Using Undergraduate Molecular Biology Labs to Discover Targets of miRNAs in Humans
Published online:
Abstract
Incorporating authentic research experiences into undergraduate labs, while shown to be particularly effective at engaging and retaining students in STEM majors, can be difficult to accomplish within the constraints of resource availability or cost, and time limitations. One area that is particularly amenable to adaptation for undergraduate lab classes is the discovery and validation of targets of microRNAs (miRs). The human genome encodes several hundred, possibly several thousand miRs, each of which is a 22 nucleotide long RNA molecule capable of regulating the expression of multiple target genes. miRs have been shown to be critical during development, for human health and disease, and are currently being investigated as both therapeutic agents, as well as possible drug targets. A lack in understanding the mechanisms by which miRs recognize their targets makes computer-based predictions of miR targets quite inaccurate, necessitating experimental verification of such predictions. In this lesson, we describe an easily adaptable lab module that can be used in existing undergraduate molecular biology lab courses to conduct authentic scientific research. Students use a variety of databases to identify likely candidate genes whose expression may be altered by a given miR, and then experimentally test their predictions in human cells. This inquiry-based module gives students a taste of real scientific research and excites them about the possibility that, even as a student, they have the potential to contribute to this cutting edge research.
Citation
Idica, A., Thompson, J., Munk Pedersen, I., and Kadandale, P. 2015. Using Undergraduate Molecular Biology Labs to Discover Targets of miRNAs in Humans. CourseSource. https://doi.org/10.24918/cs.2015.10Society Learning Goals
Bioinformatics
- DNA - Information Storage [GENOMICS]
- Where are data about the genome found (e.g., nucleotide sequence, epigenomics) and how are they stored and accessed?
- How can bioinformatics tools be employed to analyze genetic information?
Genetics
- Methods & Tools in Genetics
- What experimental methods are commonly used to analyze gene structure and gene expression?
Science Process Skills
- Process of Science
- Pose testable questions and hypotheses to address gaps in knowledge
- Plan, evaluate, and implement scientific investigations
- Interpret, evaluate, and draw conclusions from data
- Construct explanations and make evidence-based arguments about the natural world
- Address novel questions through authentic research experiences
- Modeling/ Developing and Using Models
- Recognize the important roles that scientific models, of many different types (conceptual, mathematical, physical, etc.), play in predicting and communicating biological phenomena
- Make inferences and solve problems using models and simulations
- Build and evaluate models of biological systems
- Quantitative Reasoning/ Using Mathematics and Computational Thinking
- Apply the tools of graphing, statistics, and data science to analyze biological data
- Communication and Collaboration
- Share ideas, data, and findings with others clearly and accurately
Lesson Learning Goals
- Understand the use, and limitation of biological databases and prediction algorithms
- Formulate new hypotheses
- Understand the importance of controls in experimental designs
- Understand common techniques used in molecular biology and their use to address scientific questions
- Understand the importance of quantitative measurements and reproducibility in science
Lesson Learning Objectives
- Use biological databases to generate and compare lists of predicted miR targets, and obtain the mRNA sequence of their selected candidate gene
- Use bioinformatics tools to design and optimize primer sets for qPCR
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Including authentic research experiences into the curriculum has been shown to be a powerful tool in increasing student engagement and learning,(1,2) decreasing the performance gap between majority students and that of underrepresented minorities (URMs),(1,2,3,4) and increasing the retention of students in STEM majors (3,5). However, the incorporation of such experiences is usually costly and can be severely limited by the availability of resources, especially in universities that teach classes with large student enrollments. Even labs that use inquiry-based modules often do not include real scientific research (6). In addition, those lab courses that engage students in real research are often funded by external grants,(1,2,3,4,2,7) and are consequently not sustainable in the long term. Given the limitations imposed by resources and time, the design of sustainable lab modules that can be used in large enrollment undergraduate lab is quite challenging. We describe one such module in which students in a large molecular biology lab course attempt to validate predicted human targets of microRNAs (miRs), thereby contributing to real scientific research. The module is relatively simple and sufficiently inexpensive to be used in most institutions. Further, the module has been designed so that it can be easily adapted to a number of different situations and contexts, including classes that meet for different lengths of time.
