You and Your Oral Microflora: Introducing non-biology majors to their “forgotten organ”
Published online:
Abstract
With limited time available for laboratory activities, introductory science courses for non-science majors typically use the laboratory period to reinforce material that was previously presented during lecture. This practice was true of a human biology course at the University of Minnesota, in which most lab activities centered on organ dissections. To break from this mold, we designed a new laboratory module to introduce students to their “forgotten organ”, the human microbiome. We chose this topic partly due to the explosion of recent research in this area, as well as the opportunity to expose students to online tools and techniques. Student motivation arose from the opportunity to determine the types of microbes that might be growing inside their mouth, and piqued their interest in how microbes could positively affect their health. Through a series of activities, a group of students from a large-enrollment course sampled and analyzed a subset of their oral microflora. Using the NCBI BLAST tool, they were able to tentatively identify microbial sequences amplified using primers specific for 16S rRNA. The activities greatly expanded on topics covered only briefly in lecture and provided hands-on experience with scientific techniques. This module could be adapted to fit into a number of different formats. It has been used in an introductory biology course as a multi-week activity, and as a two-day activity with high school biology teachers. It could also be modified to be an in silico activity, with instructors guiding students to public databases to obtain sequence data.
Citation
Strain, A.K. and Vang, K.B. 2014. You and Your Oral Microflora: Introducing non-biology majors to their “forgotten organ”. CourseSource. https://doi.org/10.24918/cs.2014.13Society Learning Goals
Genetics
- Nature of Genetic Material
- What are the molecular components and mechanisms necessary to preserve and duplicate an organism’s genome?
- Molecular biology of gene function
- How is genetic information expressed so it affects an organism's structure and function?
- Methods & Tools in Genetics
- What experimental methods are commonly used to analyze gene structure and gene expression?
Microbiology
- Systems
- How do microorganisms, cellular and viral, interact with both human and non-human hosts in beneficial, neutral, or detrimental ways?
- Impact of Microorganisms
- Why have the effects and potential benefits of microbial life not been fully explored?
- How can humans utilize and harness microbes and their products?
- How do microorganisms provide essential models that give us fundamental knowledge about life processes?
Science Process Skills
- Process of Science
- Pose testable questions and hypotheses to address gaps in knowledge
- Plan, evaluate, and implement scientific investigations
- Interpret, evaluate, and draw conclusions from data
- Construct explanations and make evidence-based arguments about the natural world
- Address novel questions through authentic research experiences
- Modeling/ Developing and Using Models
- Recognize the important roles that scientific models, of many different types (conceptual, mathematical, physical, etc.), play in predicting and communicating biological phenomena
- Make inferences and solve problems using models and simulations
- Build and evaluate models of biological systems
- Quantitative Reasoning/ Using Mathematics and Computational Thinking
- Apply the tools of graphing, statistics, and data science to analyze biological data
- Communication and Collaboration
- Share ideas, data, and findings with others clearly and accurately
Lesson Learning Goals
Students will:- Explore the role of the microbes in the human body, and apply this knowledge in terms of both health and disease
- Apply their understanding of biological concepts to molecular laboratory technology
- Demonstrate the ability to locate and evaluate information
Lesson Learning Objectives
Students will be able to:- Explain both beneficial and detrimental roles of microbes in human health.
- Compare and contrast DNA replication as it occurs inside a cell versus in a test tube
- Identify an unknown sequence of DNA by performing a BLAST search
- Navigate sources of scientific information to assess the accuracy of their experimental techniques
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
INTRODUCTION
The University of Minnesota offers a number of introductory biology courses in a variety of “flavors” for non-life science majors. There is a “sex” course, an “environmental” course, as well as a “human interest” course, which focuses on biology as it relates to the human body. This course, Human Biology (BIOL 1010), has traditionally been focused on human anatomy and physiology, with a laboratory emphasis on organ dissections. In keeping with the objectives of the Vision and Change report (1), we wanted to introduce more student-centered, inquiry-based learning into the laboratory activities offered to our non-majors students. We chose to focus on one of the Vision and Change goals, to provide students experience interpreting data and using large databases. Part of our reasoning behind this focus is the burgeoning amount of genome sequence data that is available to the public, allowing analysis of hundreds of thousands of organisms. Given the implications that genome sequencing holds for the future of health and medicine, our goal was to introduce students to this new wealth of information and provide them with tools for utilizing it. As HHMI-supported postdoctoral teaching fellows, we were given this challenge in the spring of 2012 and asked to develop an activity that met the following criteria: 1) it could be run as a pilot module that fall; and 2) it could be run at the same time as the other activities already being done in the lab.
The first question we had to address was how to encourage non-majors primarily interested in the human body as a whole to look instead at the genetic blueprint for building the body. Around this same time, a number of papers were published on the human gut microbiome. These papers suggested that different human populations could be characterized by the bacteria that reside in their intestinal tract (2,3). Further studies suggested that these microbial populations could potentially be altered based on factors such as diet, genetics and/or disease state (4). Other areas of the human body were also shown to have distinct microbiomes (5). We thought this might be a good “hook” for our students, in part because the research suggested these microbes played a very important role in human health. In addition, most students we spoke with seemed to be intrigued by the idea of “poop sampling”. In the end, we decided that it would be too difficult to get students to do the actual poop sampling (this was before companies started offering “do-it-yourself” sampling kits). We decided instead to focus on the microbiome of the oral cavity, as it is much easier for students to sample. This focus had the added benefit of being potentially more inquiry-based, as fewer papers had been published at the time describing the sequences of the oral microbiome.
