Teaching RNAseq at Undergraduate Institutions: A tutorial and R package from the Genome Consortium for Active Teaching
Published online:
Abstract
Next-generation sequencing is radically changing the study of biology, but there are currently few resources aimed at teaching the required laboratory and data-analysis skills to undergraduate students. This gap is especially true at primarily undergraduate institutions, where even the faculty are likely to encounter barriers related to funding, equipment, and training needed to begin research or teaching of sequencing and the associated bioinformatics. For this reason, the GCAT-SEEK network has been developed to provide training, data, and teaching materials specifically geared for general biology undergraduates and their instructors. This lesson plan was created to teach RNAseq analysis as a part of that effort. It is provided here, both in finished form and with the modifiable source code, to allow flexible adaptation to various classroom settings. In addition, we include a relevant tutorial to ease students and faculty into the R statistical environment. The associated materials are directly applicable to both faculty training and classroom settings. We expect implementation of the tutorial to strengthen bioinformatic knowledge and skills for general biology students.
Citation
Peterson, M.P., Malloy, J.T., Buonaccorsi, V.P., and Marden, J.H. 2015. Teaching RNAseq at Undergraduate Institutions: A tutorial and R package from the Genome Consortium for Active Teaching. CourseSource. https://doi.org/10.24918/cs.2015.14Society Learning Goals
Bioinformatics
- Computation in the life sciences
- What computational concepts are important in bioinformatics?
- What statistical concepts are important in bioinformatics?
- RNA - Information Transfer [TRANSCRIPTOMICS]
- How can bioinformatics tools be employed to examine transfer of genetic information?
- Computational Skills
- How do biologists employ software development as part of the scientific discovery process?
- What higher-level computational skills can be used in bioinformatics research?
Lesson Learning Goals
Students will:- understand the steps of RNAseq analysis
- appreciate the role of computers in data analysis
- understand basic statistical concepts
- learn the utility of command line based computer programming and one of its applications to biology
Lesson Learning Objectives
- From raw RNAseq data, run a basic analysis culminating in a list of differentially expressed genes.
- Explain and evaluate statistical tests in RNAseq data. Specifically, given the output of a particular test, students should be able to interpret and explain the result.
- Use the Linux command line to complete specified objectives in an RNAseq workflow.
- Generate meaningful visualizations of results from new data in R.
- (In addition, each chapter of this lesson plan contains more specific learning objectives, such as “Students will demonstrate their ability to map reads to a reference.”)
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
INTRODUCTION
The rise of next generation sequencing is fundamentally changing approaches to research in and teaching of biology [1-3]. Even many traditional non-model systems, those without substantial genomic resources, are now adding genomic tools to address a wide range of ecological questions [4-9]. As costs have fallen dramatically in recent years, sequence data are being generated more rapidly than the capacity of the available researchers and students to analyze it. Large repositories of sequence data are now publicly available from resources such as the NCBI's Gene Expression Omnibus [10] and Sequence Read Archive [10], as well as more specific resources such as MG Rast for metagenomics data [11].
The expanding potential of technologies to provide new approaches to research and teaching is particularly true for RNAseq analysis. RNAseq utilizes massively parallel next-generation sequencing technology to sequence representative copies of nearly every gene expressed in a sample. This approach allows two types of analysis. First, it allows researchers with limited (or non-existent) genomic data for their organism to focus sequencing only on the transcribed portions of the genome, increasing the amount of sequence within genes while limiting the costs spent on sequencing difficult to interpret intragenic (“junk”) DNA. Secondly, it allows researchers to estimate the expression level of each gene in a sample, making it possible to compare gene expression between groups (e.g., treatments, sexes, ages, etc.). Several reviews of RNAseq approaches may help novice users orient themselves before teaching this lesson [12-14].