miRs are 22nt (nucleotide) RNAs of different sequences, each of which is capable of regulating the expression of several target genes (8-10). Although estimates of the total number of miRs in the human genome vary widely from the hundreds (11) to the thousands,(12) their importance to normal health and disease is well established. For example, miRs are required for the regulation of normal development,(1,2,3,4,2,7,13,14) and misregulation of miRs is involved in a number of human diseases, including cancers,(1,2,3,4,2,7,13,14,15,16) mental retardation,(17) and obesity (18). miRs are actively being investigated as potential therapeutic agents (19-21) and as targets for novel drugs (22). The mechanisms by which miRs are expressed and processed is fairly well established, as is the final means by which they regulate their targets (23). However, the exact process by which miRs target specific mRNAs is still unclear and, as a result, computer algorithms to predict miR targets are notoriously unreliable (24).
The experimental validation of miR target predictions requires the use of relatively simple molecular biology techniques, including transfection of cells with miRs, RNA isolation, RT-PCR and qPCR. These techniques that are amenable for use in an undergraduate lab course and the kits required to perform these procedures are relatively inexpensive, making this project affordable and sustainable. At UC Irvine, we use this lab module in our molecular biology lab course. The course enrollment is approximately 100 students every quarter during the regular school year (for a total of 300 students in the academic year) and around 80 students in the summer quarter. Since students work in pairs, we could theoretically test ~190 predicted targets of a given miR every year, a level of effort that would make a useful contribution to the field of miR research. We have started by looking for targets of miR-128 in HeLa cells. If the lab module is more widely adopted at other institutions, a larger number of miRs could be tested in HeLa cells, or predictions for a given miR could be tested in different cell types. It's important to note that results from our labs will be made publicly available for use by others.
From the perspective of students, the possibility of making a real contribution to science, particularly in an undergraduate lab course, generates a great deal of excitement and engages students more completely in the course. The module also exposes students to an authentic research experience, requiring them to generate a set of criteria to evaluate possible research avenues, use the published literature to ultimately select one candidate gene to test, and repeat their experiment (including troubleshooting their initial attempt if it failed!) before drawing a strong conclusion from their results. Students take ownership of their project and work on "their" gene, further increasing engagement with the content of the lab. The lab module is easily adapted to a number of different questions, and can be used successfully in diverse class settings to enable undergraduate students to get a taste of research on a modern, relevant, cutting edge area.
Intended audience
The module described here is used in a large enrollment (100 students per quarter), upper division molecular biology lab course. The students meet for a common lecture by the instructor, and then are split into 5 lab sections of twenty students each. Each lab section is run by a graduate student TA, and the students work through the activities of the lab module in pairs. However, the module is easily adapted to a number of different contexts, since the technical complexity of the activities is not very high.
Learning time
The module as carried out at UC Irvine spanned three 4-hour lab sessions, with one week between consecutive lab sessions. However, there are a number of stopping points in the procedure, so that with appropriate modifications, the module can be used in shorter labs (although it would then require a greater number of lab sessions).
Pre-requisite student knowledge
A basic background in molecular biology is strongly recommended for this module. Students will need to know what RNA is, how an RNA molecule can serve as a template for synthesis of DNA by RT-PCR, and how a PCR works. Lectures during the lab course can address more advanced topics such as primer design, positive and negative controls for qPCR, and miR biology. Basic lab skills, such as dilution calculations, and pipetting will be required to successfully carry out the experiments. Basic data mining, including searching on Google and PubMed, will be required to evaluate the miR target list.
SCIENTIFIC TEACHING THEMES
Active learning
Because it involves direct student effort, a lab module is, by its nature, active. This module will specifically require students to engage with the project by taking ownership of one predicted target gene of interest, which they themselves choose, based on their exploration of a number of different databases. Students will have to use their theoretical understanding of miRs, and miR targets to evaluate their results and arrive at their independent conclusions about their candidate gene.
Assessment
Assessment involves weekly quizzes on topics relevant to the lab module and a comprehensive final exam at the end of the quarter with questions about the procedures. Questions in both the quizzes and final exam require students to apply their learning and analyze data to draw conclusions, rather than just test student knowledge of miRs and the experiments they carried out in lab. Students also submit a final lab report which is written similarly to a scientific paper, in which students must justify the rationale for their experiments, present their results and conclusions, and discuss the future directions of their research project.
