Application of a Bacterial Experimental Evolution System to Visualize and Teach Evolution in Action: A Course-Based Undergraduate Research Experience
Editor: Tammy Long
Published online:
Abstract
Concepts of evolution are typically taught through examples of extremely long timescales, which do not always resonate broadly. Here, we describe a course-based undergraduate research experience tailored for junior and senior undergraduate biology majors. Students visualize and learn in real-time how evolution can operate in bacteria in response to problems associated with a high-density lifestyle. Students directly evolve mutant strains, conduct whole genome sequencing to identify the causal mutations, carry out bioinformatics analysis to predict molecular consequences of the mutations, engineer their mutants to become antibiotic-resistant, and compete them head-to-head in a class-wide round-robin tournament to infer the properties of natural selection. The presented format is designed for a full semester, but the modular structure of the course allows instructors to make simple modifications for a shorter duration. A substantial portion of this course also focuses on scientific communication. Each student prepares a lab report structured as an original research article to gain experience in writing a publication quality manuscript. Individual components of their reports are prepared throughout the semester and are followed with instructor- and peer-based draft edits. Finally, students are tasked with working as a team to deliver an oral presentation, which drives them to come to a consensus on the interpretation of their group’s data. Such a comprehensive research experience is difficult for a student to acquire without securing a research position in a faculty lab, but this course allows a large group of students to directly experience and actively contribute to open-ended and hypothesis-driven research.
Primary image: A graphical abstract of the course’s relationship to the CURE framework. Individual lab components encourage students to collaborate with peers and use genuine scientific practices in an iterative process to uncover novel results with broad relevance.
Citation
Kessler C, Mazza W, Christopher K, Kim W. 2022. Application of a bacterial experimental evolution system to visualize and teach evolution in action: A course-based undergraduate research experience. CourseSource. https://doi.org/10.24918/cs.2022.24Society Learning Goals
Biochemistry and Molecular Biology
- Information storage and flow are dynamic and interactive
- What is a genome?
- How does the nucleotide sequence of the gene lead to biological function?
- How do genomes transmit information from one generation to the next?
- Discovery requires objective measurement, quantitative analysis and clear communication
- What is the scientific process?
- What skills are needed to access, comprehend and communicate science?
- What constitutes a scientific community of practice?
- Evolution
- What is the significance of evolution?
- What are the mechanisms of evolution?
- How is natural selection a key evolutionary mechanism?
- What is the molecular basis of evolution?
Bioinformatics
- DNA - Information Storage [GENOMICS]
- How can bioinformatics tools be employed to analyze genetic information?
- Computational Skills
- How do biologists employ software development as part of the scientific discovery process?
- What higher-level computational skills can be used in bioinformatics research?
Cell Biology
- Evolutionary History of Cells
- How does evolutionary history explain the similarities and differences among cells?
Genetics
- Molecular biology of gene function
- How is genetic information expressed so it affects an organism's structure and function?
- Genetic variation
- How do different types of mutations affect genes and the corresponding mRNAs and proteins?
- Genetics of model organisms
- How do the results of molecular genetic studies in model organisms help us understand aspects of human genetics and genetic diseases?
Microbiology
- Evolution
- Considering the immense variety of microenvironments, how have mutations and horizontal gene transfer selected for a huge diversity of microorganisms?
- Metabolism
- How can the growth of microorganisms controlled by physical, chemical, mechanical, or biological means?
- How are the interactions of microorganisms among themselves and with their environment determined by their metabolic abilities?
- Information Flow and Genetics
- How can cell genomes be manipulated to alter cell function?
- How do genetic variations impact microbial functions (e.g., in biofilm formation, pathogenicity, and drug resistance)?
Lesson Learning Goals
Goals in this course are divided into practical laboratory techniques as well as broader skills that will contribute to a deeper understanding of science, communication, and experimental design. The identified learning goals and objectives are evaluated through aligned assessments detailed in Table 1. Nature of Science: By the end of the semester, students should be able to:- Explain and analyze primary scientific literature
- Develop testable hypotheses and predict outcomes of experiments
- Design experiments to address hypotheses
- Conduct experiments in a technically sound manner and explain how technical errors could generate noise in the experimental data
- Record and interpret results, including an evaluation of positive and negative controls
- Explain the importance of reproducibility and statistical analysis
- Maintain a meticulous lab notebook for potential publication
- Carry out aseptic techniques and understand their principles
- Culture bacteria for diverse processes, including:
- isolating individual genotypes from mixed cultures
- mixing different genotypes
- harvesting cells from liquid and solid media
- diluting and plating cells to enumerate populations
- utilizing specific antibiotics to maintain respective plasmids
- freezing cells for long term storage
- Engineer antibiotic-resistant bacteria
- Engineer fluorescent bacteria
- Experimentally evolve bacteria and isolate individual mutants from a population
- Design an experiment to test functional consequences of mutations
- Extract and quantify genomic DNA
- Analyze genome sequence data
- Apply bioinformatics tools to predict functional consequences of mutations
Lesson Learning Objectives
Students will be able to- use reasoning and deductive methods to develop a testable hypothesis and predict expected results.
- gather empirical and measurable evidence through observation and experimentation.
- assess cellular processes and mechanisms that lead to physiological functions.
- describe the principles of evolutionary theory and the mechanisms leading to genetic diversity.
| Learning Objective | Aligned Assessment(s) |
|---|---|
| Students will be able to use reasoning and deductive methods to develop a testable hypothesis and predict expected results. |
|
| Students will be able to gather empirical and measurable evidence through observation and experimentation. |
|
| Students will be able to assess cellular processes and mechanisms that lead to physiological functions. |
|
| Students will be able to describe the principles of evolutionary theory and the mechanisms leading to genetic diversity. |
|
| Students will be able to explain and analyze primary scientific literature. |
|
| Students will be able to design experiments to address hypotheses. |
|
| Students will be able to maintain a meticulous lab notebook for potential publication. |
|
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Undergraduate biology students are taught about evolution early in their academic careers, but few get the opportunity to personally witness it in real time. While many institutions teach evolutionary courses through visual comparisons of macro-structures in vertebrates (e.g., Darwin’s finches, flightless birds, fossils, etc.), the timescale for many undergraduates remains abstract and underappreciated. Compounding this issue, not all students receive equal exposure to evolutionary concepts in high school, which presents many challenges. Recent polling of our incoming biology first year class shows that nearly one fourth claim they were never taught evolution in high school (1). Moreover, even college students that have already completed introductory biology courses exhibit high rates of misconceptions about evolution and natural selection (2). Many traditional teaching strategies, even when specifically designed to address these preconceived conceptual misunderstandings, have fallen short of accomplishing this task, emphasizing the need for interactive instructional designs (3, 4). How then can we appreciably implement an evolutionary course that illustrates the power of natural selection and its molecular consequences? Previous evidence suggests that inquiry-driven teaching strategies may be particularly well-suited to helping students confront and dispel their own misconceptions (5, 6). Thus, we designed an original course-based undergraduate research experience (CURE) with the goal of engaging students in authentic inquiry into evolutionary processes. In our course, junior and senior undergraduate biology majors directly evolve bacteria to observe natural selection in real time and quantitatively assess the evolved molecular differences of mutant organisms. Collection of authentic data allows students to come to meaningful conclusions that challenge their preconceptions in a way that traditional lecture-based evolutionary courses cannot.