At the same time that these large sequence approaches are emerging, there is a renewed push within the Life Sciences to emphasize authentic research experiences for undergraduate education [15]. While the availability of sequence data presents ample opportunity for such experiences, the analysis tools needed for these approaches are radically different from those generally used by both students and faculty. Even after obtaining these tools, there are substantial up-front costs and training challenges that must be overcome before the tools can be effectively implemented in undergraduate classes [1]. To address these needs, the GCAT-SEEK network has been formed and is developing educational resources such as this tutorial and sample data for use in undergraduate classrooms [16].
Here, we present a tutorial (Supplemental Materials S4), complete with sample next-generation sequence data (Supplemental Materials S1) that can be used to introduce students and faculty alike to the concepts and competencies required to begin RNAseq bioinformatic analyses. This tutorial focuses on RNAseq, a leading interest of instructors in the GCAT-SEEK network [17], which was historically focused on microarray technology [18]. Many other educational modules are currently available on the GCAT-SEEK web page (http://lycofs01.lycoming.edu/~gcat-seek/workshops.html), which also contains information on joining the network and future workshops.
This tutorial relies heavily on an R package (rnaseqWrapper) developed specifically to ease novel computer users into bioinformatic analysis. The package provides simple functions to encompass many of the complicated steps of RNAseq analysis, along with improved and user-friendly functions for visualization and output of data analysis. These wrappers include simple, easy to use, but limited functions for the packages DESeq [19], topGO [20], seqinr [21], gplots [22], and ecodist [23]. As such, the package should be of use to undergraduate students and to researchers at all stages. Installation instructions and usage examples with the provided sample data are included in chapters 4 through 6 of the tutorial. The package is under continuous improvement, so bug reports, feature requests, and contributions are actively sought at our code repository (bitbucket.org/petersmp/rnaseqwrapper). To further ease the learning of R, we also include a brief introduction to using R (Supplemental Materials S3) along with sample data files (Supplemental Materials S6).
Intended Audience: This tutorial can be effective for a wide variety of audiences, including undergraduates and biology faculty who are novices to RNAseq analysis. Here, we focus on the implementation of this tutorial in an experimental undergraduate course in bioinformatics at a small liberal arts school (Juniata College). This course included sophomore, junior, and senior biology majors, all of whom had taken at least two other biology courses. In the discussion, we describe some alternative implementations. This tutorial will likely work best in a lab section or class sessions with no more than 10 to 15 students per instructor (i.e. professor or well-trained TA), as questions inevitably arise that require intervention from the instructor.
Because, each chapter focuses on accomplishing specific bioinformatic endpoints, rather than on teaching the related genome biology, the specific student backgrounds are flexible. Brief comments within the text and links to further information serve as starting points for in-class lectures and discussions (such as those included in the online repository with the tutorial), rather than ends unto themselves. In this way, the tutorial can be geared to match student needs. These differences may be related to student background (e.g., a 100 or 200 level biology class will need much more background than a graduate level class) or student interests (e.g., a course for computer scientists will focus more on the Linux implementations than the underlying biology that may interest a group of biologists). In this way, this tutorial is designed to serve as a technical supplement for a course rather than as a syllabus for a standalone course.
Learning time: Each chapter takes approximately one to two hours of work to complete, depending on the student background. The exception is the first chapter, which includes links to supplemental instruction that will take additional time to complete. These additional pieces can either be completed in class (if all students will require them), or outside of class (if only a few students require the remediation).
We spent approximately half of the semester working through this material. We covered roughly one chapter per class week, devoting the equivalent of one three-hour lab time to work on the tutorial. This lab was supplemented with additional concept-oriented lectures and discussions that are not included in this tutorial. Samples of these materials are available in the online repository with the tutorial.
The workflow of a standard unit began with a brief (roughly ten minute) overview of the concepts and goals for the lesson. This introduction was generally in a discussion format without prepared slides and often centered on a primary research article including those describing the tools we were about to use (as referenced in the manual) and other cutting edge articles relevant to the interests of the class. (See the samples listed in the online repository.) Later in the semester, students were assigned to lead these discussions. After the introduction, the students then worked individually and at their own pace through the tutorial. As problems and questions arose, they worked together to solve them with guidance from the instructor as needed. At the end, students compared their results and discussed any differences they noted. Later chapters require the students to use data generated by all of the other students, so they are generally motivated to help make sure the work goes smoothly for everyone.
