics" course at California State University San Marcos. There were 19 students in the course, all of whom were senior-level Biological Sciences or Biotechnology majors. The tutorial was also used in a "Genes and Genomes" course at the College of Wooster. In this case, the tutorial was modified for use in the analysis of the instructor's own RNA-seq data from yeast. The offerings at Wooster had nearly fifty students total, sophomores to seniors majoring in Biology, Cellular Neuroscience, or Biochemistry and Molecular Biology. Overall, this tutorial is best suited for upper division biology, biotechnology, and/or biochemistry students who have taken at least one basic cell and molecular biology course previously. It could easily be integrated into laboratory sections of upper division genetics and/or molecular biology courses.
Required Learning Time
The Lesson is designed for three laboratory periods (~ 8 hours in total). Each session begins with an instructor overview of key concepts and data analysis steps to be performed, using the included PowerPoints (Supporting File S4: RNA-seq Annotated Instructor PowerPoints). This overview takes about 30-45 minutes. For the remainder of the laboratory period, students use the detailed tutorials to carry out the computational analyses of the RNA-seq data (Supporting Files S1-S3: RNA-seq Student Tutorials I-III). Each student carries out the computational work independently. The instructor circulates to assist students who encounter problems, and students are encouraged to help each other. A series of questions are embedded in the tutorial to evaluate student understanding of key concepts and data outputs. Students answer these questions in writing either during or after the laboratory session, and they are submitted the following week for grading. The Lesson can be carried out in any computer laboratory with internet access, since all analyses are web-based and computational steps are performed in the cloud. Students may also use their own computers, since no special software is installed and there are no specific computer hardware requirements.
Pre-Requisite Student Knowledge
Computationally, very little student background is required. All analysis programs have graphical user interfaces and are essentially "point and click." During one step in the tutorial, a list of genes is exported into Microsoft Excel and sorted. While some basic background in Excel data manipulation is required, students without this background are often assisted by Excel-savvy classmates.
To fully understand the RNA-seq technique and each computational analysis step, several concepts should be introduced prior to the Lesson. Background on high-throughput DNA sequencing in general, and Illumina sequencing in particular, is important prerequisite knowledge. Likewise, the basic molecular biology techniques of RNA isolation and cDNA synthesis should also be introduced, since RNA-seq is essentially high-throughput sequencing of cDNA. Students clearly must be familiar with the basics of molecular biology, including DNA structure and the process of gene expression. We typically have students read two relevant publications: A brief overview of the Galaxy platform (15), and the primary article describing the A. thaliana nematode infection experiments (14). These articles can be reviewed with students via a round table discussion prior to starting the tutorial.
Pre-Requisite Teacher Knowledge
Again, no specific computational background is required other than basic data analysis in Excel. However, the instructor should work carefully through the tutorial prior to teaching with it (Supporting Files S1-S3: RNA-seq Student Tutorials I-III). The appearance and organization of the Galaxy platform is regularly updated, so instructors may need to modify instructions or screenshots to keep the exercise "up to date" and minimize student confusion.
The instructor should have a firm grasp of high-throughput sequencing and RNA-seq data analysis concepts. Instructor PowerPoints are annotated, however additional background reading is suggested for faculty who have little background with high throughput sequencing (7,15,18-20).
SCIENTIFIC TEACHING THEMES
Active Learning
As a laboratory exercise, this Lesson is inherently "hands on" active learning. Each student individually downloads data files from public repositories, computationally analyzes the data to identify differentially expressed genes, and considers the biological relevance of their findings. Students are also encouraged to help one another and discuss/compare their findings.