In response to calls for student-centered opportunities for inquiry in biology education, CUREs have become an increasingly popular method to engage larger numbers of students in authentic inquiry than possible through individual mentored research internships (7–9). Under the framework developed by Auchincloss and colleagues, CUREs are structured to encourage students to collaborate with peers, using genuine scientific practices in an iterative process to uncover novel results in a field with broad relevance (9). Our course satisfies the parameters of a CURE by enabling groups of students to propose hypotheses, carry out experiments, and analyze their own data to discover and characterize novel mutant bacterial strains. Numerous studies have found that students that participate in CUREs report greater self-efficacy and ownership over their laboratory projects, demonstrate increased persistence in STEM fields, and use more effective scientific thinking processes as compared to those taking traditional lecture or laboratory courses (10–12). Moreover, it has been proposed that use of CUREs supports a diverse and inclusive learning environment, potentially increasing the proportion of students from underrepresented groups able to participate in scientific research and thus develop self-efficacy and scientific identity, which ultimately promote retention in the sciences (13–15). Recent evidence suggests that gains in scientific identity and student ownership of the research project are most pronounced when students analyze data they have collected themselves (16), as is the case in our course.
Our course utilizes an experimental system that allows students to observe and analyze mutation-driven adaptations in real time. Through experimental evolution, students isolate mutant strains that arise through methods of adaptation that also give rise to clinically significant pathogens (17–19). In this course, students begin with colonies of Pseudomonas fluorescens comprised of billions of densely packed cells. One week after plating the bacterial cells, students observe the emergence of strains that have acquired secretion mutations, allowing them to physically push away their ancestral neighbors, rise to the surface of the colony, and escape the densely crowded population. As shown in Figure 1, these mutants are observable to the human eye and are phenotypically reminiscent of an algal plume rising from a pond. The molecular changes that give rise to these phenotypes also result in fitness differences among the isolated mutants. This presents the students with an opportunity to learn how random mutations can provide a competitive advantage, a necessary step in allowing an individual to stand the test of time under the lens of natural selection. Students then compete their own evolved strains against those of their peers head-to-head in a round-robin tournament format to determine a relative fitness rank and postulate how single nucleotide changes alter molecular function to ultimately manifest the phenotypic changes they observe.
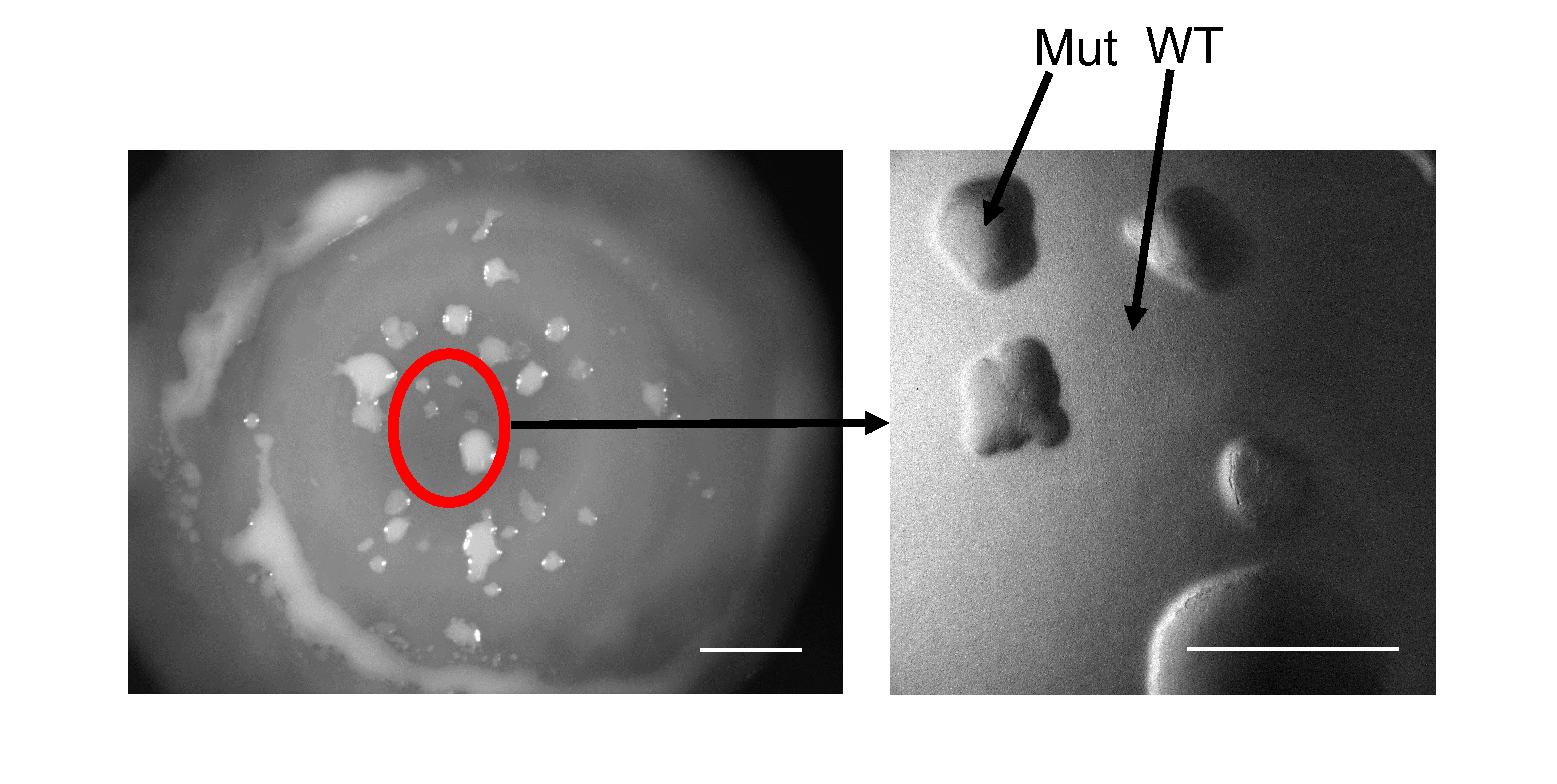
This experimental system is based on a model utilized in the Kim lab where diverse mutations in a single gene called rsmE repeatedly arise in P. fluorescens colonies (18). The RsmE protein is a translational repressor that prevents the translation of multiple mRNAs that contribute to the production of diverse extracellular secretions. These secretions are predicted to work together to push away the neighboring cells in a densely populated environment, a powerful evolutionary solution to address the problem of crowding. However, many questions remain, including how RsmE physically interacts with diverse mRNAs, what the RsmE-regulated secretions are, and how these secretions actually function to create space. Through this experimental system, students isolate and characterize their own unique rsmE mutants that deregulate the native function of RsmE across a spectrum of repressive activity and phenotypes. In a simple sense, RsmE normally functions like a bottle cap to prevent the inner contents from spilling out; thus, a mutated RsmE becomes less effective at carrying out this repressive function. Mutations that terminate RsmE’s function through frameshift (insertions or deletions) are predicted to result in the production (de-repression) of many secretions, which provide a substantial fitness benefit observable by the naked eye (18). Students also capture missense mutations caused by a single nucleotide substitution, which alter the amino acid sequence of RsmE at different residues. Cells with missense mutations present a range of phenotypes, from those comparable to the frameshift mutants (loss of all function – all secretions are de-repressed) to those with partial loss of function (some secretions remain repressed); the former exhibits greater fitness gain than the latter since all secretions are necessary to create optimal spacing. In this course, students isolate rsmE mutants and are empowered to formulate hypotheses based on their own phenotypic assay data. Students then carry out a competition experiment and analyze their own data to determine which rsmE mutations provide the greatest fitness. This entirely novel data allows students to discover and characterize important functional domains of the RsmE protein. Our main goal for students in this course is to assess how these mutations alter cellular processes that lead to the observed competitive phenotypes, thereby achieving the parameters of the CURE as described above. In addition, P. fluorescens possesses two RsmE paralogs (RsmA and RsmI) that are generalized to perform overlapping functions. However, no mutations in these genes have been observed in any of our experiments, which suggests that these paralogs possess divergent functions. This observation allows students to understand how a duplication event and subsequent mutations can lead to the evolution of functionally distinct paralogs.