At the conclusion of this tutorial, students had developed sufficient skills to engage in a student-led data-mining research project during the second half of the semester. The students were given a brief introduction to the GEO and Sequence Read Archive [2] and asked to propose a potential project for the class. The proposals were then presented informally to the class, with instructor-provided feedback on the design and feasibility. The students then voted on a project to pursue for the rest of the semester. The chosen project involved analysis of NCBI Gene Expression Omnibus data from two different species with X monosomy (Turner Syndrome); the results of the research project are currently in preparation for publication. A similar approach could be utilized to find projects of appropriate scope and difficulty for just about any class. Making this goal explicit also helped to motivate the students to learn the data analysis skills: they weren't just doing the lessons because they were told to, but rather to develop the skills they would need to address a research project of their own choosing.
Pre-requisite student knowledge: This tutorial focuses on teaching the computer competencies necessary to conduct an RNAseq analysis. A basic working knowledge of computers is required (e.g., how to install programs on your own computer). Since most of the actual computation is performed on super computers, only basic computers are needed to complete the tutorial. Only three computer programs need to be installed on the computers used by the students: R, RStudio, and a program to connect to other computers via ssh connections. (Linux and Mac computers should already have an ssh connection program in default installations.) No background in Linux, R, or computer programming is assumed, since these skills are taught to the extent that they are required. It is important to note that this lesson is no substitute for an actual computer programming class.
Together with a basic working knowledge of computers, students should have a basic background knowledge of statistics and biology, such as would be gained in a one semester introductory course in each discipline. The specific biology and statistics concepts necessary for the learning goals of a particular course could be taught alongside this tutorial without impeding student progress on the computer work. Students with more extensive backgrounds in biology, statistics, or computer science will find much of the content easier to understand, though a lack of that knowledge will not prevent them from completing the lessons
This tutorial also assumes that the instructor has a strong background in these fields. It is strongly recommended that the instructor using these materials be comfortable with the general concepts being taught, and with the requisite skills and tools (e.g., Linux, R, and general principles computer programming). Working through the tutorial completely may be sufficient. In addition, there are a number of options to develop those skills, including workshops offered by GCAT-SEEK each summer where this tutorial is used to teach faculty.
SCIENTIFIC TEACHING THEMES
This tutorial was implemented as part of a Course-Based Undergraduate Research Experience (CURE). CURE's are emerging as an effective way to directly engage large numbers of students in the research process and networks are developing to share the resources necessary to conduct them [24]. As such, this tutorial is primarily a hands-on tool, pushing students to directly interact with the data and including all members of the class in the analysis. The end goal for this tutorial was to conduct an independent research project that uses the tools and concepts learned.
Active Learning
All students move through the tutorial on their own, with instructor guidance available when they hit problems. Additional instruction, such as lectures or discussions on the underlying biology, may be necessary, but the primary learning goals of each chapter are actively explored and accomplished directly by the student. In addition, students will troubleshoot problems as they arise, particularly when a second student encounters similar errors. These troubleshooting steps are not intentionally inserted, but students tend to make small errors when running the commands that cause errors until they are identified.
Also contributing to active learning, student peers help with the troubleshooting process, which appeared to help both the helper and the struggling student to learn in more depth. Instructors, particularly in larger classes, may wish to formalize this teamwork approach using the “Jigsaw” technique [25], which stresses the individual contribution of each team member to a larger goal. This approach may also alleviate the load on instructors in larger classes. Finally, the entire analysis, from raw sequence data to downstream interpretation, is carried out directly by the students, creating substantial buy-in and individual investment. This buy in can be further increased by utilizing data for a research question that reflects the student’s interest; many data sets, representing a broad variety of data, are freely available online for the students to use to answer their own questions.