A few inquiry-based projects using genomic sequencing technology have been reported in the literature, although many of these have focused on introducing biology or other STEM majors to sequencing and bioinformatics. For example, Banta et al. report on a number of modules developed as part of the Genomics Collaboration to introduce undergraduate science majors to sequencing technology (6). In one of the modules, upper-level biology majors amplified and analyzed sequences of a polymorphic olfactory receptor gene in canines (6). Other modules in the Genomics Collaboration include metagenomic analysis of Winogradsky columns, and using BLAST searches to construct phylogenetic trees (see http://serc.carleton.edu/genomics/activities.html for examples). Similar activities for biology majors have been previously described by Boomer et al. (7), who developed a ten-week module to analyze the microbial diversity present in Yellowstone National Park. Fewer examples have been published that focus on introducing non-biology majors to genomic research. One program that has had some success with non-STEM majors is the SEA-PHAGE course described by Caruso et al. (8). Students enrolled in the SEA-PHAGE program learned to “hunt” for bacteriophage in the environment. They collected and characterized soil phages, ultimately isolating and sequencing the viral genomes. This project has been used with both majors and non-majors, and was recently described in an article that lists many of the undergraduates as co-authors (9). Closer to home, a non-majors introductory biology course at the University of MN was recently modified to utilize a genomic database of the microbes present in the Mississippi River in the laboratory portion of the course (the M3P project, Minnesota Mississippi Metagenomics Project) (10). This course has been successfully implemented over several semesters, introducing students to functional and sequence-based metagenomics (Brian Gibbens, personal correspondence and unpublished results). We wanted to continue expanding the introduction of bioinformatics tools to non-majors, and chose to focus our approach on microbes found in the oral cavity of students.
The second question we had to address was how to fit this activity into the lab structure that was already in place. This challenge was partly because only a small percentage of the class of ~200 would be participating in the module. These 48 students still had to complete all the other required lab activities that their classmates were doing in their two-hour lab periods. Choosing to focus on the oral microbiome allowed us to incorporate many of the initial experimental steps into activities that were already being performed in the lab (e.g. microscopy, loading a gel). For the later steps, it provided additional hands-on experience with scientific methods that students had only read about previously (PCR, DNA sequences).
Our ultimate goal was to expose students to the wealth of information available to them within genomic sequencing databases, and to have students ask questions that interested them about this data. With the number of DNA sequences available for analysis rising exponentially and the increasing interest in applying this knowledge to the treatment of human disease, it is clear that even students outside of science majors should have an understanding of the types of information available to them and how these sequences could be analyzed. The activity we describe here was designed to address all of these criteria, and has been presented to a number of different audiences. It can be expanded or pared down as needed, but is a good starting point for introducing students to the online resources available to them (especially genomic and research databases).
Intended Audience
This module is intended for an introductory biology course. It is primarily targeted at non-life science majors, typically first or second year students.
Learning Time
This activity is written as a seven-week activity with appropriate breaks in between. However, the number of days could be changed to meet course needs. The specific length of time needed for each activity is provided in Table 1.
Prerequisite Student Knowledge
The module does not have any specific prerequisite knowledge, as we have attempted to provide the necessary information throughout the activity. However, the activity fits best in a course in which students have been introduced to the following biological concepts: central dogma of molecular biology (DNA, RNA, protein); eukaryotic and prokaryotic cells; cell structure; DNA replication. Although microbiology is not typically covered extensively in introductory courses, microbial cells are typically mentioned in comparison to eukaryotic cells. In addition, this activity could be used as an opportunity to use microbial examples for introducing taxonomy, a topic that is typically covered in introductory biology course. Additional guidance may be needed to analyze sequence data, depending on your specific course goals, but students can also perform the necessary BLAST searches with the instructions provided in the activity. We have included a short introduction to 16S rDNA in the laboratory handout for students (Supplemental File S1). However, this is not meant to be an exhaustive primer. For more information on this subject, we recommend additional background reading on 16S rRNA from a book such as “Introduction to Genomics” by Arthur M. Lesk (11), or a minireview on using 16S rRNA by Janda and Abbott (12).
Teaching Assistant Training
All of the introductory biology courses at the University of MN are large-enrollment courses, with over 200 students in each lecture section. These students are divided into at least twelve lab sections with 20 students each. We typically rely on teaching assistants (TAs) to take the lead on instruction for the laboratory-based aspects of the course. The TAs for our course were either upper level science majors at the top of their class in ranking, or graduate students hired as teaching assistants. In either case, the TAs must have successfully completed an anatomy and physiology course and show competence in anatomy and physiology. In addition, our TAs typically have at least one semester of research experience.
For this module, although the authors of the activity (AKS, KBV) were primarily responsible for presenting the biological concepts, it was done in conjunction with the TAs who took the lead in running the laboratory sections. The TAs in the laboratory sections undergo a weeklong training session with the course instructor and lab manager prior to the start of the semester. During the semester, they attend weekly training meetings lead by an experienced head TA, who was also one of the TAs involved in teaching the Oral Microflora module. These weekly training meetings include a discussion of upcoming activities and any potential problems that might arise. The authors of this activity worked with the two TAs leading the Oral Microflora sections at least one week in advance of the activity being presented to students to train them on the experimental techniques and answer any questions or safety concerns for the TAs. This preparation provided the TAs with a level of comfort with the activity but did not necessarily make them experts in all aspects of microbiology. Senior lab staff was always available during laboratory sections to lend assistance for troubleshooting technical problems that TAs were unable to handle themselves (usually involving focusing on bacteria).
A Word on Safety
This activity requires students to work with bacteria cultured from inside the surface of their mouths. The safety policies of the University of MN allow students to handle bacteria isolated from their own bodies as long as they observe appropriate safety precautions. This assumes that the bacteria isolated from their body surfaces are part of their own personal microflora and should not pose a threat to them. The precautions required include wearing personal protective equipment such as disposable gloves, goggles, lab coats, long pants and closed-toed shoes whenever they are working with cultured bacteria. If biological safety is a concern, this activity could be modified by skipping the culturing steps and providing students with sequences to analyze from the database. We would still recommend doing a cheek swab on day one and staining the slide with methylene blue to compare the bacterial cells to cheek epithelial cells. We did not observe any safety issues during the running of this module in the fall of 2012, or again when the activity was presented in the summer of 2013 to secondary-school science teachers.