Through this course, students witness and identify genetic changes that result in the emergence of a newly evolved phenotype and empirically evaluate the consequences of this phenotypic change. This short-timescale evolutionary course generates a first-hand collection of authentic data that provides directly observed evidence of natural selection. Importantly, the selective pressure involves the readily relatable problem of overcrowding, which can be solved by simply creating more space. Students are also empowered to directly relate their findings to real-world problems like the threat of antibiotic resistance, which could emerge through the same processes of random mutations and selection in a short timescale. Such understanding can be appreciated by all people regardless of their background and provides the seed that will challenge students’ preconceptions of evolutionary topics that they will encounter throughout their careers. While numerous novel CUREs have been reported in the last decade, our CURE specifically aims to use this teaching strategy to help students explore the molecular bases of evolution by natural selection through a hands-on wet-lab experience.
Intended Audience
This upper-level molecular and cell biology course was taught to undergraduate biology majors within their third or fourth year. Techniques used in this lab require little to no laboratory experience and have been tested with success on high school students in an internship setting. However, the theoretical content assumes a basic understanding of molecular and cellular biology, and therefore lecture materials would need to be modified with additional background information if these exercises were used for an introductory level course.
Required Learning Time
Lab times did not exceed 2.5 hours per session. This time can be reduced by providing materials already prepared by the instructor. Out-of-class preparation and assignments are estimated to be 3-6 hours per week for the students as a 4-credit lab.
Prerequisite Student Knowledge
Students must first complete an introductory cell and molecular biology course and be familiar with the central dogma (DNA > RNA > Protein) as this is the backbone of the course. More specialized background knowledge on the experimental system is obtained in pre-lab lecture(s) given by the instructor or through reading the associated primary literature.
Prerequisite Teacher Knowledge
The instructor must be conceptually sound with core principles of molecular biology, biochemistry, and evolution, in addition to being technically familiar with growing and maintaining bacterial cell cultures, genetic engineering, genome sequencing, and bioinformatics. In addition, a basic understanding of statistics and sequence analysis should be sufficient for the successful implementation of this course. The instructor should review and have a working knowledge of the source material as presented in Kim et al. (18) as it will serve as the main text for this course. The key research findings of this paper that are relevant to this course are summarized in the Lesson Plan.
Scientific Teaching Themes
Active Learning
Given its setting as an inquiry-driven laboratory, this course fundamentally includes many active learning techniques. Each lab session begins with a brief demonstration and/or lecture-based orientation to the day’s activities, but the vast majority of time in class is spent engaged in hands-on laboratory research or small-group data analysis and discussion. Students gain firsthand experience with all lab techniques, providing valuable skills that they will carry forward in their careers. Students collect, graph, and analyze their data. Generation and evaluation of de novo research data stimulates independent literary research to enable the student to explain and teach their findings to their peers. Students work in groups of three or four to develop skills in teamwork, effective communication, and critical thinking, consistent with the collaborative nature of CUREs (9).
Students are also given multiple opportunities to develop skills in scientific communication. To strengthen their ability to analyze and interpret primary journal articles, students engage in guided discussions of key papers related to the project, using a published data-focused strategy (20). These exercises also help the students to learn the structure and organization of an original research article, and students directly translate this knowledge by preparing the key components (introduction, materials and methods, results, discussion) based on their own research outcomes (Supporting File S1. Bacterial Evolution – Course Manual with Teaching Guides, Student Protocols, and Assessments). Each component is independently prepared by weaving together all lab sessions within a few pages, challenging students to differentiate essential and non-essential information and helping them practice concise scientific argumentation. Detailed feedback is provided by the instructor, then the students compile and integrate all components to write a final lab report, which essentially culminates in a manuscript describing original research. In addition, the students communicate their research findings to their peers and the instructor in a graded oral presentation. Multiple rounds of peer and instructor feedback throughout these processes allow revision of hypotheses, goals, and written work, both allowing for formative assessment and emphasizing the iterative and collaborative characteristics of science. In addition to developing communication skills, the summative written and oral assessments encourage the student to revisit their obtained knowledge, which is paramount in skill acquisition (21).
Assessment
Student achievement of learning objectives is assessed in a variety of ways, consistent with best practices in inclusive pedagogy. The graded components are listed below, and associated rubrics are provided (Supporting File S1. Bacterial Evolution – Course Manual with Teaching Guides, Student Protocols, and Assessments). Because this course was originally developed to satisfy a Duquesne University intensive writing curriculum requirement, a substantial portion of the work centers around scientific communication:
- Writing and Paper Interpretation Assignments (50%): This portion of the grade is split between formative assessments of early drafts of each individual section of the laboratory report, written analyses of primary research articles, and – as a form of summative assessment – the final complete draft of the lab report, which is structured in the format of a primary journal article. Students receive extensive feedback from instructors and also engage in peer review to model authentic science practices.
- Team Oral Presentation (10%): Although the writing assignments in the course are completed on an individual basis, each group of students is tasked with working as a team to generate and deliver an oral presentation. This collaboration drives them to come to a consensus on their interpretation of their group’s data. Students are also assessed on their ability to ask thoughtful questions during their peers’ presentations. Students are awarded both group and individual grades on the presentation to foster collaboration while also maintaining fairness.
- Quizzes (20%): Throughout the semester, several short-answer quizzes are administered. These focus largely on application, providing an additional opportunity for students to demonstrate their ability to interpret the kinds of data they are gathering. Quiz questions also ask students to show that they understand the rationale of the experimental approaches they are taking in the lab.
- Lab Notebook (10%): Students keep detailed lab notebooks, consistent with learning authentic scientific practices. Notebooks are assessed on the basis of adherence to formatting, comprehensiveness, and accuracy.
- Lab Performance and Exit Tickets (10%): Students are assessed on their lab citizenship (including appropriate attire, collaborative behavior, and responsible clean-up) as well as their ability to answer the Exit Ticket “Questions To Think About” found at the end of each lab session guide. By answering these questions before departing each lab, students confirm that they understand the rationale behind the work they are doing and are able to take ownership of the project and make decisions about upcoming experiments.
Inclusive Teaching
This course includes varied types of instruction (discussion, hands-on activities, open-ended inquiry, mini-lectures) and multiple modes of assessment as noted above, both of which are inclusive teaching strategies that provide students with a variety of ways to learn and demonstrate competence (22, 23). Numerous opportunities for low-stakes formative feedback are provided early on through drafts of written work and paper analyses (24). Furthermore, all assignment instructions are designed using principles of transparency shown to improve sense of belonging and success for students from marginalized backgrounds (25). In keeping with transparency practices, detailed rubrics and guidelines are provided for all assessments to ensure that students are aware of what is expected of them.
Additionally, the instructors serve as guides and research mentors rather than coming from a place as the absolute holders of knowledge and authority in the classroom. For example, students are empowered with the agency to make their own decisions about experiments, such as choosing which mutant strains to work with. Because analyses of primary journal articles and student-generated data take place through small-group discussion, peer instruction is emphasized over instructor-led lectures.