Assessment
For an active learning tutorial such as this, our major direct assessment was successful completion of the required task. Students were allowed as much time as they needed to obtain the result, and were provided with assistance as needed. Additional course assessments – including quizzes, homework, syllabus, and reports – were structured primarily around the concepts being taught alongside the tutorial, but competencies students developed in the tutorial were used to answer the questions.
We used open-everything (notes, internet, etc.) quizzes roughly weekly to assess student knowledge and ability, including asking them to troubleshoot deliberately created problems. For example, after being introduced to R, the students were given a quiz that asked them to modify an existing script so that it would work with a new data format, a difficult process for novice users. This quiz, along with other assignments, is posted as an example along with several other assignments that were utilized in the course (Supplementary Materials S8; we will continue to add additional examples on our website https://bitbucket.org/petersmp/publishedtutorials/).
The tutorial also requires students to create a large number of files that can be readily evaluated for completion and accuracy. The final portion of grading was participation, providing a solid approach to reward students who were most actively involved, including those who assisted others in troubleshooting. Our participation grade was a holistic, subjective assessment of the degree to which students were engaged during class time with particular focus on their effort to overcome challenges and assist other students.
Inclusive Teaching
The successful completion of this tutorial requires every student to complete their assigned portion of the analysis, enforcing inclusion and encouraging cooperation in overcoming hurdles.
In addition, as with most STEM disciplines, it is important to draw explicit connections to the diversity in the field. According to the non-profit group ScriptEd, only 1 in 10 high schools in America teaches computer science and this number is even lower in minority serving schools (www.scripted.org). This deficiency increases the value of engaging undergraduates in the computer programming side of bioinformatics, where there can be a large diversity gap. We strongly recommend that instructors pay special attention to the diversity of those who have contributed to the field. From Ada Lovelace, the woman who wrote the first computer program, through the tragic arc of Alan Turing's life and punishment for his sexuality, to the current diversity of contributors in the field of bioinformatics, there are many examples to choose from. Such discussions fall outside the scope of the technical approach in our tutorials, but are likely to be a great way to draw in the human-interest side of many students.