Inclusive teaching
Students get to pick a candidate gene that interests them and they do so using a variety of criteria that reflect their individuality and diversity. Since students work in pairs (students randomly pair up at the beginning of the quarter, and keep the pairing for all lab sessions), they get to scientifically debate the merits and issues with picking different candidates, and must arrive at a consensus choice.
LESSON PLAN
The entire module takes three lab sessions of 4 hours each. Table 1 depicts the timeline of activities in each lab session.
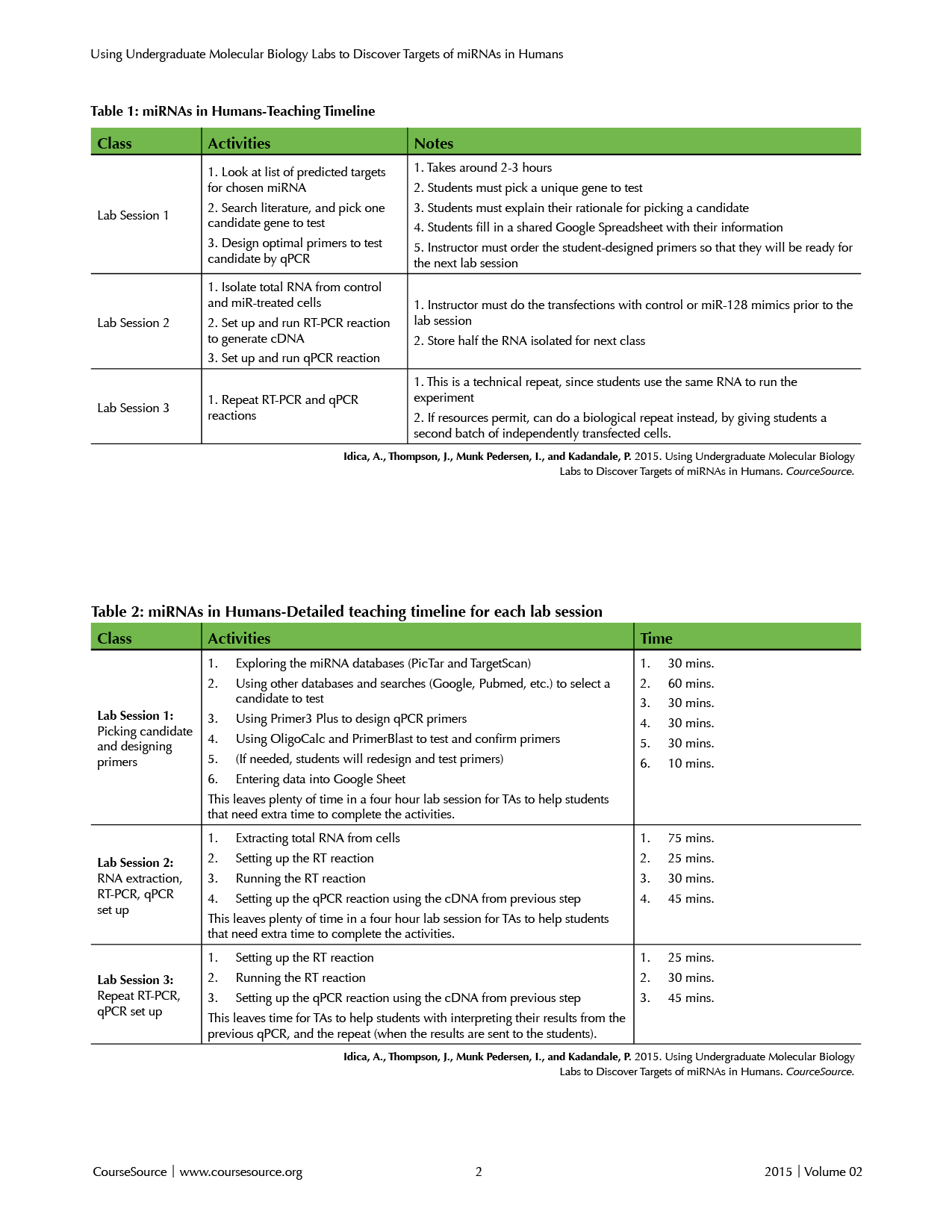
Tables 1 and 2. miRNAs in Humans-Teaching Timelines
LAB SESSION 1
1.A. Instructor Preparation for Lab Session 1
Supplemental File S1 details the two databases (PicTar - http://pictar.mdc-berlin.de/ and TargetScan - http://www.targetscan.org/) and the procedure to generate a list of predicted targets for a specific miR. We recommend that the instructors familiarize themselves with the two databases prior to teaching the students to use it. By design, we do not provide students with details about how to use the two databases. The online resources explain the databases and their outputs quite well, and we expect students to explore these resources on their own and figure out what the information available on the websites means. This approach mirrors the research experience, when scientists are forced to use unfamiliar tools to obtain data relevant to their projects.
