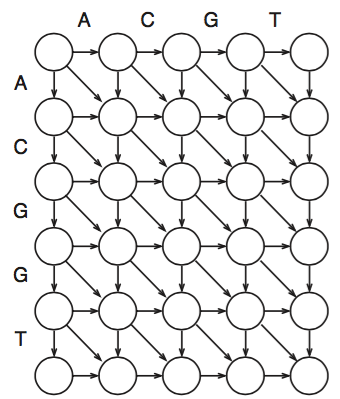
Needleman - Wunsch Algorithm Exercise
Author(s): Michael Sierk1, Sam S Donovan2, Neal Grandgenett, Bill Morgan3, Mark A. Pauley4, Elizabeth F Ryder5
1. Saint Vincent College 2. University of Pittsburgh 3. The College of Wooster 4. University of Nebraska at Omaha 5. Worcester Polytechnic Institute
1350 total view(s), 188 download(s)
Bundle(ZIP | 346 KB)
- License terms
Description
This exercise is used in a sophomore-junior level bioinformatics course. The algorithm is introduced to the students who then spend around 15 minutes filling out the diagram. Students have typically been doing some sequence alignment exercises, such as BLAST searches, and have used different scoring systems to score different short alignments, so they are familiar with sequence alignments and the concept of scoring the alignments, but they have not been exposed to how to generate an alignment.
Notes
The publication was updated to include the most recent materials and authorship list.
Cite this work
Researchers should cite this work as follows:
- Michael Sierk, Donovan, S. S., Grandgenett, N., Morgan, B., Pauley, M. A., Ryder, L. F. (2020). Needleman - Wunsch Algorithm Exercise. NIBLSE Incubator: Needleman-Wunsch Algorithm Exercise, (Version 4.0). QUBES Educational Resources. doi:10.25334/H7KQ-F202