By its nature, this course makes a mentored research experience accessible to all enrolled students and enables participation in scientific inquiry, which together can help students develop a sense of belonging in science (13). All materials and resources for this course were provided by Duquesne University, granting equal access to learning and experimental procedures regardless of the background or need of our students. Adaptations to this protocol may be made to accommodate students with disabilities or students at an increased risk of infection from microorganisms. This course is taught under the context of microevolution and does not infringe on religious beliefs related to macroevolution. Finally, by discussing how diversity provides source material upon which natural selection can act, the course content fosters appreciation for the value of existing diversity in a population for the overall fitness of that species.
Lesson Plan
We have developed a 16 week, 21 lab-session course manual that includes teaching guides for each section, allowing the instructor to complete the class preparations and master the biology themes and skills needed to successfully implement this course (Supporting File S1. Bacterial Evolution – Course Manual with Teaching Guides, Student Protocols, and Assessments). The student protocols have been developed and refined after teaching this course for three consecutive years. An answer key is provided for the questions within the lectures and student protocols that test students’ knowledge. Finally, we have included the original lecture slides given during the course and divided the materials by the sessions identified in Table 2 (Supporting File S2. Bacterial Evolution – Prelab Lecture Teaching Slides). While this course was taught as one collection, the lab manuals provided can be adapted for standalone experiments that can be integrated into existing courses. Throughout the course, students work with naturally evolved mutant strains and then quantify how these mutations affect fitness. As shown in Table 2, we have divided this course into five parts focusing on the identified society learning goals.
Table 2. An overview and timeline of the course, divided into five parts that could be modified for incorporation into another existing course or function as a standalone course.
| Activity | Description | Time | Assignments |
|---|---|---|---|
| Lecture | Introductory lecture to provide background and course goals. | 2 hours | |
| Part 1: Functional Consequences of Mutation (Competitions Round 1) | |||
| Session 1 | Mystery mutant competition setup and surfactant assays. | 2.5 hours | |
| Session 2 | Collect and analyze surfactant assay data and seed count data. | 1 hour | Paper Analysis Worksheet #1 |
| Session 3 | Harvest competitions (Day 7) and prepare enumeration plates. | 2 hours | |
| Part 2: Natural Selection as a Mechanism of Evolution | |||
| Session 4 | Begin experimental evolution of mucoid variants and collect and analyze competition data from Part 1. | 2.5 hours | |
| Session 5 & 6 | Isolate and propagate evolved mucoid variants. | 1 hour / session | |
| Session 7 & 8 | Prepare and analyze surfactant assays for each evolved mutant and generate cell pellets for DNA extractions. | 1 hour / session | Mini Lab Report |
| Session 9 | DNA extraction of the Session 7 evolved mutant cell pellet and propagation of cells for Session 10. | 2.5 hours | |
| Session 10 | Conjugation of evolved mucoid variants for round 2 competitions | 2 hours | Introduction Draft Due |
| Session 11 & 12 | Isolation and confirmation of antibiotic resistant mucoid variants engineered in Session 10. | 1 hour / session | Lab Notebook Check #1 |
| Session 13 & 14 | PCR and gel visualization of the Tn7 antibiotic resistance cassette to confirm successfully engineered strains. | 1.5 hours / session | Paper Analysis Worksheet #2 |
| Part 3: Functional Consequences of Mutation (Competitions Round 2) | |||
| Session 15 & 16 | Class-wide competition of mucoid variants selected by students from Part 2 and subsequent enumeration of competition seed mixtures. | 2 hours / session | Methods Draft Due |
| Session 17 & 18 | Harvest competitions (Day 7) and enumeration followed by analysis of the collected data. | 2 hours / session | |
| Session 19 | Calculate relative fitness and run statistical analyses on the round 2 competition data. | 2.5 hours | Results Draft Due |
| Part 4: Evaluating the Molecular Basis of Evolution and its Consequences | |||
| Session 20 | Analysis and discussion of previously generated competition data to evaluate the role of mutations on protein function | 2 hours | Lab Notebook Check #2 |
| Session 21 | Bioinformatics on returned sequence data from the Session 10 extractions to determine the mutation of the evolved mutant studied in Parts 2 and 3. | 2 hours | Discussion Draft Due |
| Part 5: Experimental Conclusions and Assessment | |||
| Writing workshop | Students participate in a peer-review workshop to revise the results and discussion sections of their lab reports. | 1 hour | |
| Concluding presentations | Group presentations of the data collected from Parts 1-4 and student interpretations of how the identified mutation in their evolved mutant resulted in the change of fitness observed in the round 2 of competitions. | 2.5 hours | Final Lab Report Due |
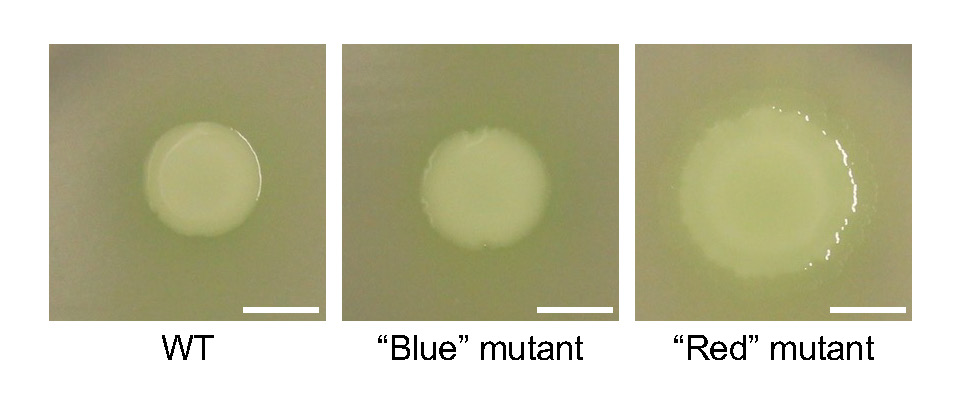
In our simple experimental system using agar plates, mutations randomly occur in colonies of the bacterium Pseudomonas fluorescens, and some of these mutations provide a fitness advantage over the wild-type (WT) ancestor. The experimental system is described in detail in a publication by Kim et al. (18), which serves as the main reference for the course. These mutant cells evolutionarily solve the problem of restricted growth in densely populated colonies by physically creating new space at the microscopic level, which manifests as readily observable mucoid patches on the colony surface at the macroscopic level (Figure 1). Remarkably, isolating and sequencing individual mutants from over 500 independent mucoid patches reveal that this repeatedly emergent phenotype is exclusively caused by diverse single mutations (i.e., one unique mutation per cell) in the regulator of secondary metabolites gene rsmE. RsmE protein natively represses secretions by binding to cognate mRNAs and preventing translation; thus, the acquired mutations act to reduce this inhibitory interaction. In Kim et al. (18), rsmE mutants are classified into two categories based on readily visible secretions: mutants that produce both a mucus and a surfactant are categorized as “red” mutants, while those that appear to only produce the mucus are considered “blue” mutants. Both red and blue mutants outcompete the WT, but the red mutants have a significantly higher fitness advantage over the WT compared to the blue mutants. This observation indicates that the combination of both the mucus and surfactant (red mutants) provides a greater competitive advantage compared to producing the mucus alone (blue mutants). Unaltered RsmE represses the production of both the mucus and surfactant under standard experimental conditions. RsmE is predicted to control the production of additional secretions that are not visible to the naked eye and remain unidentified.