LESSON PLAN
The best preparation for teaching this tutorial is to work through it yourself. Many of the common pitfalls will become apparent and the additional readings will prepare you for many of the questions the students are likely to raise during the class. Before working on the tutorial, you will need to: i) obtain access to a Linux computer cluster; ii) download the data; iii) confirm that all of the steps work at your location; and iv) change any necessary steps in the tutorial to reflect your particular Linux system. Each of these steps is detailed below (for an overview, see Table 1).
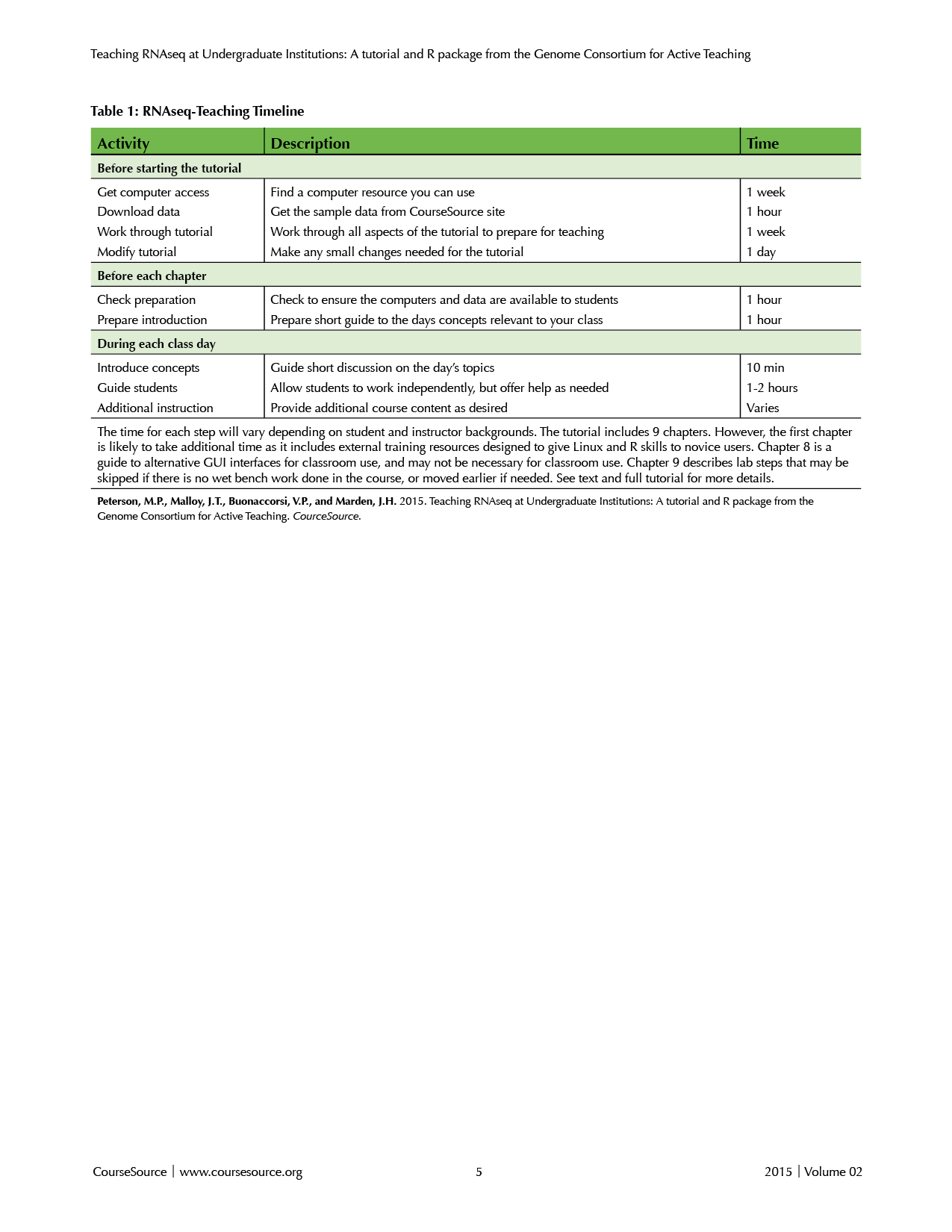
Table 1. RNAseq-Teaching Timeline
Apart from resources that may be available at one’s home institution, many systems provide free public access to Linux supercomputer resources for education, including the National Center for Genome Analysis Support (NCGAS), the iPlant collaborative, and the GCAT-SEEK network. Contact one of those groups for more information on how to meet the specific needs of your classroom.
Once you have access to a Linux computer cluster, download the data (Supplemental material S1) to the system, either directly using wget or download to your local machine and transfer using scp (discussed in chapters 1 and 2 of the tutorial). Unpack the data (e.g., tar -zxvf fileName.tar.gz) and you are ready to get started.
Because each Linux system is a bit different, you will need to work through the tutorial on your particular system to ensure that all of the steps work properly, particularly the installation steps. To assist in the analysis, the online repository contains information on how to conduct the analysis in parallel for all of the samples. This parallel analysis will allow an instructor to see the final results expected from each student. In addition, a sample of a completed R tutorial is included in the bitbucket repository with the source code. If any of these steps cause problems, please contact the corresponding author or the GCAT-SEEK network for help. For any steps that do not work smoothly on your system, the information in the manual pages linked within the tutorial, combined with online searches, should solve the issues you may be having. It will be important for you to edit the manual to reflect the differences that you discover, because novice users (like the intended student audiences) rely heavily on very specific directions.