SCIENTIFIC TEACHING THEMES
Active Learning
Students were actively engaged by hands-on activities that included: swabbing their mouth, growing bacterial colonies on LB plates and isolating and assessing the fidelity of their microbial DNA samples. In addition, students analyzed their Sanger sequencing results using BLAST and reported their bacterial strain(s). The results of their inquiry based research projects were completely unknown.
Assessment
Our primary teaching assessment tool was the BLAST search results exercise (see Supplemental File S1 and Supplemental File S4). Through a series of questions, we asked students to determine whether their BLAST search yielded a reasonable sequence result. Student learning was also assessed through questions asked on the mid-term lab practical exam (see Supplemental File S6). More importantly, students self-evaluated their learning at a number of stages. They assessed their learning of technical skills by determining whether they had growth on their colony plates, whether appropriate bands could be visualized in their gel after PCR, and if they had obtained DNA sequences to analyze. They were able to assess learning of computational skills through their BLAST searches and the ability to find information about the organisms they identified.
Inclusive Teaching
Students worked in pairs during the lab sessions. This pairing helped them work together to solve the more technical aspects of the experiment as outlined in the module (Supplemental File S1). We anticipated that some students would choose not to swab their own mouth and were prepared to provide them with a previously isolated DNA sample. Using this DNA sample, they could participate in the BLAST search and analysis. Had students chosen not to swab their oral cavity, they could make observations from a fellow classmate’s plates, provided they did not handle an opened plate themselves. This is in compliance with the biological safety policy at the University of MN. Students are allowed to handle cultured samples of their own body microflora because it is presumably part of their own normal flora. However, they must still wear personal protective equipment when handling cultured bacteria, including long pants, closed-toed shoes, disposable gloves, eye protection and a laboratory coat. We were pleased to note that all of our students participated and were able to identify a diversity of microbes in their mouths. In order for students to more fully appreciate this diversity, the de-identified results were pooled and shared between the two sections that participated in the pilot study. Students were able to observe the differences between the two sections and think about how this difference might impact human health and disease.
LESSON PLAN
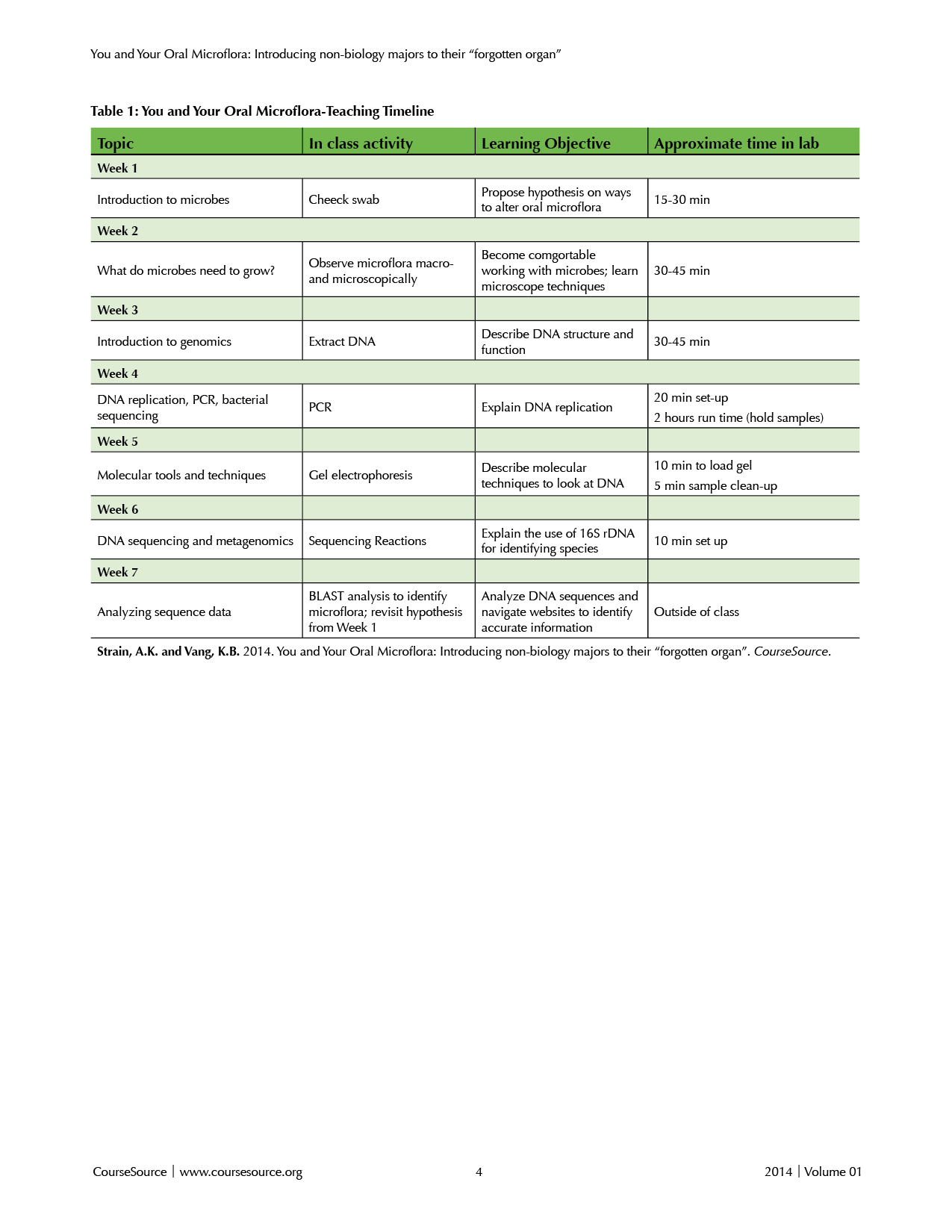
Table 1. You and Your Oral Microflora-Teaching Timeline
Laboratory Notebooks
The Oral Microflora module is a “wet-lab” activity, so preparation is needed to ensure that all the necessary materials and supplies are available to students at the appropriate times. We strongly recommend piloting the Oral Microflora module to make sure all the kinks are sorted out (Supplemental File S1). In addition to familiarizing you with the module, a pilot run will yield supplementary BLAST sequences for any students who choose to opt out of swabbing their oral cavity. It also gives you the chance to show students a small sample of what has been growing in your own mouth! The materials needed for the module can be found in Supplemental File S1, with a weekly breakdown of the major components needed provided in Supplemental File S2. Once the materials have arrived, basic lab-prep activities need to be done: prepare reagents according to the manufacturer directions, prepare LB plates, autoclave any necessary components, program the thermocycler (provided in Supplemental File S1), and contact the sequencing facility of choice (if doing this step). After the reagents have been prepared you may want to aliquot the reagents to partition them into as many laboratory sections as required. We made master mixes and reagents for two to four students to share.