If the loss RsmE function provides such a competitive advantage, then why is the rsmE gene evolutionarily conserved? This question is already addressed by Kim et al. (18), but a simple answer is that the production of multiple extracellular secretions is costly and RsmE’s native repressive activity ensures an optimal cost-benefit balance. Our experimental system utilizes a nutrient-rich medium, which readily supports the cost of producing extracellular secretions. Not surprisingly, rsmE mutations are frequently observed in nutrient rich environments like the rhizosphere. Conversely, when an rsmE mutant colonizes a nutrient-poor environment, then the WT rsmE gene will sweep through the population. Our experimental system thus favors the selection of diverse mutations in rsmE by reducing the metabolic cost to produce the extracellular secretions that provide high benefit through creating space in a very crowded environment. As described by Kim et al. (18), no other mutation outside of rsmE has been observed in more than 500 independently evolved isolates, which indicates that the fitness advantage is exclusively obtained through altered RsmE activity. P. fluorescens possesses two additional paralogs of RsmE (RsmA and RsmI), which are generalized in the literature to be functionally redundant, as there is very little sequence divergence and also share the same predicted secondary structures. However, the absence of mutations in rsmA or rsmI suggests that RsmE regulates unique mRNA targets. The functional domains and essential residues that differentiate RsmE’s function from that of RsmA and RsmI remain unknown, and the students participating in this course are provided with an exciting opportunity to help resolve this important question.
This novelty provides the perfect backdrop for a CURE. Here, students evolve and isolate mutants in real time, carry out phenotypic assays to assess the de-repression of the mucus and/or surfactant to categorize them as either red or blue, and they choose a blue mutant from their own collection of evolved strains to compete in a class-wide round-robin tournament. Blue mutants are chosen for competition, because all mutated forms of RsmE in the red category are generalized to have entirely lost the native function of inhibiting mRNA translation (i.e., all secretions are now de-repressed), while individual RsmE mutations in the blue category are predicted to have partially lost function across a spectrum (i.e., one blue mutant de-represses more or less individual secretions compared to another). The key hypothesis here is that RsmE mutants that de-repress more secretions (i.e., produce more molecular tools to create space) will be more fit in a head-to-head competition. Students also extract DNA from their own mutants for genome sequencing, map their mutations to the primary RsmE sequence and identify affected functional domains, and predict consequences of their mutations on secondary and tertiary protein structure. Students statistically rank the relative fitness of all mutants in the tournament by integrating their own competition results with those generated by their peers, then correlate the outcome to the mutation data. Students also learn that the individual RsmE mutants ranked across the relative fitness spectrum (e.g., most fit, medium fit, and least fit) will serve as new subjects for future research to characterize distinct profiles of mRNA binding. Overall, our goal for this course is to expose students to a line of scientific inquiry where they can make potentially pioneering discoveries that are novel to the students, instructors, and the greater scientific community.
Part 1: Functional Consequences of Mutation (Competitions Round 1)
A fundamental component of a CURE is to engage students in meaningful and iterative work by which they are exposed to a question and repeat and refine their skills for their research to progress and address the question. With this in mind, we have the students carry out two rounds of competitions to assess the fitness of isolated mutants. For the first competition, students are provided with two “mystery” rsmE mutants that were previously isolated and characterized (18), and the instructor will know which strain is red and which is blue, but the students are blinded to this information. It is the students’ task to determine whether their mutants should be classified as either a red or blue mutant through a surface-movement assay. Students then confirm their prediction by carrying out a competition between their mystery strains. We use this first round of competitions to allow students to develop proficiency in the key concepts, tools, and techniques needed to successfully complete their independent projects in the second-round competitions as detailed in Part 3.
First, students inoculate the mystery rsmE mutants on a polycarbonate membrane to assess their ability to produce secretions. We use a specific Whatman filter paper as described in Kim et al. (18), which has a dull side and a shiny side. On the dull side, the cells get trapped within microscopic surface irregularities but continue to produce the secretions. As the secretions build up, a visible ring is formed around the colony. On the smooth shiny side, produced secretions reduce surface tension as expected by a surfactant, allowing the dividing cells to spread out (hence the term, “surface-movement” assay; Figure 2). Visualization of the ring on the dull side is time-sensitive and the ring eventually spreads out entirely, which could lead to misinterpretation. Therefore, we focus only on the shiny side for the surface-movement assay, where surface-spreading of the inoculated cells serves as a direct and quantifiable proxy for surfactant production. The results from the surface-movement assay provide the students with the basis to hypothesize whether their mutants should be categorized as red (spreading) or blue (little to no spreading). Students then test their hypothesis by competing their mystery mutants, where the red mutant (which produces more secretions) is expected to outcompete the blue mutant as described above.
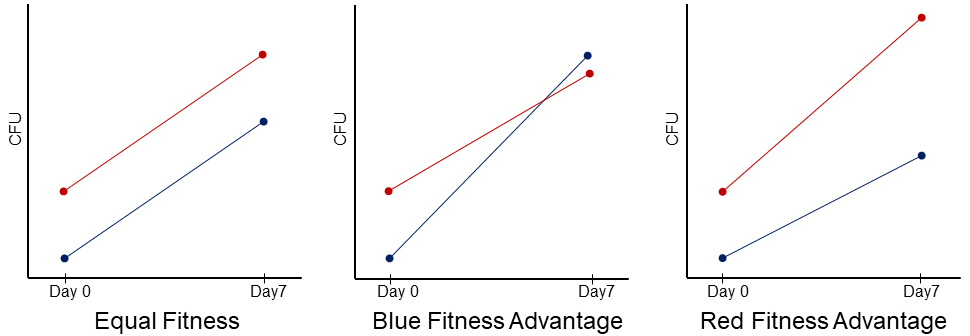
To compete the strains, students mix two different cell types in liquid and spot the mixtures on agar plates, then enumerate the number of each cell type at the beginning and end of the competition. To differentiate the competing cell types, each mystery mutant is individually labeled with distinct antibiotic resistance markers (either kanamycin or streptomycin resistance genes). The students then mix the kanamycin resistant and streptomycin resistant strains in each competition and later tell them apart by utilizing the respective antibiotic supplemented plates. For each competition mixture, we have students initially underrepresent the kanamycin resistant strain because this has been shown to best capture the competition dynamics (18). Regardless of the initial relative frequencies of the competing strains, the slope of the growth curves remains the same if the two strains are equally fit (Figure 3). The kanamycin resistant strain exhibits a shallower slope if it is less fit than the streptomycin resistant strain, and a steeper slope if it is more fit (Figure 3). Students enumerate the population size of each competing strain on Day 0 (seed) and Day 7. The seed data represents the population sizes in each liquid mixture spot at the start of the competition. On Day 7, students physically scrape off the resulting colonies into liquid, perform serial dilutions, and then plate out the dilutions on both kanamycin and streptomycin plates. Students then count the number of colony forming units (CFUs) in the following lab session to calculate the final population size of each strain within the competitions and carry out statistical tests to determine the outcomes.
Since the instructors can anticipate the outcome of the mystery strain competitions, they can evaluate whether or not the students obtained the expected results where the red mutants outcompete the blue mutants. Contradictions to these results could indicate poor pipetting skills, inaccurate serial dilution technique, possible contaminants, or incorrect data analysis. These potential issues are addressed for each lab group to ensure that students refine their skills for their research to progress. We expect the red mutants to outcompete the blue mutants due to the fitness advantage gained from surfactant production. In red strains, the mutations within rsmE appear to function as knockouts (i.e., non-functional), while blue strains generally appear to possess knockdown (i.e., semi-functional) mutations. Since blue strains manifest a spectrum of phenotypic severity depending on the location of the mutation within the coding sequence, this provides a unique opportunity to study which regions of RsmE likely have the greatest functional contribution. Students generate novel blue mutants in Part 2 and then carry out blue versus blue competitions in Part 3 to determine if there is a range of fitness among the isolated blue strains. This data will rank the relative fitness of semi-functional RsmE mutants and determine which regions of RsmE likely have the greatest functional contribution.