Assessment
A series of detailed questions is embedded within the tutorials. Some questions test basic student understanding (e.g. What is the purpose of this computational step?), some require students to report specific data outputs (e.g. screenshots), some require students to analyze and summarize complex data, and some ask students to place their findings in a broader biological context. The questions cover every level of Bloom's taxonomy (21). We expect students to answer these questions as they move through the tutorial, forcing them to slow down and think about the analysis and what their data outputs mean. Written answers to these questions are handed in to the instructor one week after completing the laboratory exercise. We try to return the graded assignments within two days, allowing the students to assess their understanding before the next group of tutorial questions is due. Answer keys for all questions are provided among the instructor resources (Supporting File S5: Instructor Grading Key). At the College of Wooster, students were also required to submit a laboratory report written in the format of a scientific paper one week after the entire module was completed.
Inclusive Teaching
At California State University San Marcos, students worked through the tutorial exercises independently but were encouraged to work together when they encountered problems and to discuss their findings. At Wooster, students worked through the tutorial in permanent teams of three students. The Lesson does not explicitly reference diversity in science, but it has been successfully implemented with diverse student audiences at multiple institutions, and all students successfully completed the exercise.
LESSON PLAN
Preparation to teach this Lesson begins with the instructor acquiring a Galaxy user account (as described in Supporting File S1: RNA-seq Student Tutorial I) and downloading the appropriate data files from NCBI SRA into the user History. At that point, the instructor can work through the complete tutorial at her/his own pace, identifying any potential changes to the Galaxy website and/or instructions that may need to be clarified. The instructor should also carefully review the annotated PowerPoint presentations (Supporting File S4: RNA-seq Annotated Instructor PowerPoints) and potentially do background reading on high throughput sequencing (19,20), RNA-seq (7,18), and/or Galaxy (15). The Lesson is designed for a computer laboratory over three lab sessions (~8 hr total). Students should be introduced to high throughput sequencing and the concept of RNA-seq (e.g., in the lecture section of the course) before starting the tutorial.
The first lab session has the longest instructor presentation, since both the key experimental background and the Galaxy platform are reviewed. Before the lab, students are assigned to read Afgan et al. (2016), which introduces Galaxy, and Shanks et al. (2016), which introduces the Arabidopsis/nematode experimental system (14,15). There is typically enough time to do a brief (~30 minute) round table discussion of the Afgan et al. (2016) article before students begin to work on the tutorial (Supporting File S1: RNA-seq Student Tutorial I). We randomly assign half of the students to work with an "infected replicate 1" file (RNA isolated from A. thaliana roots infected with H. schachtii; NCBI SRR2221834), and the other half work with the "control replicate 1" file (RNA from uninfected A. thaliana roots; NCBI SRR2221833). Students then individually follow the tutorial, which provides detailed and illustrated instructions on registering for a Galaxy user account and uploading the appropriate sequence file from NCBI SRA and the A. thaliana genome annotation file from Ensembl Plants into their Galaxy History. Thus, by the time students reach the end of Tutorial I, they will have established a Galaxy user account and acquired all of the files that they will need to "hit the ground running" during the next lab session. Throughout the first tutorial, students will encounter eight questions, which review basic experimental design concepts and ask them to explore their data. We encourage students to answer the questions as they work through the tutorial, and hand in their written answers at the end of the lab period. Alternatively, instructors could ask students to answer some or all of the questions orally or via an instant response polling system as they work through the exercise, allowing "just in time" feedback to clear up potential areas of confusion.
During the second laboratory session, students perform read quality control (using FastQC), read trimming (using Trimmomatic), and read mapping (using HISAT2) (22-24) (Supporting File S2: RNA-seq Student Tutorial II). After performing these steps "manually," they create a computational workflow using Galaxy's "Create Workflow" function. This allows the students to automatically download and analyze two additional RNA-seq data files ("infected replicates" 2+3 or "control replicates" 2+3) without additional hands-on time. Before the students begin the tutorial, the instructor presents the concepts behind each of these computational steps using the provided PowerPoint slides (Supporting File S4: Annotated Instructor PowerPoints). Again, a series of embedded questions keeps the students conceptually "on track" to assure that the tutorial does not simply become a mindless "point and click" exercise. When the second laboratory is complete, students will have generated Counts Tables from three "control" or three "nematode infected" samples. Again, all files are stored on each student's individual Galaxy History (250 gigabyte capacity), so no on-site storage is needed.