Initial Preparation
In keeping with the format for other sections of this course, the students in our lab sections were not required to record their results and observations in a “certified” laboratory notebook. Because keeping track of their data is important for the success of certain aspects of this activity, we provided adequate space for them to record their observations in the laboratory handout. Although note-taking was not a graded activity, we noted that most students recorded and kept track of relevant information. We would like to stress that the purpose of this activity was not to have students perform exactly like scientists. Even though the module was technical in detail, our goal was for students to focus on the big picture of microbial diversity in the mouth, and the positive and negative effects of these microbes on health. As such, this lesson provides students the opportunity to isolate and analyze bacteria from their mouth and explore scientific databases in order to identify their isolates.
Lab Day 1: Sample collection
Teacher preparation: Although a familiarity with bacteria is useful for this activity, it is not required that you be a microbiology expert in order to complete this activity. You can familiarize yourself with some background reading on microbes in general or for the oral cavity, specifically, by looking through an introductory textbook or checking out the excellent resources online (e.g. the American Society for Microbiology provides great information at MicrobeWorld (13). We presented a short introduction to students on Day 1 of the module that could be used as a short refresher (Supplemental File S3). On this day, a tray should be prepared with all required reagents for students to use (see Supplemental File S1 and S2).
Student preparation: We will only emphasize it once, but students should be reminded that they should come to the laboratory prepared for whatever experiments they have planned for the day. Also, we expect them to complete the background reading beforehand. Questions about the material are always welcome, but students should have a working knowledge of the day’s experimental procedure.
In class summary/laboratory: We start by introducing students to microbes, some of their characteristics, the sheer number of microbes that have been identified and their potential roles in human health and disease. It is important to highlight that most microbes are beneficial. For example the “good” microbes may out-compete the “bad” microbes for colonization of surface areas such as the mouth or the gut. (Supplemental File S3 is the presentation file used for Day 1.) Students are encouraged to write their thoughts on a piece of paper in response to two questions (see Supplemental File S1).
Laboratory Activity:Students select a sampling site in their oral cavity. Students have the option of choosing a combination of multiple sites such as teeth and tongue, or can choose one site. In addition, students could choose sites in response to an activity such as before and after mouth washing. For a detailed look at the laboratory procedures see Supplemental File S1. Table 1 provides an overview of the teaching timeline.
Once a sampling strategy has been determined, students sample their oral cavity with sterile cotton swabs, streak labeled agar plates with their cotton swabs, and then incubate the plates for 2 days at 37ºC.
NOTE: During this activity, we emphasized sterile technique and that students should handle only their own samples, in compliance with standard safety protocols in place at the University of MN as noted above.
After lab 1: The instructor or TA should incubate plates for 24-48 hours at 37°C. It is helpful to check after 24 hours to determine the size of colony formation. If the colonies are small, incubate an additional 24 hours at 37°C. If the colonies are very large, they can be removed from the incubator and stored, wrapped in plastic, at 4°C until the next lab period.
Lab Day Two: Observing and describing bacteria
Teacher preparation: The most important aspect of this lab day is for students to observe the variety of different colonies that form after incubation, and to develop their microscopy skills. We emphasized the fact that the colonies on the plate can be formed only by organisms that can grow on the medium and under the conditions provided, so they only represent a small fraction of the microbes in their mouths. Students filled out a colony morphology worksheet (bacterial colony morphologies (size, shape, color, height, etc.) are described in Supplemental File S1). An example of some of the student plates from our course are shown in Figure 1A.
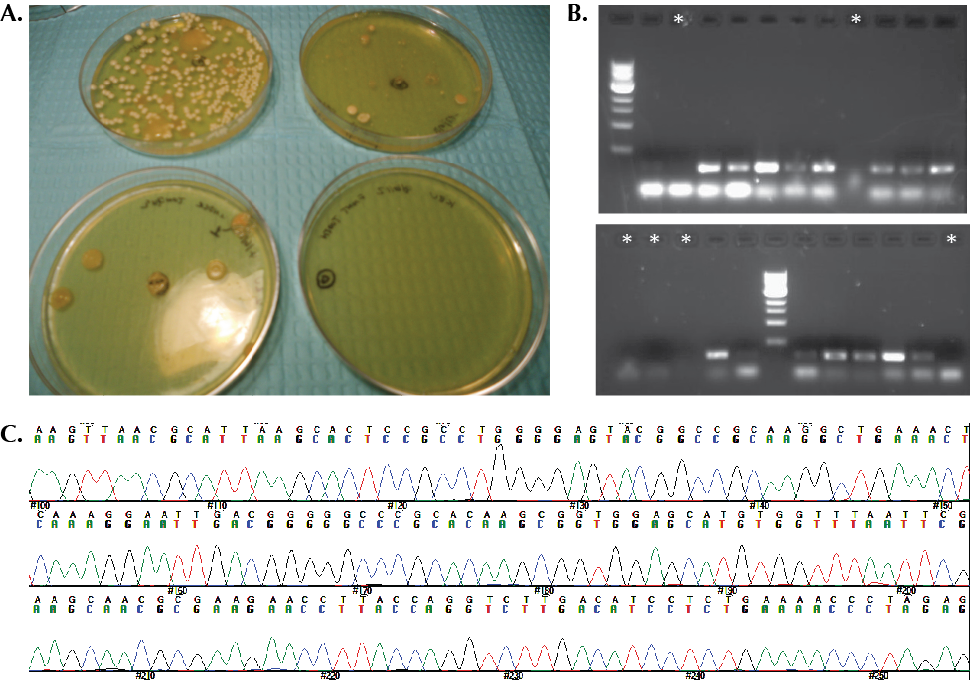
Figure 1. Sample student data. A. Oral swabs grown on LB plates. Plates shown were incubated at 37°C for 36 hours. B. Gel electrophoresis of 16S rDNA PCR reactions (~75% of student reactions worked; for those students for whom none of the reactions worked, we provided a sample for them to use in the following lab activities). Asterisks indicate lanes in which NO product was visible. C. Result of one student’s sequencing reaction, used to obtain BLAST search results.