Part 2: Natural Selection as a Mechanism of Evolution
Students begin with a liquid culture of WT Pseudomonas fluorescens and observe its growth on solid media. Since mutations randomly occur due to errors in DNA replication and the number of cells in a bacterial colony surpasses 109 after several days of growth, it is likely that every single gene is mutated at the population level (18). A deleterious mutation would cause death or reduced replication, so such mutation would be never or rarely observed. In contrast, a beneficial mutation (e.g., that allow cells to better access/utilize resources) would result in more rapid replication, so such mutation would be more readily observed. Indeed, after several days of growth, students readily observe the emergence of many mucoid patches in the colony (Figure 1). A resource that is not frequently considered among students is the physical space that bacterial cells need to replicate. When physical space is constrained, an organism may not be able divide or it may grow very slowly. In this experiment, the emerging mucoid patches are produced by random mutations in the rsmE gene, resulting in the production of extracellular polymers and surfactants that physically push away the non-secreting WT cells. Since the rsmE mutants can sequester space, they can grow unimpeded and exploit the newly available niche, ultimately resulting in the mucoid patches the students observe. In all instances that we have tested, we find that the mucoid variants have acquired the ability to produce a variety of extracellular secretions as determined by the mucoid colony morphology and the surface-movement assay. Genome sequencing of all tested mucoid variants exclusively identifies a mutation in rsmE. Knowing this, the students isolate mutants from their WT colonies and set up the surface-movement assay to qualitatively assess the secretions their mutants produce. Students observe from the surface-movement assay that there is a spectrum of secretion among the mutants. We discuss how this wide range in phenotypes can serve as an indicator that different molecular changes have occurred within these cells as a product of natural selection and that we can quantify these changes through competition. Students identify their blue mutants and then grow liquid cultures to carry out genomic DNA extractions. Extracted DNA is then submitted to the Microbial Genome Sequencing Center (MiGS) in Pittsburgh for WGS (whole genome sequencing), and the respective data are evaluated in Part 4.
To quantify fitness differences of their blue mutants, students first label their isolated strains with either a kanamycin or streptomycin resistance gene through conjugations using a Tn7 transposon system. This technique allows the students to grow two mutant strains together in a competition and subsequently enumerate the cells using the respective antibiotics as they already learned in Part 1. Students then confirm that their strains were successfully labeled by growing them in the presence of the respective antibiotics and also by antibiotic resistance gene-specific PCR to identify correct gene insertion. After the successful completion of bacterial conjugations, students begin to set up the second round of competitions.
Part 3: Functional Consequences of Mutation (Competitions Round 2)
Students discovered in the Part 1 red versus blue competitions that the red strains significantly outcompete the blue strains. This observation is due to the production of both the mucus and surfactant in the red strain compared to the blue strain that only produces the mucus. This dichotomy oversimplifies the categories of mutants, since we expect the native RsmE to repress additional non-visible, as-yet-unidentified secretions. This is also reflected in the diversity observed in the surface movement assay of blue strains in Part 2. The key hypothesis here is that the blue mutants with unique RsmE mutations and competitive abilities de-repress a spectrum of these non-visible secretions, ultimately resulting in variable fitness. To test this hypothesis, students carry out blue versus blue competitions using their evolved strains from Part 2. RsmE mutants that students identify to be more or less fit than others will serve as key protein models to characterize differential mRNA binding and currently unknown secretions in the near future. This information, coupled with the mutation data, will elucidate the regions of RsmE that are functionally important and will determine which residues are essential in the regulation of select mRNA products – information that is novel to the students, instructors, and the greater scientific community.
For the blue versus blue competition, each student group selects what they believe to be their best blue competitor based on the surface-movement assay and begins a round-robin tournament with their classmates. They repeat the same competition steps from Part 1 with the new strains to generate CFU counts on Days 0 and 7. This reiterates the skills obtained in Part 1 which is an essential component of a CURE and greatly increases new skill retention. Since each student group is using the same set of unique blue mutant strains with reversed antibiotic resistance markers, students can compare their own competition data with those generated by other groups to assess both the quality and noise at the class level to ultimately identify and exclude potential outliers for efficient data interpretation. Students run statistical analyses of their competition data to determine the relative fitness (w) of each strain and to determine if their calculated fitness differences are statistically significant through a one-way ANOVA followed by a Tukey’s HSD test. Both the students and the instructors are blinded to the mutations that these strains carry since whole genome sequencing (WGS) takes several weeks to be completed. In Part 4, students evaluate the WGS data of their blue mutants to bioinformatically determine why one blue strain may have outcompeted another blue strain.
Part 4: Evaluating the Molecular Basis of Evolution and its Consequences
Until this point, we have generalized all red mutations to be functionally identical to an rsmE knockout mutation – assuming all red mutations to render RsmE to be completely functionless. One reason for making this assumption is that all red mutants studied to date equally produce both the mucoid polymer and surfactant. However, given that we expect to see functional variation across the blue mutation spectrum, we may also observe such variations among the red mutations. This hypothesis could be directly tested by conducting a red versus red tournament, but this requires two additional weeks in which students would essentially repeat the same techniques as in Parts 1 and 3. Instead, we instructed students to engage in a discussion-based in silico experiment (Supporting File S3. Bacterial Evolution – Red versus Red Dataset) where they critically evaluate how different red mutations may impact RsmE function. As part of this thought experiment, they review a provided dataset of a small-scale red versus red competition where the strains exhibit comparable relative fitness. The final takeaway by the students is that while there may be subtle variations in the relative fitness of red mutants, we expect them to mostly act as a knockout mutant since the majority of red mutations are either non-conservative missense or nonsense mutations. This exercise also primes students with essential concepts to successfully analyze their WGS data.
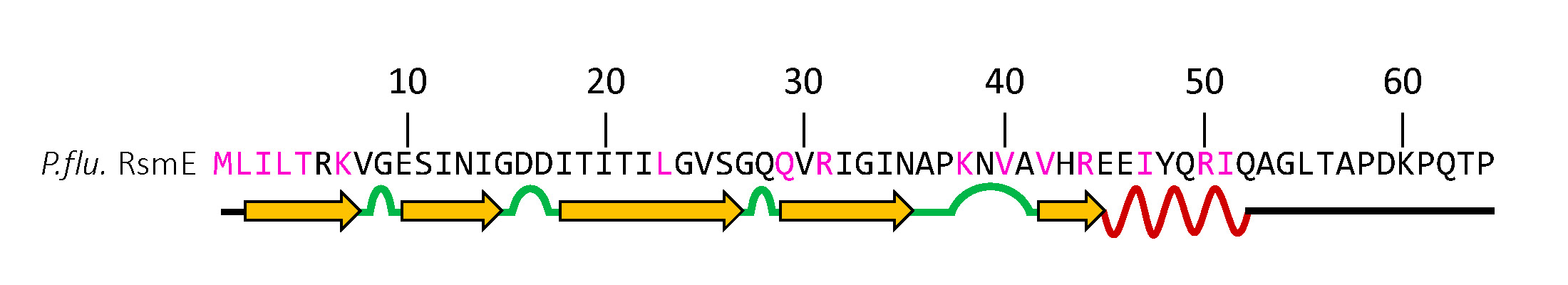
Students evaluate their WGS data for the blue mutants used in the competitions under Part 3. Detailed instructions for the instructor are provided (Supporting File S1. Bacterial Evolution – Course Manual with Teaching Guides, Student Protocols, and Assessments). After receiving the paired-read data from the sequencing center, we generated a FASTA file for each student using the nucleotide sequence. Students then used BLAST and ClustalW to compare this file to the WT rsmE to identify which nucleotide(s) are changed and the translational consequences of these mutations. A schematic of the currently predicted functional regions of RsmE (Figure 4) is provided in the protocol as a guide to determine if their mutations occur in an RNA binding site, a dimerization domain, or in other regions of unresolved function. Using their own mutation data, students actively learn how and why mutations naturally occur and the phenotypic effect that a mutation may cause. Novel mutants identified by the students are flagged for further study in the Kim lab to identify previously unknown mRNA and secretions regulated by RsmE.