We have initially chosen to identify the targets of miR-128, since the Munk Pedersen lab is currently studying its role in disease and transposon activation, and most of the targets of miR-128 are not known. We note here, though, that the choice of specific miR is relatively unimportant. Instructors can choose any miR for which there is scant information about targets.
Supplemental File S2 explains how to bioinformatics tools to design optimal primers for measurement of mRNA levels by qPCR (Primer3Plus - http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi, OligoCalc - http://www.basic.northwestern.edu/biotools/OligoCalc.html and PrimerBLAST - http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The Pre-lab Lecture should include details about primer design, so students can understand the meaning of the output from these tools.
1.B. Lecture Prior to Lab Session 1
You will need to cover several concepts in lecture prior to students entering lab. (Example PowerPoint slides used to cover these concepts are Supplemental File 7 & 8. In addition, the authors will provide podcasts of their lectures upon request.) Ideally, students will have much of this information in their lab manuals as well, so they have multiple sources to use to understand the background and methods. Lecture should cover the fundamentals of what miRs are, and their functions and importance in living systems, and methods to discover miR targets, as listed below:
- Fundamentals of miR
- What are miRs?
- What are the functions of miRs?
- What is the involvement of miRs in human diseases?
- Why are computer algorithms that predict miR candidates not reliable?
- Using TargetScan and PicTar to generate lists of potential miR targets
- If two different algorithms predict that a particular gene is likely to be regulated by a given miR, does that improve the accuracy of the prediction?
- If predictions are unreliable, of what use are they?
- How could you use overexpression of a miR in human cells to identify its targets?
- What predictions would you make about a gene that is regulated by a miR if cells are overexpressing that miR?
- From a list of candidate genes generated by TargetScan or PicTar, what criteria can we use to pick the most likely targets?
- Designing PCR primers
- Qualities of a primer that are important to consider during design
- Use of bioinformatics tools (Primer3Plus, OligoCalc and PrimerBLAST) to design and optimize PCR primers
1.C. Lab Sesson 1 Activities
Students will follow instructions given in lecture to generate lists of putative miR targets, obtain information about some of these targets, discuss with their lab partner, and finally choose one gene to test by qPCR.
Having chosen one target gene to test, students will then design gene specific primers for qPCR of this gene. Students will fill out a Google Sheet (Supplementary figure S3) with information on the gene that they chose, and the primer sequence that they designed. It is not essential to use Google Sheets specifically; any method by which the information can be obtained and shared will work.
1.D. Instructor Follow-Up After Lab Session 1
Immediately following the lab section, the instructor will need to order the primers that the students designed. Since the students are directly pasting the sequences from the Primer design program's output and are testing the primers using OligoCalc and PrimerBLAST, the instructor does not need to double check the quality of the primer sequences. Make sure to use a primer ordering service that will deliver the primers in time for the next lab session.
LAB SESSION 2
2.A. Instructor Preparation for Lab Session 2
The instructor should be familiar with RT-PCR and qPCR, including the various controls that should be included in such experiments to get interpretable results. Supplemental File S4 lists the setup of tubes used in our labs. Instructors should also understand the meaning of the threshold cycle and the calculations used to convert threshold cycle values to fold-expression values (Supplemental File S5). The RNA isolation, RT-PCR and qPCR kits used at UC Irvine are listed in Supplemental File S6. There is no reason to use these kits specifically; any RNA isolation, RT-PCR and qPCR kit will do the job. The only considerations in choosing the kits to use are cost, simplicity, and time required to complete the experiments using the kit.