This observation activity was done in conjunction with learning about human cells and cell structures, so students also used microscopes to look at their bacterial cells. Instructors should be familiar how to set up compound microscopes and how to prepare a wet mount slide (Supplemental File S1). The activity emphasizes the difference in size between prokaryotic and eukaryotic cells. We did not use any stains to visualize the cells; you may include this if desired, but students were able to find some cells without stains. An optional extension to this activity would be to have students prepare a simple cheek swab with a methylene blue stain. This allows visualization of the large, nucleated epithelial cells and the tiny bacterial cells that can be seen scattered amongst the larger cheek cells. This provides a nice comparison of the contrast in size and shape of these two distinct cell types. Protocols for methylene blue-stained cheek swabs can be found online as well as in standard laboratory protocols (see http://www.microbehunter.com/heat-fixing-and-staining-human-cheek-cells/ for one possible example).
Student preparation: Students should read Day 2 activities and respond to the questions asked as their “ticket for entry to lab” for the day.
In class summary/laboratory:Students observe the bacterial colonies on their plates. They record their observations: color, shape, height and they count the number of colonies on a worksheet (Supplemental File S1). Students are encouraged to take photos of their plates and/or draw their observations. We then ask several questions:
Based on this information, how many different kinds of bacterial colonies do you have on your plate?
Does any particular colony morphology seem to predominate?
Next, students make a wet mount slide preparation of some of their colonies and observe their slides under the compound microscope. Here we emphasize how and why wet mount slides are made (Supplemental File S1) and have students make basic comparisons between prokaryotic and eukaryotic cells. The purpose is to help students develop a better understanding of the differences in size and scale between microbes and human cells and tissues.
After lab 2: Once done, plates are collected and stored at 4°C until the next lab period, and students clean up using 10% bleach.
Lab Day Three: Extracting DNA
NOTE: Depending on your course goals and resources, you could skip the following three lab days and provide students with access to sequences from the Human Oral Microbiome Database (14) for the BLAST search activity (see Lab Day Seven).
Teacher preparation: Students will be extracting DNA from some of their bacterial colonies today. The materials we used for this are described in Supplemental File S1. You or your lab staff will need to prepare these reagents ahead of time. It was helpful for us to aliquot the Chelex beads and sterile water. You may want to read the introduction in Supplemental File S1 on the size of the bacterial genome. In addition, we give a brief overview of what the 16S rRNA gene is and why we are interested in using it in their PCR reactions (Supplemental File S1).
Student preparation: Students should read through the protocol for the day. For our course, this week aligned with discussions in lecture about cellular macromolecules. This gave students a starting point for understanding the work with the 16S rRNA gene.
In class summary and laboratory:SAFETY NOTE: Students are now working with organisms isolated from the human body. It is important to stress the need for lab safety this week. Students MUST wear gloves, lab coats and eye protection this week when working with their samples, especially after steps involving the boiling water bath or centrifugation. Samples must be allowed to cool on ice before being opened, and students must wait at least 30 seconds before opening the centrifuge to allow potential microaerosols to settle.
Students select three bacterial colonies to sample, draw a circle around each colony, and label the circle (1, 2, or 3), so they can refer back to this information in later weeks. Students use sterile toothpicks to transfer a small amount of a colony from their plates into a microcentrifuge tube containing sterile water, using one toothpick and one tube per colony. They pellet the cells in the microcentrifuge.
Using the Chelex method of DNA extraction, students isolate their bacterial DNA (Supplemental File S1). On a technical note, Chelex beads settle quickly, so remind students to mix the beads well between each of their samples. Remind students to use a new tip between each addition of Chelex to a microcentrifuge tube.
After lab 3: Students should place their tubes containing their DNA samples into a microcentrifuge rack or freezer box at the end of the lab. Instructors or TAs should store the DNA samples at -20°C until the next lab period.
Lab Day Four: DNA amplification using PCR
Teacher preparation: Prepare the reagents and equipment for PCR. If you need a refresher on PCR and why we selectively amplified the 16S rRNA gene (16S rDNA) of their isolated bacterial colonies, see Supplemental File S1.
Student preparation: Students should read through the lab activity for the day. Our students had covered cellular DNA replication by this time, but a review of DNA replication as it occurs in the cell would be useful here.
In class summary/laboratory:Students set up their PCR reactions. We included a discussion of experimental controls, and why these are important. This discussion was especially important because we provided controls rather than having each student prepare their own.
After lab 4: We collected all the students’ PCR tubes in a single ice bucket, prepared appropriate controls, and ran the thermocycler using the protocol indicated in Supplemental File S1. We had two thermocyclers available for use, so were able to stagger the runs. Once the thermocycler run was completed, the PCR samples were stored at 4°C until the next lab period. We placed their original DNA samples back into the freezer; you may choose to hold onto the original DNA or dispose of it at this time.
Lab Day Five: Visualizing DNA
Teacher preparation: Read through the activity for the day to be sure you are prepared with gels and gel boxes. You will need to prepare the appropriate number of 2% agarose gels needed for your students. Examples of results from our students are shown in Figure 1.