To change the tutorial to reflect any installation issues and to edit directory names to match the computer you end up using for your class, you will need to download and edit the LaTeX source files for the tutorial (Supplemental Materials S5). For this purpose, you will need a version of LaTeX for your local computer, but no knowledge of LaTeX should be necessary to make your changes. Install a LaTeX flavor based on the instructions from a Google search for “install LaTeX on XXXXX” where XXXXX is the type of computer you have (Ubuntu, Red Hat, Debian, Fedora, Mint, Mac, Windows, etc.).
Making changes in LaTeX files is simple, as the files are plain text. The data directory is defined as a variable in the main LaTeX file “sequenceAnalysis.tex” near the top, with comments explaining what to change. Simply change the directory to match yours, and run the conversion to pdf again. Any other changes will occur within the files for the given chapters, but should be easy to find and modify with any plain text editor or LaTeX IDE (for a partial list of options, see http://tex.stackexchange.com/questions/339/latex-editors-ides ). Make your changes and re-run the pdf conversion on the main document.
It is important to consider that the preparation for teaching the course will take several days or even weeks, as setting up accounts on new systems is not always an easy process (and often each student will need an account), so give yourself plenty of time. If you are planning to work through this tutorial in class, I would expect the first chapter, which includes two rather long computer tutorials (for Linux and R), to take approximately two weeks of class time (assuming at least 3 hours of lab time per week devoted to the tutorial). After that, working through at a pace of about a chapter per week (in a two or three hour lab) should provide sufficient time for the tutorial with additional instruction in the lab and lecture periods.
TEACHING DISCUSSION
In the end, students appeared to enjoy this lesson and learn a lot from it. Out of four students completing the CURE in which early versions of this tutorial were used, the three juniors secured summer research fellowships working with bioinformatics data and have continued on in similar research. The goals of the tutorial and the parallel instruction appear to be reflected in the student evaluations. One undergraduate from the course remarked that “This was by far the most exciting, technically difficult class I had this semester … It has been a boost of confidence in a skill I could never imagine myself learning.”
Of the faculty who attended one of the two workshops in which this set of tutorials was taught, many were amazed at how quickly they picked up the computer programming portions, despite initial hesitation at learning a new operating system and the R computer language. All of the faculty have, to the best of our knowledge, continued on to analyze sequence data from their own research projects.
In spite of the ultimate positive outcomes, initial misgivings and push back are likely to be common when this tutorial is used at any level. The transition from graphical user interfaces (GUI) to command line can be intimidating for many novice computer users. However, by helping students to slowly build those skills, and by reminding them why they are learning the skills, the instructor can help students gain confidence. In fact, many who have used this tutorial have begun to use their new skills even beyond the original scope in which they were taught. For example, several students are now leveraging the RNA-seq training to explore population genomics and other big data projects.
Alternative Implementations: In addition to the undergraduate course, this tutorial has been used in other contexts and could be readily extended to more. One of the primary motivations of the GCAT-SEEK group, for which this tutorial was developed, is to train faculty to bring next generation sequencing tools to undergraduate classrooms. To this end, this tutorial is also being used as part of the GCAT-SEEK faculty workshops. Each chapter can also stand on its own if an instructor wants to use just a small portion. The tutorial also includes a chapter describing graphical interfaces that may be better suited to implementation in classes less heavily focused on bioinformatics training. These implementations each present unique challenges and opportunities.
The faculty workshops are much shorter than the course, lasting only a week, and thus necessarily provided much less information outside of the tutorials. As such, the focus was on learning the general process and how to find the way to analyze your own data. Faculty evaluation comments reflected that approach, stating that the workshop “was immersive ... I don't know if I will be able to replicate all of the steps we took, but I do feel that I will be able to find the right information for future analyses.” Another participant responded that “Overall, I feel more confident in my knowledge and ability to execute genomics datasets, and that I can perform my own work to look up the appropriate information.” A similar approach has also been used in training summer undergraduate researchers, providing a short, intense immersion in these tutorials, followed by development of skills and concepts needed for the research project at hand.