Prior to Lab Session 2, the instructor will need to transfect cells with either a control scrambled miR, or the miR of interest (For our labs, we transfected miR-128 into HeLa cells, which are easy to obtain and easy to transfect). The specific protocols for are in Supplemental File S5, and the transfection protocol that we use is outlined in Supplementary S11 but again, it is not essential to use these exact methods - any transfection protocol that works for you is fine. You must transfect enough cells for all your students to use; the transfected cells can aliquoted, pelleted, and stored at -80oC for use by the students. It is also highly recommended that these transfections are carried out well in advance of the lab, and that the transfected cell "batches" are tested using qPCR of a known target of the miR (the positive control) to ensure successful transfection.
2.A. Lecture Prior to Lab Session 2
Lecture should cover the principle of RNA isolation from cells, use of RT-PCR to make cDNA, and quantification of gene expression using qPCR. (The authors will provide podcasts of their lectures upon request. Example PowerPoint slides used to cover these aspects are Supplemental File 7 & 8). Important points to address are listed below:
- How can RNA be isolated from cells?
- Why can't we directly measure RNA levels in cells?
- Why do we use poly-T and random hexamer primers for the RT-PCR?
- The principle of qPCR, and the concept of the threshold cycle
- Calculating changes in expression levels based on the threshold cycle
2.C. Lab Session 2 Activities
Students will follow the protocols for the specific kits used to isolate RNA, make cDNA using RT-PCR, and then set up the qPCR reaction. Immediately after RNA isolation, students should freeze half of their RNA sample at -20oC to run a repeat in the next lab session.
2.D. Instructor Follow-Up After Lab Session 2
Once the qPCR run is complete, the instructor emails the qPCR results to the students.
LAB SESSION 3
Repeat the RT-PCR and qPCR experiments of Lab Session 2 using the frozen RNA from lab session 2. This replication provides a technical repeat of the experiment. Because of budget constraints, we were unable to ask students to do a complete repetition of the experiment. This technical repetition provides some appreciation for the need for reproducibility, but the instructor may want to review why a better experiment would be to repeat the entire experiment from start to finish. Needless to say, if resources permit, one can have students repeat the entire experiment. If doing so, it would be appropriate to make two sets of independently transformed cells available to the students, so they can perform a true biological repeat.
AFTER THE LAB SESSIONS
Have students calculate changes in expression levels of their predicted target gene using the qPCR data. Students must write a lab report in the form of a scientific paper, describing the rationale for the experiment, justification for picking their candidate gene, qPCR results, their conclusions, and a discussion of their results. The general rubric handed out to students for their lab reports is provided in Supplemental File S10. In addition, examples of the assessment questions used to gauge student understanding of the experiments are provided in Supplemental File S9. We used these questions in weekly quizzes and a cumulative final exam, to provide both formative and summative assessment of student learning.
TEACHING DISCUSSION
The main goal of this module is to give students an authentic research experience. We have run this module in three different quarters over the regular school year (Fall 2013-Spring 2014), and in 4 sections taught by two different instructors over the summer quarter (Summer 2014). The successful implementation of the modules by the different instructors in the summer demonstrates that this module is now sufficiently mature to be implemented by others. In addition, the cost of implementation is sustainable. The software tools used in this lab module are all freely available online and the reagents used for the experiments are relatively inexpensive and readily available. The only expensive piece of equipment required for this lab is access to a qPCR machine.
Based on student feedback, students appear to have a much higher level of engagement with this lab module, compared to the other, more "cookbook" type modules used in the rest of the molecular biology lab course. Students start referring to candidate genes as "my gene," and are very excited about the possibility of discovering something new. Many students even start asking about whether they will be authors on a paper, and display a newfound curiosity about scientific discovery. We have, however, not yet conducted a formal assessment to measure changes in student learning or attitudes due to this module.