Student preparation: Students should read through the activities for the day before coming to lab.
In class summary/laboratory: The focus of this day was for students to explain the PCR reaction of the previous week in terms of DNA replication. We discussed ways to visualize DNA, and the method used in the lab was agarose gel electrophoresis. Students predicted what their gels should look like based on the samples loaded, and then compared their predictions to the actual results.
Students checked the fidelity of their PCR reactions by gel electrophoresis. This also provided a qualitative assessment of how much DNA they would need to use the following week, as they only ran a small amount of their PCR reaction.
After lab 5: The remainder of the student PCR reactions should be returned to storage at 4°C until the next lab period.
NOTE: This day included a lab practical exam on other activities performed up to this point (Oral Microflora questions were not asked on this exam). Students in our two lab sections were required to stay for the full length of the lab period in order to analyze their DNA gel results.
Lab Day Six: DNA Sequencing
Teacher preparation: You should gather all the appropriate reagents for Sanger sequencing (Supplemental Files S1 and S2), depending on the requirements of your sequencing facility. A review of Sanger sequencing can be found in Supplemental File S1.
Student preparation: Students should complete the reading for Lab Day Six in Supplemental File S1.
In class summary/laboratory:We reviewed the results of the gel electrophoresis from the previous week and then provided an introduction on Sanger sequencing (see Supplemental File S1 for a more detailed discussion).
Students selected the sample which produced the best result (i.e. a single bright band) and added the 16S primer (and water, if necessary) to the corresponding microcentrifuge tube to prepare it for sequencing (Supplemental File S1). Students provided a personal code for their sample so that the data would remain anonymous.
After lab 6: The samples were sequenced by the Biomedical Genomics Center (University of Minnesota) (www.bmgc.umn.edu). To keep everything running smoothly, we transferred the student samples into a 96-well plate, as needed by the sequencing facility. Sequencing at our facility typically takes less than 72 hours. An example of student sequence results is shown in Figure 1C. Remaining student PCR reactions were collected and stored at 4°C mainly for later use. They could be disposed at this point if you do not foresee a use for them.
Lab Day Seven: Identifying Your Bacteria- What was that in your mouth?
Teacher preparation: For our students, we chose to do the “clean up” of the sequence results for them, trimming away the ends of the sequences that were not readable. This decision was partly due to time constraints and partly because it was the process they would be least likely to repeat in the future, because sequences in databases are already trimmed. On this day, students learn how to analyze their data. If you have not used BLAST or are unfamiliar with at looking at sequencing results, we advise you to check out the Supplemental File S1. There is also an excellent tutorial available for using the BLAST tool (15) including several YouTube videos (http://www.ncbi.nlm.nih.gov/books/NBK1734/; http://www.youtube.com/playlist?list=PLH-TjWpFfWrtjzMCIvUe-YbrlIeFQlKMq). For the sequence analysis, we used the free trial version of a sequence analysis tool called Geneious (http://www.geneious.com/), which allowed us to do the minimal sequence clean up required. We then uploaded the trimmed sequence files to the course website so students could access their data at home. They used this data to complete the BLAST assignment worksheet provided in Supplemental File S4. An answer key with suggested responses and an example of actual student responses is provided in Supplemental File S5.
Student preparation: Students should read through the Lab Day Seven description before class and come prepared with questions if anything seems confusing. You could also assign the BLAST tutorial videos for them to watch in preparation for class.
In class summary/laboratory: To tie all of these activities together, we provided a review of the entire experience: analyzing colonies, extracting DNA, running PCR, and sending the samples out for sequencing. We showed some examples of their results and discussed what the chromatograms and peaks mean (Supplemental File S1). We emphasized that each student could potentially obtain unique results, so if they compared their sequence to another student’s it would likely not be the same. To further expand on the theme of diversity, time could be taken during the laboratory period to introduce students to the Human Food Project (aka American Gut http://americangut.org/ and the Human Microbiome Project (http://commonfund.nih.gov/hmp/index), both projects which seek to identify the diversity of microbes that live on and in the human body. We would also recommend using the “Diseased/Altered State” jigsaw activity described below in the “Immunology Extension” section as a way to assist students in looking for scientific articles that report differences in the types microbes that can be isolated from the gut based on a person’s diet or health status.
NOTE: Depending on the level of your course, you may choose to have an in-depth discussion on the sequence analysis program “Geneious,” or you may choose to just give a brief overview of how it may be used. We introduced students to the process of performing a BLAST search (Supplemental File S1), which was needed to complete the assignment provided in Supplemental File S4.
Students use the BLAST algorithm to identify the sequence in the database that is most similar to the sequence of their bacterial sample. This “top hit” will provide them the identity of one of the microbes found in their mouth. For a more in-depth discussion on BLAST, see Supplemental File S1 and http://blast.ncbi.nlm.nih.gov/Blast.cgi.
After identifying this organism, students research their bacterial strain using scientifically sound websites. (If you have not already covered this in your class, this would be a good time to introduce students to how to find scientifically accurate sources of information).
TEACHING DISCUSSION
Effectiveness of Lesson: The Oral Microflora activity was effective in achieving our learning goals and objectives: students were able to isolate and identify one species of bacteria from their oral swabs. Students performed commonly used molecular and microbial techniques in their inquiry experiments, something that had not been previously done in this course. In addition, students analyzed their results and reported the importance of their bacterial strains in health and disease prevention. The activity was especially effective in helping students develop the ability to locate and evaluate DNA sequence information. Each of these learning objectives was assessed by student responses to the BLAST worksheet (Supplemental File S4). In addition, this activity reinforced course material on DNA replication and PCR which students originally encountered during the lecture section. We assessed this learning objective by asking students to respond to questions on a lab practical exam given at the end of the semester. For students in our two lab sections, this exam included both the Oral Microflora material and the concepts also covered in all other sections. The Oral Microflora questions asked on this exam are provided in Supplemental File S6, along with expected answers. These questions were asked using a multiple-choice or short answer format, in keeping with the format used for the other portions of the laboratory exam.