It is during the final laboratory session that students begin to see the biologically relevant "payoff" of the RNA-seq experiment: A list of all genes that are significantly up- or down-regulated in A. thaliana roots in response to nematode infection (Supporting File S3: RNA-seq Student Tutorial III). First, students must share Counts Table data with a "partner" (using Galaxy's Share History function) so that they have three "control" replicates and three "nematode infected" replicates in their History (at Wooster, each three-person team processed all six raw data files on their own). Differentially expressed genes are then identified using DESeq2 (25). The differentially expressed gene list from DESeq2 is exported to Excel and sorted. Finally, over-represented functional categories are identified amongst the up- and down-regulated genes using the online Panther Gene List Analysis tool (26). Again, the conceptual basis and operation of all of these tools are introduced in the instructor "pre-lab" presentation. Because students can now begin to glean biological meaning from their data, the questions embedded in the third tutorial are more frequent and detailed, and students typically need to spend time outside of the laboratory session to thoroughly answer them.
A timeline of the Lesson is provided in Table 1, below. In addition, all associated materials are provided as Supporting Files. Specifically, student tutorials are provided as Supporting Files S1-S3 (Supporting File S1: RNA-seq Student Tutorial I; Supporting File S2: RNA-seq Student Tutorial II; Supporting File S3: RNA-seq Student Tutorial III). The three-part Instructor PowerPoint and a grading key for the tutorial questions are also provided (Supporting File S4: Annotated Instructor PowerPoints; Supporting File S5: Instructor grading key). Finally, a document containing additional instructor background and instructions on identifying alternative RNA-seq data sets on NCBI SRA is included (Supporting File S6: Additional Instructor Background).
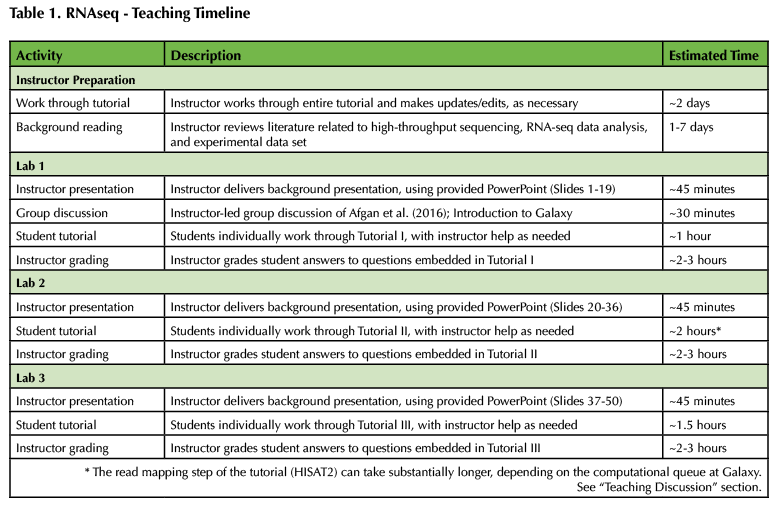
Table 1. RNAseq - Teaching Timeline
TEACHING DISCUSSION
As mentioned previously, this Lesson has been utilized three times, once with a group of 19 senior-level Biology and Biotechnology majors at California State University San Marcos, and twice with ~50 students total at the College of Wooster. At CSU San Marcos, this exercise made up the last three weeks of the semester-long course lab for BIOL 503: Modern Molecular Biology and Genomics. The first ten weeks of the BIOL 503 course lab consisted of an independent genome annotation exercise on Drosophila ficusphila using the educational materials developed by the Genomics Education Partnership (27). As such, it was somewhat difficult to interpret student comments on formal course evaluations- most comments spoke more generally to the organization and outcomes of the labs. However, it is notable that the percentage of students that were "very interested" in genomics increased from 29% before the course lab to 77% after taking the course lab. Informally, students indicated that they appreciated the clarity of the tutorials and had greatly increased their understanding of computational analysis of high-throughput sequence data. They also appreciated the relatively "low stakes" regular assessments (answers to tutorial questions) compared to the large lab report and oral presentation that was the assessment metric for the genome annotation exercise.