Suggestions for adaptations
Overall, this is a very flexible lab module that can be easily changed for use in a number of different contexts and environments. The timeline described in Table 1 can be easily modified, since there are a number of stopping points within the different protocols. For example, in lab sessions that are less than four hours, the timeline could be adapted so that lab session 2 and 3 could be split into four separate sessions. Once the RNA is isolated or the cDNA is produced, either can be stored at -20C and used in the next lab session, instead of directly proceeding to the qPCR reaction. The first lab session is a "dry lab" in which students identify a gene of interest to test and design primers to measure expression of that gene; this lab session could even be used as a standalone module.
For labs with more resources, student work can be expanded. For example, students can transfect the cells with the miRNA mimic themselves. In addition, instead of testing only one candidate gene per group, students can work with two or more genes, testing them simultaneously. The module can also be expanded to have students test candidate gene expression in different cell types.
SUPPLEMENTAL MATERIALS
- Supplemental File S1. miRNAs in Humans:Using databases to generate lists of candidate targets of miRs.
- Supplemental File S2. miRNAs in Humans:Designing primers to test your candidate gene. Use of free online tools to design primers for a candidate gene.
- Supplemental File S3. miRNAs in Humans:Example spreadsheet for noting different candidate genes, and primer sequences for each.
- Supplemental File S4. miRNAs in Humans:An example setup of qPCR tubes that is used in our labs.
- Supplemental File S5. miRNAs in Humans:Calculating changes in fold expression based on qPCR data.
- Supplemental File S6. miRNAs in Humans:List of reagents used, and links to the manufacturers' product pages and manuals.
- Supplemental File S7 and S8. miRNAs in Humans:PowerPoint presentations used to give students the theoretical background underlying the lab module.
- Supplemental File S9. miRNAs in Humans:Example questions used for assessment of student learning.
- Supplemental File S10. miRNAs in Humans:General rubric provided to students for their lab reports.
- Supplemental File S11. miRNAs in Humans:Transfection protocol used for transfecting miR-mimic and control into HeLa cells.
ACKNOWLEDGMENTS
The authors would like to acknowledge the entire UCI instructional support group for their help and patience during the development of this lab module. We would also like to thank the summer lab instructors Nadja Dvorkin and Cathie Overstreet for bravely being the first "outside" implementers of this lab module.
References
- Gasper BJ, Gardner SM. Engaging Students in Authentic Microbiology Research in an Introductory Biology Laboratory Course is Correlated with Gains in Student Understanding of the Nature of Authentic Research and Critical Thinking. J Microbiol Biol Educ. 2013;14(1):25-34. doi:10.1128/jmbe.v14i1.460.
- Jordan TC, Burnett SH, Carson S, et al. A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. MBio. 2014;5(1):e01051-13. doi:10.1128/mBio.01051-13.
- Villarejo M, Barlow A. Encouraging minority undergraduates to choose science careers: career paths survey results. CBE-Life Sci .... 2008;7:394-409. doi:10.1187/cbe.08-04-0018.
- Shaffer CD, Alvarez CJ, Bednarski a. E, et al. A Course-Based Research Experience: How Benefits Change with Increased Investment in Instructional Time. Cell Biol Educ. 2014;13(1):111-130. doi:10.1187/cbe-13-08-0152.
- Harrison M, Dunbar D, Ratmansky L, Boyd K, Lopatto D. Classroom-based science research at the introductory level: changes in career choices and attitude. CBE Life Sci Educ. 2011;10(3):279-86. doi:10.1187/cbe.10-12-0151.
- Rissing S, Cogan J. Can an inquiry approach improve college student learning in a teaching laboratory? CBE-Life Sci Educ. 2009;8:55-61. doi:10.1187/cbe.08-05-0023.
- Hatfull GF, Pedulla ML, Jacobs-Sera D, et al. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet. 2006;2(6):e92. doi:10.1371/journal.pgen.0020092.
- Banerjee D, Slack F. Control of developmental timing by small temporal RNAs: a paradigm for RNA-mediated regulation of gene expression. Bioessays. 2002;24(2):119-29. doi:10.1002/bies.10046.
- Ke X-S, Liu C-M, Liu D-P, Liang C-C. MicroRNAs: key participants in gene regulatory networks. Curr Opin Chem Biol. 2003;7(4):516-23. Available at: http://www.ncbi.nlm.nih.gov/pubmed/12941428. Accessed August 6, 2014.