We observed that students initially experienced minor challenges with some of the techniques required of them throughout the seven week activity. This was because the Oral Microflora module was unfamiliar territory for non-majors. Some things we noticed that were challenging were technical issues such as using a pipette or adjusting the microscope in order to focus on their microbes. Other areas included recognizing that they actually had enough reagent volume to perform PCR. This was because we provided students with “master mixes” (in microliters) of their PCR reagents and some students mistakenly thought the micro centrifuge tubes were empty. These issues were readily addressed by either the TAs or the instructors during the laboratory period. Indeed, at the conclusion of the activity, we found that the students produced a body of work that was of good quality.
As detailed in Table 1, this lab module is laid out over a seven week period, with the amount of time dedicated to module-related activities varying weekly. The length of time spent on each activity can be modified to suit the purposes of any introductory course. Our students were required to complete all of the activities related to the Oral Microflora module as well as the lab work required of other sections. Not surprisingly, given the need to complete two lab activities within a single lab period, the primary negative comment from students was that the module required “too much time”. However, we note that for most lab periods the time required for Oral Microflora-related activities involved less than 30 minutes of the two hour lab period. The techniques and protocols often overlapped with activities already being done (e.g. using microscopes, looking at cell structure, using a pipet), and could have simply replaced the other lab activity. We have also presented this module as a part of a week-long workshop for high school science teachers (16) and were able to complete all the activities except for sequencing newly-isolated DNA within the more compressed time frame. Our experience working with non-science majors suggests that the times listed in Table 1 represent the minimum amount of time needed for students to achieve reasonable results from the experiment. We would recommend spending additional time if possible focusing on the concepts involved in the activity of each day. We feel this time would be well-spent and could stimulate further discussions in and out of the classroom. Indeed, several of our students indicated that they would have appreciated additional time to focus on the Oral Microflora material.
Due to the relatively small number of students that have completed this learning module, we cannot say definitively whether the inquiry nature of this module was more successful than another laboratory without the module. However, we observed that the students who performed the Oral Microflora module were engaged in the activity throughout the process. This was especially true when they were able to culture and observe their own microbes. The idea that these microbes came out of their mouths was at once disgusting and somehow exciting! Students were also very interested in seeing the results that they got back from the sequencing facility. Several students indicated that it was an interesting and enjoyable activity. One student commented in particular that “in the end, I understood a lot more about microorganisms and oral microflora than I did before”.
An important aspect to consider when implementing any new laboratory activity is the cost required in order to offer sufficient supplies and reagents. With the help of the Lab Manager and Coordinator for the College of Biological Sciences Teaching Laboratories, we determined the cost per student for purchasing reagents and consumables for the Oral Microflora activity to be approximately $19, when the lab is performed as described. This includes the cost of the consumables listed in Supplemental File S2 such as tape, tubes, agar and sequencing plates, as well as miscellaneous items such as paper towels, soap and bleach. It does not include the cost of purchasing pipettes or major equipment such as micro-centrifuges.
Authentic research as a possible extension: The Oral Microflora activity could be used as the start of an authentic research activity by having students revisit their data and propose a hypothesis about how they could alter the bacteria found in their mouths. Students could then design an experiment to test their hypothesis, conduct the experiment, and analyze their own data. This extension would likely require starting the lab activities early in the semester, continuing activities throughout the course of the semester.
An alternative approach for incorporating authentic research would be to have students compare their data to sequences found in the Human Oral Microbiome Database (http://www.homd.org/). This publically accessible genomic database provides information on the many prokaryotic species found in the human oral cavity. The database could be used before students begin the activity, as information that can enable them to predict what they may find with their sequences, or to propose hypotheses for ways to alter the bacteria found in their mouth.
To emphasize the importance of scientific communication, you could require students to present their Oral Microflora experiments and/or results via a poster session or oral presentation. Students (particularly majors) could also write a grant proposal or a journal article on their work.
Immunology Extension: The Oral Microflora activity could be used as a transition into the immune system. Some topics to consider are: infections and pathogenesis, autoimmunity, and adaptive versus innate immunity. Along these lines, we have developed a jigsaw activity that extends the activity to learning about gut microflora and the role these microbes have been proposed to play in human health and disease (see Supplemental File S7). In this jigsaw, each student would be assigned to a particular “altered” state (such as diabetes, obesity, irritable bowel syndrome, or even just alternative diet plans) and they find an article about the topic to report back to their group during the following class period. Students answer a series of questions, and the articles they identified are turned in along with their group’s completed jigsaw worksheet. The questions asked could range from the particular bacterial populations that predominate in a given “altered state”, to the immune response to the bacteria. This jigsaw activity was developed to accompany the laboratory module but we were unable to test it in the classroom. This activity was field-tested at a workshop for middle- and high-school science teachers to introduce them to the scientific literature on the human microbiome. The teachers found it to be a useful activity and proposed using it with their students, perhaps modifying it to permit the use of reviews of scientific articles from secondary sources (e.g. articles on gut enterotypes reviewed in The New York Times (17)).
SUPPLEMENTAL MATERIALS
Table 1. You and Your Oral Microflora-Teaching Timeline
Figure 1. You and Your Oral Microflora-Sample Student Data
Supplemental File S1: The Oral Microflora Laboratory Manual:The entire project is divided into a seven week activity. Each week, a short introduction on laboratory related topics is followed by a detailed laboratory procedure. Week one includes a primer on microbes. Week two provides students the opportunity to explore the different microbes. In week three, students are introduced to genomics and they learn to extract DNA. During week four, students learn about PCR and perform their PCR reactions. In week five, students use gel electrophoresis to determine the fidelity of their PCR results. In week six, students learn about 16S rRNA and they prepare and send out their DNA for sequencing. The final in-lab activity, week seven, culminates in data analysis and a BLAST search of their resultant strain. Much of the work in week seven can be done outside of class time.