Part 5: Experimental Conclusions and Assessment
The course concludes with the preparation and assessment of individually written reports and group oral presentations that integrate research findings throughout Parts 2 to 4. Each student group actively carries out their own discussions during class to converge on a unified story board and conclusions for the oral presentation. They reflect on why the rsmE gene is exclusively targeted by natural selection under the experimental conditions and formulate a clear hypothesis. They present their competition data and discuss how they utilized the class-wide data to ultimately justify their relative fitness ranking. Students also discuss their mutations found from genome sequencing, how they may change the secondary and/or tertiary structure, and their molecular consequences by reflecting on RsmE’s native function.
Teaching Discussion
Observations in Achieving Learning Goals and Objectives
The described course has been successfully implemented in the Spring semesters of 2019, 2020, and 2021, even in the midst of the global COVID-19 pandemic. Coursework for 2019 and 2020 was completed within eight weeks due the modular nature of the course. Students completed many of the lab modules in parallel, and we concluded the Spring 2020 session prior to COVID lockdown procedures. The course presented here was lengthened to 16 weeks in 2021 to account for changes in social distancing and room capacity guidelines. Across the three iterations, all classes successfully completed their experiments, showcasing the predictability of this experimental evolution system – though as might be expected, individual groups occasionally made missteps on some of the experimental protocols. Rather than detracting from the experience, this course provided students with a genuine glimpse into the technical noise of real data and the iterative nature of science, as students were encouraged to repeat and refine experiments as needed. In general, we observed that students were highly engaged in the material, likely due to an invested interest in the mutant strains they directly evolved and isolated, consistent with the sense of “emotional ownership” developed when students gather their own data (16). During an informal class discussion at the beginning of Part 3, students voiced that they could see the whole picture and truly understand the real-world applications of the research they were conducting. This observation was also reflected in improved responses to quiz questions focused on the underlying evolutionary principles. Upon completion of the course and throughout the student presentations, we observed that most of the students had mastered both the technical lab content and the more philosophical perspectives of evolution. By the end of the semester, student responses to open-ended assessment questions no longer reflected serious misconceptions about natural selection, which were common at the start of the course.
Dispelling such misconceptions is integral to achieving the stated learning objectives of the course, given our heavy emphasis on building understanding of the fundamental principles of natural selection. Common misconceptions among our students at the start of the course – and indeed, among any general population of non-experts – included the assumptions that populations start out with very little variation and that specific, purposely beneficial new mutations are driven by the needs of the organisms (27). In our course, this was best demonstrated by the incorrect idea that a crowded environment would directly stimulate – rather than simply select for – mutations in rsmE. We challenged such misconceptions through a multi-pronged, iterative approach including direct instruction on the mechanism of random mutation as well as guided observation and analysis of data from our experiments showing the existence of variation within starting populations and the events of natural selection in action. Students are also able to observe that when mutations occur randomly (such as the spontaneous antibiotic resistance we aim to eliminate in our conjugations), they can have both beneficial and deleterious effects for the organism. Furthermore, we frequently asked students probing questions that drove them to use their own data to confront and supplant their existing misconceptions regarding the mechanism of evolution, including through small group discussions, full-class “clicker questions,” and individual quiz questions. By using their own observations and collected data, students were able to self-generate convincing evidence that argued against previous misunderstandings; we hypothesize that the higher-order cognitive tasks involved in this process will be more effective at maintaining this understanding long-term as compared to lecture-based strategies that simply “tell” students that their misconceptions are wrong.
We also suspect that the students who participated in this course have gained a deeper understanding of experimental design and hypothesis testing. Throughout this procedure, students carried out statistical analyses of their captured data, practiced their acquired molecular biology skills, and used bioinformatics to assess their compiled data to evaluate the molecular implications of their research. This course highlights the new era of multidisciplinary molecular biology and provides a foundation for the collaborative research efforts that all new graduates will require to succeed. Importantly, this course provides the instructor with the flexibility to incorporate select modules into existing courses or to carry out a module as a standalone lesson. Overall, we found that students had an increased appreciation for the society learning goals, such as i) the significance, mechanisms, and molecular basis of evolution, ii) the importance of natural selection as the key evolutionary mechanism, and iii) how evolution that occurs through mutation in a nucleotide sequence of a gene can alter biological functions.
Instructor Reactions to the Lesson and Possible Adaptations
We have found that the greatest limitation to this course is time. Students commonly make experimental mistakes, but this course requires them to be immediately corrected in order to progress through the downstream lab exercises. To adjust for this limitation, affected students were required to come into the lab between the regularly scheduled lab periods. Our teaching assistants were also required to work behind the scene and assist students between labs to ensure that research progressed seamlessly. In particular, we observed during the surface-movement assay lab that a few students had accidentally re-isolated WT cells rather than mutants.
To avoid having to repeat the entire evolution experiment, which would otherwise require an additional week of time, we had each student group isolate multiple mutant strains and freeze them down. We also had the teaching assistants set up extra evolving colonies in parallel so that students had immediate access to new samples when necessary.
This course could be readily modified to be offered over a shorter duration if necessary. Solving the identity of the mystery mutants in Part 1 could be an independent module on its own or be removed entirely. For Parts 2-4, students could also evolve and phenotypically characterize their own rsmE mutants without genome sequencing or independently labeling new mutants with antibiotic resistance genes. In this case, the instructor could conceptually bridge the gap without physically working with newly evolved strains by providing previously isolated and labeled rsmE mutants for students to carry out the competitions and conduct bioinformatics analysis. The conceptual red versus red competition experiment described in Part 4 could also be physically carried out as an additional or independent module using the same protocols described for the blue versus blue competitions. However, we believe that the course format as currently laid out is optimal for providing a genuine research experience to teach students about evolution and its molecular consequences on a scale that students can directly observe and analyze. Directly evolving and characterizing their own mutant strains through a series of interdisciplinary experiments provides a powerful learning medium. Indeed, when we carry out Part 3: Functional Consequences of Mutation (Competitions Round 2), students are excited to compete their own blue strain against their peers’ blue strains to determine the most fit competitor. We believe that our modular lesson design, simple experimental platform, and relatively low-cost materials requirement collectively permit this course to be readily adopted by other institutions to provide a unique and hypothesis-driven research experience. We are also committed to working with other instructors to help them acquire the supplies and strains needed to conduct this course.