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522-31. doi:10.1038/nrg1379.
- Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299(5612):1540. doi:10.1126/science.1080372.
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32(Database issue):D109-11. doi:10.1093/nar/gkh023.
- Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350-5. doi:10.1038/nature02871.
- Kloosterman WP, Plasterk RHA. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11(4):441-50. doi:10.1016/j.devcel.2006.09.009.
- Zhang W, Dahlberg JE, Tam W. MicroRNAs in tumorigenesis: a primer. Am J Pathol. 2007;171(3):728-38. doi:10.2353/ajpath.2007.070070.
- Wiemer EAC. The role of microRNAs in cancer: no small matter. Eur J Cancer. 2007;43(10):1529-44. doi:10.1016/j.ejca.2007.04.002.
- Singh SK. miRNAs: from neurogeneration to neurodegeneration. Pharmacogenomics. 2007;8(8):971-8. doi:10.2217/14622416.8.8.971.
- Peng Y, Yu S, Li H, Xiang H, Peng J, Jiang S. MicroRNAs: Emerging roles in adipogenesis and obesity. Cell Signal. 2014;26(9):1888-1896. doi:10.1016/j.cellsig.2014.05.006.
- Van Rooij E, Kauppinen S. Development of microRNA therapeutics is coming of age. EMBO Mol Med. 2014;6(7):851-864. doi:10.15252/emmm.201100899.
- Chen Y, Gao D-Y, Huang L. In vivo delivery of miRNAs for cancer therapy: Challenges and strategies. Adv Drug Deliv Rev. 2014. doi:10.1016/j.addr.2014.05.009.
- Tay FC, Lim JK, Zhu H, Hin LC, Wang S. Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv Drug Deliv Rev. 2014. doi:10.1016/j.addr.2014.05.010.
- Gounaris-Shannon S, Chevassut T. The Role of miRNA in Haematological Malignancy. Bone Marrow Res. 2013;2013:269107. doi:10.1155/2013/269107.
- Gurtan AM, Sharp PA. The role of miRNAs in regulating gene expression networks. J Mol Biol. 2013;425(19):3582-600. doi:10.1016/j.jmb.2013.03.007.
- Akbari Moqadam F, Pieters R, den Boer ML. The hunting of targets: challenge in miRNA research. Leukemia. 2013;27(1):16-23. doi:10.1038/leu.2012.179.
Article Files
Login to access supporting documents
Using undergraduate molecular biology labs to discover targets of miRNAs in humans(PDF | 145 KB)
Supplemental File S1. miRNAs in Humans-Using databases to generate lists of candidate targets of miRs.docx(DOCX | 915 KB)
Supplemental File S2. miRNAs in Humans-Designing primers to test your candidate gene.docx(DOCX | 1 MB)
Supplemental File S3. miRNAs in Humans-Example spreadsheet for noting different candidate genes and primer sequences for each.docx(DOCX | 139 KB)
Supplemental File S4. miRNAs in Humans-An example setup of qPCR tubes that is used in our labs.docx(DOCX | 18 KB)
Supplemental File S5. miRNAs in Humans-Calculating changes in fold expression based on qPCR data.docx(DOCX | 27 KB)
Supplemental File S6. miRNAs in Humans-List of reagents used and links to the manufacturers product pages and manuals.docx(DOCX | 19 KB)
Supplemental File S7. miRNAs in Humans-PowerPoint presentations used to give students the theoretical background underlying the lab module.pptx(PPTX | 2 MB)
Supplemental File S8. miRNAs in Humans-PowerPoint presentations used to give students the theoretical background underlying the lab module.pptx(PPTX | 2 MB)
Supplemental File S9. miRNAs in Humans-Example questions used for assessment of student learning.pdf(PDF | 468 KB)
Supplemental File S10. miRNAs in Humans-General rubric provided to students for their lab reports.pdf(PDF | 216 KB)
Supplemental File S11. miRNAs in Humans-Transfection protocol used for transfecting miR-mimic and control into HeLa cells.docx(DOCX | 18 KB)
- License terms

Comments
Comments
There are no comments on this resource.