At the College of Wooster, students in the second offering of the "Genes and Genomes" course were asked to rate and identify the strengths and weaknesses of the RNA-seq module. Given that the students had diverse interests (the course is an elective for Biology and Neuroscience majors, but a requirement for Biochemistry & Molecular Biology majors) and experience (sophomores to seniors), it was not surprising that there was a great deal of variation in responses to the tutorial. Some students, likely with limited molecular biology background, found the exercise confusing; many others found the tutorial instructions clear. Several students recommended more opportunities for reflection should be incorporated. Since more than two dozen reflection questions are already integrated into the tutorial, it may be necessary for the instructor to emphasize that students must address the reflection questions at the same time as they are carrying out the exercise, thus minimizing a "race to the finish" approach. Wooster students were also asked if they agree with the statement "I understand and can carry out the computational steps involved in analyzing RNA-seq data." Again, there was a range of responses: 44% agreed or strongly agreed, 22% were neutral, and 33% disagreed or strongly disagreed (N = 27). The range of responses from this diverse group of students suggests that this tutorial might be best suited for upper-level undergraduates with some experience and interest in molecular genetics and computational biology.
While the use of Galaxy's cloud computational resources simplifies instructor setup and minimizes necessary in-house computational infrastructure, there are some downsides. Computationally-intensive data analysis steps are placed in a computational "queue" by Galaxy, which means that wait times can vary substantially, depending on the activity of other users. In this Lesson, the issue is relevant in Laboratory 2 during the read mapping step (HISAT2). We have found that this step can take as little as ten minutes to more than three hours, with no predictable pattern to the wait time. As such, instructors may wish to split Laboratory 2 into two shorter laboratory sessions (~1.5 hr each), allowing a "gap" for read mapping compute time. It should be noted that once a computational step is initiated in Galaxy, it will run to completion in the cloud, even when the student user logs out.
As demonstrated by the implementation of the Lesson with instructor-provided RNA-seq data at The College of Wooster, the provided tutorials are sufficiently flexible to allow instructors to analyze a variety of different data sets. One alternative data set in NCBI SRA is outlined in the Additional Instructor Background file, and instructions allowing users to identify their own appropriate data sets by searching SRA are also included (Supporting File S6: Additional Instructor Background). Obviously, any alternative data sets would need to be tested using the tutorial before implementation in the classroom, and specific details in the instructor PowerPoint and the tutorials would need to be modified. (For larger datasets, computational time and the Galaxy user account 250 gigabyte capacity should be considered.)
While this Lesson was originally developed for use in "dry-lab" (computational) laboratory courses, it is certainly possible to use it with student-generated RNA-seq data from a wet-lab course. Given the Lesson's modest time commitment (three lab sessions), it may be possible to pair it with RNA isolation, RNA quality assessment, sequencing library preparation, and high throughput sequencing exercises, thus generating the RNA-seq "input data" in the same course. Given the rapidly decreasing costs of Illumina sequencing, these types of end-to-end, wet lab to dry lab genomics approaches are increasingly feasible in upper division undergraduate Biology course laboratories.
SUPPORTING MATERIALS
- S1. RNA-seq - Student tutorial I
- S2. RNA-seq - Student tutorial II
- S3. RNA-seq - Student tutorial III
- S4. RNA-seq - Annotated instructor PowerPoints
- S5. RNA seq - Instructor grading key
- S6. RNA-seq - Additional instructor background
ACKNOWLEDGMENTS
We thank the Network for Integrating Bioinformatics into Life Sciences Education (https://qubeshub.org/community/groups/niblse) for coordinating a faculty incubator which led to the development of this Lesson. We also thank Dr. Sam Donovan (University of Pittsburgh) for his advice in early stage development of the tutorial.