SUPPORTING MATERIALS
- Supporting File S1. Bacterial Evolution – Course Manual with Teaching Guides, Student Protocols, and Assessments
- Supporting File S2. Bacterial Evolution – Prelab Lecture Teaching Slides
- Supporting File S3. Bacterial Evolution – Red versus Red Dataset
Acknowledgments
We thank the students who participated in this course and provided constructive feedback for making improvements. This work was supported by the Department of Biological Sciences at Duquesne University and the National Institute of General Medical Sciences of the NIH 1R15GM132856 (W.K.).
References
- Evolution in Pennsylvania. National Center for Science Evolution. 2013, April 13. Retrieved June 5, 2020, from https://ncse.ngo/evolution-pennsylvania.
- Nehm RH, Reilly L. 2007. Biology majors’ knowledge and misconceptions of natural selection. BioScience. 57(3):263-272. doi: 10.1641/B570311.
- Demastes SS, Settlage J, Good R. 1995. Students’ conceptions of natural selection and its role in evolution: Cases of replication and comparison. Journal of Research in Science Teaching. 32(5):535-550. doi: 10.1002/tea.3660320509.
- Nelson CE. 2008. Teaching evolution (and all of biology) more effectively: Strategies for engagement, critical reasoning, and confronting misconceptions. Integrative and Comparative Biology. 48(2):213-225. doi: 10.1093/icb/icn027.
- Robbins JR, Roy P. 2007. The natural selection: Identifying & correcting non-science student preconceptions through an inquiry-based, critical approach to evolution. American Biology Teacher. 69(8):460-466. doi: 10.2307/4452205
- Abraham JK, Meir E, Perry J, Herron JC, Maruca S, Stal D. 2009. Addressing undergraduate student misconceptions about natural selection with an interactive simulated laboratory. Evolution: Education and Outreach. 2:393-404. doi: 10.1007/s12052-009-0142-3
- American Association for the Advancement of Science. 2011. Vision and change in undergraduate biology education: A call to action. Washington, D.C.
- Wei CA, Woodin T. 2011. Undergraduate research experiences in biology: Alternatives to the apprenticeship model. CBE Life Sciences Education. 10(2):123-131. doi: 10.1187/cbe.11-03-0028.
- Auchincloss LC, Laursen SL, Branchaw JL, Eagan K, Graham M, Hanauer DI, Lawrie G, McLinn CM, Pelaez N, Rowland S, Towns M, Trautmann NM, Varma-Nelson P, Weston TJ, Dolan EL. 2014. Assessment of course-based undergraduate research experiences: A meeting report. CBE Life Sciences Education. 13(1):29-40. doi: 10.1187/cbe.14-01-0004.
- Hanauer DI, Frederick J, Fotinakes B, Strobel SA. 2012. Linguistic analysis of project ownership for undergraduate research experiences. CBE Life Sciences Education. 11(4):378-385. doi: 10.1187/cbe.12-04-0043.
- Brownell SE, Hekmat-Scafe DS, Singla V, Chandler Seawell P, Conklin Imam JF, Eddy SL, Stearns T, Cyert MS. 2015. A high-enrollment course-based undergraduate research experience improves student conceptions of scientific thinking and ability to interpret data. CBE Life Sciences Education. 14(2):1-14. doi: 10.1187/cbe.14-05-0092.
- Martin A, Rechs A, Landerholm T, McDonald K. 2021. Course-based undergraduate research experiences spanning two semesters of biology impact student self-efficacy but not future goals. Journal of College Science Teaching. 50(4):33-47.
- Bangera G, Brownell SE. 2014. Course-based undergraduate research experiences can make scientific research more inclusive. CBE Life Sciences Education. 13(4):602-606. doi: 10.1187/cbe.14-06-0099.
- Estrada M, Burnett M, Campbell AG, Campbell PB, Denetclaw WF, Gutiérrez CG, Hurtado S, John GH, Matsui J, McGee R, Okpodu CM, Joan Robinson T, Summers MF, Werner-Washburne M, Zavala ME. 2016. Improving underrepresented minority student persistence in stem. CBE Life Sciences Education. 15(3):1-10. doi: 10.1187/cbe.16-01-0038.
- Rodenbusch SE, Hernandez PR, Simmons SL, Dolan EL. 2016. Early engagement in course-based research increases graduation rates and completion of science, engineering, and mathematics degrees. CBE Life Sciences Education. 15(2):1-10. doi: 10.1187/cbe.16-03-0117.
- Cooper KM, Knope ML, Munstermann MJ, Brownell SE. 2020. Students who analyze their own data in a course-based undergraduate research experience (CURE) show gains in scientific identity and emotional ownership of research. Journal of Microbiology & Biology Education. 21(3):1-11. doi: 10.1128/jmbe.v21i3.2157.
- Cooper VS, Staples RK, Traverse CC, Ellis CN. 2014. Parallel evolution of small colony variants in Burkholderia cenocepacia biofilms. Genomics. 104(6): 447-452. doi: 10.1016/j.ygeno.2014.09.007.
- Kim W, Racimo F, Schluter J, Levy SB, Foster KR. 2014. Importance of positioning for microbial evolution. Proceedings of the National Academy of Sciences. 111(16):E1639–E1647. doi: 10.1073/pnas.1323632111.
- Lenski RE. 2017. Experimental evolution and the dynamics of adaptation and genome evolution in microbial populations. ISME Journal. 11:2181-2194. doi: 10.1038/ismej.2017.69.
- Round JE, Campbell AM. 2013. Figure facts: Encouraging undergraduates to take a data-centered approach to reading primary literature. CBE Life Sciences Education. 12(1):39-46. doi: 10.1187/cbe.11-07-0057.
- Chism NVN, Angelo TA, Cross KP. 1995. Classroom assessment techniques: A handbook for college teachers. The Journal of Higher Education. 66(1):108-111. doi: 10.2307/2943957.
- Orr AC, Hammig SB. 2009. Inclusive postsecondary strategies for teaching students with learning disabilities: A review of the literature. Learning Disability Quarterly. 32(3):181-196. doi: 10.2307/27740367.
- Tanner KD. 2013. Structure matters: Twenty-one teaching strategies to promote student engagement and cultivate classroom equity. CBE Life Sciences Education. 12(3):322-331. doi: 10.1187/cbe.13-06-0115.
- Meer NM, Chapman A. 2014. Assessment for confidence: Exploring the impact that low-stakes assessment design has on student retention. International Journal of Management Education. 12(2):186-192. doi: 10.1016/j.ijme.2014.01.003.
- Winkelmes M-A, Bernacki M, Butler J, Zochowski M, Golanics J, Weavil K. 2016. A teaching intervention that increases underserved college students’ success. Peer Review. 18.
- Mercante J, Suzuki K, Cheng X, Babitzke P, Romeo T. 2006. Comprehensive alanine-scanning mutagenesis of Escherichia coli CsrA defines two subdomains of critical functional importance. Journal of Biological Chemistry. 281(42):31832-31842. doi: 10.1074/jbc.M606057200.
- Gregory, T. 2009. Understanding natural selection: Essential concepts and common misconceptions. Evolution: Education and Outreach. 2:156-175. doi:10.1007/s12052-009-0128-1.
Article Files
Login to access supporting documents
Kessler-Application of a bacterial experimental evolution system.pdf(PDF | 412 KB)
S1. Bacterial Evolution-Course Manual with Teaching Guides Student Protocols and Assessments.docx(DOCX | 20 MB)
S2. Bacterial Evolution-Prelab Lectures Teaching Slides.pptx(PPTX | 23 MB)
S3. Bacterial Evolution-Red vs Red Dataset.xlsx(XLSX | 11 KB)
- License terms

Comments
Comments
There are no comments on this resource.