Supplemental File S2: Materials and Supplies for the Oral Microflora: This supplement outlines the topic of the week for the seven weeks described in the Oral Microflora module, along with the equipment and preparation needed for the activities.
Supplemental File S3: Day 1 PowerPoint slides for the Oral Microflora Laboratory: This presentation includes a short primer about microbes for Day One of the Oral Microflora activity, introducing the learning objectives and learning goals. We provide suggested links for images to include in an introduction to microbes.
Supplemental File S4: You and Your Oral Microflora BLAST Assignment: This supplement is the BLAST assignment given to students for analyzing their DNA sequences.
Supplemental File S5: Answer Key: You and Your Oral Microflora BLAST Assignment: This supplement provides suggested responses to the Oral Microflora BLAST assignment. An example of actual student responses is provided.
Supplemental File S6: Lab Practical 2, Oral Microflora Questions: This supplement presents the lab practical exam questions asked of students who participated in Oral Microflora module. Answers are marked in red.
Supplemental File S7: Altered States Jigsaw Activity: This supplement provides a proposed extension activity for both introducing students to the impact of the microbiome on human health and extending the discussion of microbial diversity beyond the human mouth.
ACKNOWLEDGMENTS
The authors thank Melissa Palmer for providing the opportunity to test this module in her Human Biology course at the University of Minnesota. We thank Melissa Palmer, Jane Phillips and Mary Williams for critical review of this manuscript. We thank Brian Gibbens for the helpful PCR figure used in the lab handout. Anna K. Strain and Kieng B. Vang are supported by a grant from the Howard Hughes Medical Institute awarded to Robin L. Wright.
References
- Woodin T, Carter VC, Fletcher L. 2010. Vision and change in biology undergraduate education: a call for action-initial responses. CBE Life Sci Educ. 9:71-73.
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J; MetaHIT Consortium, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Mérieux A, Melo Minardi R, M'rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P.2011. Enterotypes of the human gut microbiome. Nature. 473:174-80.
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science. 334:105-8.
- Lepage P, Leclerc MC, Joossens M, Mondot S, Blottiere HM, Raes J, Ehrlich D, Dore J. 2013. A metagenomic insight into our gut's microbiome. Gut. 62:146-58.
- Wade WG. 2013. The oral microbiome in health and disease. Pharmacol Res. 69:137-43.
- Banta LM, Crespi EJ, Nehm RH, Schwarz JA, Singer S, Manduca CA, Bush EC, Collins E, Constance CM, Dean D, Esteban D, Fox S, McDaris J, Paul CA, Quinan G, Raley-Susman KM, Smith ML, Wallace CS, Withers GS, Caporale L. 2012. Integrating genomics research throughout the undergraduate curriculum: a collection of inquiry-based genomics lab modules. CBE Life Sci Educ. 11:203-8.
- Boomer SM, Lodge DP, Dutton BE. 2002. Bacterial Diversity Studies Using the 16S rRNA Gene Provide a Powerful Research-Based Curriculum for Molecular Biology Laboratory. Microbiol Educ. 3:18-25.
- Caruso SM, Sandoz J, Kelsey J. 2009. Non-STEM undergraduates become enthusiastic phage-hunters. CBE Life Sci Educ. 8:278-82.
- Jordan TC, Burnett SH, Carson S, Caruso SM, Clase K, DeJong RJ, Dennehy JJ, Denver DR, Dunbar D, Elgin SC, Findley AM, Gissendanner CR, Golebiewska UP, Guild N, Hartzog GA, Grillo WH, Hollowell GP, Hughes LE, Johnson A, King RA, Lewis LO, Li W, Rosenzweig F, Rubin MR, Saha MS, Sandoz J, Shaffer CD, Taylor B, Temple L, Vazquez E, Ware VC, Barker LP, Bradley KW, Jacobs-Sera D, Pope WH, Russell DA, Cresawn SG, Lopatto D, Bailey CP, Hatfull GF. 2014. A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. MBio. 5:e01051-13.
- Minnesota Mississippi Metagenomics Project. College of Biological Sciences, University of MN http://www.mississippi-metagenome-project.umn.edu/. Accessed 2014 June 20.
- Lesk AM. 2012. Introduction to Genomics. p. 397. Oxford University Press, New York, New York.
- Janda JM, Abbott SL. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol. 45:2761-4.
- American Society for Microbiology. 2014 February 21. About Microbiology. . <http://www.microbeworld.org/about-microbiology>. Accessed 2014 February 21.
- The Forsyth Institute. Human Oral Microbiome Database. The Forsyth Institute <http://homd.org/>.
- Wheeler D, Bhagwat M. 2007. BLAST QuickStart: example-driven web-based BLAST tutorial. Methods Mol Biol. 395:149-76.
- M3P. 2013 June 17. Metagenomics workshop for high school teachers. College of Biological Sciences, University of Minnesota http://www.mississippi-metagenome-project.umn.edu/. Accessed 2014 June 17.
- Zimmer C. 2011. Bacterial ecosystems divide people into 3 groups, scientists say. The New York Times.
Article Files
Login to access supporting documents
You and Your Oral Microflora-Introducing non-biology majors to their forgotten organ(PDF | 831 KB)
Supplemental File S1. You and Your Oral Microflora-Laboratory Manual(PDF | 797 KB)
Supplemental File S2. You and Your Oral Microflora-Materials and Supplies List(PDF | 161 KB)
Supplemental File S3. You and Your Oral Microflora-Day 1 PowerPoint Presentation(PPT | 177 KB)
Supplemental File S4. You and Your Oral Microflora-BLAST Assignment(PDF | 120 KB)
Supplemental File S5. You and Your Oral Microflora-BLAST Assignment Answer Key(PDF | 123 KB)
Supplemental File S6. You and Your Oral Microflora-Lab Practical 2 Questions(DOCX | 424 KB)
Supplemental File S7. You and Your Oral Microflora-Altered States Jigsaw Activity(PDF | 144 KB)
- License terms

Comments
Comments
There are no comments on this resource.