References
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y-J, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Ho CH, Irzyk GP, Jando SC, Alenquer MLI, Jarvie TP, Jirage KB, Kim J-B, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376-380.
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IMJ, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DMD, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham JA, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Chiara E Catenazzi M, Chang S, Neil Cooley R, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fuentes Fajardo KV, Scott Furey W, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, Hoschler K, Hurwitz S, Ivanov DV, Johnson MQ, James T, Huw Jones TA, Kang G-D, Kerelska TH, Kersey AD, Khrebtukova I, Kindwall AP, Kingsbury Z, Kokko-Gonzales PI, Kumar A, Laurent MA, Lawley CT, Lee SE, Lee X, Liao AK, Loch JA, Lok M, Luo S, Mammen RM, Martin JW, McCauley PG, McNitt P, Mehta P, Moon KW, Mullens JW, Newington T, Ning Z, Ling Ng B, Novo SM, O'Neill MJ, Osborne MA, Osnowski A, Ostadan O, Paraschos LL, Pickering L, Pike AC, Pike AC, Chris Pinkard D, Pliskin DP, Podhasky J, Quijano VJ, Raczy C, Rae VH, Rawlings SR, Chiva Rodriguez A, Roe PM, Rogers J, Rogert Bacigalupo MC, Romanov N, Romieu A, Roth RK, Rourke NJ, Ruediger ST, Rusman E, Sanches-Kuiper RM, Schenker MR, Seoane JM, Shaw RJ, Shiver MK, Short SW, Sizto NL, Sluis JP, Smith MA, Ernest Sohna Sohna J, Spence EJ, Stevens K, Sutton N, Szajkowski L, Tregidgo CL, Turcatti G, Vandevondele S, Verhovsky Y, Virk SM, Wakelin S, Walcott GC, Wang J, Worsley GJ, Yan J, Yau L, Zuerlein M, Rogers J, Mullikin JC, Hurles ME, McCooke NJ, West JS, Oaks FL, Lundberg PL, Klenerman D, Durbin R, Smith AJ. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456:53-59.
- Bronner IF, Quail MA, Turner DJ, Swerdlow H. 2014. Improved Protocols for Illumina Sequencing. Curr Protoc Hum Genet 80: 18.2.1-18.2.42.
- Barabaschi D, Tondelli A, Desiderio F, Volante A, Vaccino P, Valè G, Cattivelli L. 2016. Next generation breeding. Plant Sci 242:3-13.
- Shyr D, Liu Q. 2013. Next generation sequencing in cancer research and clinical application. Biol Proced Online 15:4.
- Boughner LA, Singh P. 2016. Microbial Ecology: Where are we now? Postdoc J 4:3-17.
- Wang Z, Gerstein M, Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57-63.
- Galbraith DA, Yang X, Niño EL, Yi S, Grozinger C. 2015. Parallel epigenomic and transcriptomic responses to viral infection in honey bees (Apis mellifera). PLoS Pathog 11:e1004713.
- Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. 2009. Transcriptome sequencing to detect gene fusions in cancer. Nature 458:97-101.
- Peterson MP, Malloy JT, Marden JH, Buonaccorsi VP. 2015. Teaching RNAseq at Undergraduate Institutions: A tutorial and R package from the Genome Consortium for Active Teaching. CourseSource. 10.24918/cs.2015.14.
- Campbell CE, Nehm RH. 2013. A Critical Analysis of Assessment Quality in Genomics and Bioinformatics Education Research. CBE Life Sci Educ 12:530-541.
- Maloney M, Parker J, LeBlanc M, Woodard CT, Glackin M, Hanrahan M. 2010. Bioinformatics and the Undergraduate Curriculum Essay. CBE Life Sci Educ 9:172-174.
- Attwood TK, Blackford S, Brazas MD, Davies A, Schneider MV. 2017. A global perspective on evolving bioinformatics and data science training needs. Brief Bioinform bbx100, https://doi.org/10.1093/bib/bbx100.
- Shanks CM, Rice JH, Zubo Y, Schaller GE, Hewezi T, Kieber JJ. 2016. The Role of Cytokinin During Infection of Arabidopsis thaliana by the Cyst Nematode Heterodera schachtii. Mol Plant-Microbe Interact 29:57-68.
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, Grüning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J. 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44:W3-W10.
- Makarevitch I, Frechette C, Wiatros N. 2015. Authentic Research Experience and "Big Data" Analysis in the Classroom: Maize Response to Abiotic Stress. CBE Life Sci Educ 14. 10.1187/cbe.15-04-0081.
- Makarevitch I, Martinez-Vaz B. 2017. Killing two birds with one stone: Model plant systems as a tool to teach the fundamental concepts of gene expression while analyzing biological data. Biochim Biophys Acta 1860:166-173.
- Wolfien M, Rimmbach C, Schmitz U, Jung JJ, Krebs S, Steinhoff G, David R, Wolkenhauer O. 2016. TRAPLINE: a standardized and automated pipeline for RNA sequencing data analysis, evaluation and annotation. BMC Bioinformatics 17:21. 10.1186/s12859-015-0873-9.
- Levy SE, Myers RM. 2016. Advancements in Next-Generation Sequencing. Annu Rev Genomics Hum Genet 17:95-115.
- Goodwin S, McPherson JD, McCombie WR. 2016. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 17:333-351.
- Adams NE. 2015. Bloom's taxonomy of cognitive learning objectives. J Med Libr Assoc 103:152-153.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114-2120.
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357-360.
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15. 10.1186/s13059-014-0550-8.
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. 2017. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45:D183-D189.
- Shaffer CD, Alvarez C, Bailey C, Barnard D, Bhalla S, Chandrasekaran C, Chandrasekaran V, Chung H-M, Dorer DR, Du C, Eckdahl TT, Poet JL, Frohlich D, Goodman AL, Gosser Y, Hauser C, Hoopes LLM, Johnson D, Jones CJ, Kaehler M, Kokan N, Kopp OR, Kuleck GA, McNeil G, Moss R, Myka JL, Nagengast A, Morris R, Overvoorde PJ, Shoop E, Parrish S, Reed K, Regisford EG, Revie D, Rosenwald AG, Saville K, Schroeder S, Shaw M, Skuse G, Smith C, Smith M, Spana EP, Spratt M, Stamm J, Thompson JS, Wawersik M, Wilson BA, Youngblom J, Leung W, Buhler J, Mardis ER, Lopatto D, Elgin SCR, Wakimoto B. 2010. The Genomics Education Partnership: Successful Integration of Research into Laboratory Classes at a Diverse Group of Undergraduate Institutions. CBE--Life Sci Educ 9:55-69.
Article Files
Login to access supporting documents
Tackling "Big Data" with Biology Undergrads: A Simple RNA-seq Data Analysis Tutorial Using Galaxy(PDF | 136 KB)
S1. RNA-seq - Student tutorial I.docx(DOCX | 2 MB)
S2. RNA-seq - Student tutorial II.docx(DOCX | 3 MB)
S3. RNA-seq - Student tutorial III.docx(DOCX | 2 MB)
S4. RNA-seq - Annotated instructor powerpoints- open source.ppt(PPT | 9 MB)
S5. RNA-seq - Instructor grading key.docx(DOCX | 31 KB)
S6. RNA-seq - Additional instructor background.docx(DOCX | 697 KB)
- License terms

Comments
Comments
There are no comments on this resource.