It is also possible to work through any single chapter of this tutorial for an individual class. To make this use easier, there are scripts (with clear directions in an included README file) to run all of the analysis steps in bulk (Supplemental material S7). Running these scripts up to the point where you expect students to take over will provide all of the files they need to continue on. In addition, these scripts can serve as a guide to running bulk analysis of RNAseq samples for research purposes, or as part of an independent project in class. We also include the final output files from the read mapping and SNP analysis portions (Supplemental material S2), if you would prefer students started on the analysis in R, rather than generating the files themselves.
The final chapter of the tutorial reviews several GUI-based approaches for RNAseq analysis, describing their strengths and limitations. It is important, especially for researchers or students intending to pursue careers involving bioinformatics, to learn the Linux command line and R-based tools that underlie nearly all next-generation sequence analysis. However, these GUI approaches may provide an appropriate introduction to these methods in a class with less bioinformatics focus, so we encourage readers to explore them. Having completed the tutorial in Linux will provide the preparation to address issues that arise within those GUI-based programs.
Changes to the tutorial: Several things about this tutorial have changed since it was initially developed. Perhaps the largest is the de-emphasis of GUI tools, which may at first seem counterintuitive. In the first implementation of this tutorial, many participants latched onto the GUI tools, and so were less motivated to overcome the challenges of learning Linux. However, after completion of the course and their attempts to use the GUI tools for full-scale analysis, they faced sufficient challenges to return to the Linux approaches. In addition, a number of changes were made as it became clear which challenges students faced in learning the material. If you encounter any similar hurdles, either for your own work of that of your students, please contact us so that we may update this instrument to serve as wide an audience as possible.
SUPPLEMENTAL MATERIALS
In addition to the versions archived with this publication, we will continue to update these tutorials at https://bitbucket.org/petersmp/publishedtutorials.
- Supplemental File S1. RNAseq–Sequence data files (tar.gz, a little over 100 MB) will be provided separately before publication. We have included sample data from 16 dark-eyed juncos, 8 males and 8 females (each sex labeled from A to H). All samples are from the livers of captive animals, though all samples are limited down to a subset of expressed genes to increase the speed of data processing steps. These data are a good data sample set, though we encourage other researchers to utilize data that is directly relevant to their classrooms (e.g., sequence data from your own research or retrieved from a public repository for an organism, disease state, or treatment condition that is of interest to the class and/or students).
- Supplemental File S2. RNAseq–dataForStartingAtR.zip is a zipped directory of the data files from the read mapping and SNP analysis, for use starting with R
- Supplemental File S3. RNAseq–learningR.pdf is the R tutorial PDF
- Supplemental File S4. RNAseq–rnaseqTutorial.pdf is the RNAseq tutorial PDF
- Supplemental File S5. RNAseq–tutorialSourceFiles.zip is a zipped directory with the LaTeX source files for the tutorial, along with scripts for parallelization.
- Supplemental File S6. RNAseq–data.zip is a zipped directory with the data files for the R tutorial as comma-separated values (csv) files
- Supplemental File S7. RNAseq–parallelScripts.zip is zipped directory containing scripts to run all analyses in parallel intended to prepare data files for starting at a later stage in the tutorial, showing the instructor what outputs to expect, and/or show researchers how to apply these approaches to their own work.
- Supplemental File S8. RNAseq–sampleTeachingMaterials.zip is a zipped directory containing sample assessments and keys for the completion of tutorial steps. Additional resources will continue to be added to our website at https://bitbucket.org/petersmp/publishedtutorials/
ACKNOWLEDGMENTS
This work was funded by the National Science Foundation (DBI-1248096; DEB-1120476; and IOS-1354667), a grant to Juniata College from the Howard Hughes Medical Institute through the Precollege and Undergraduate Science Education Program, and the Huck Institutes of the Life Sciences, at Pennsylvania State University. There are no potential conflicts of interest. We would also like to thank the other instructors at the GCAT-SEEK workshops, Nancy Trun, Jeff Newman, Tammy Tobin, and Regina Lamendella, for their support in development of the program and this tutorial.
References
- Boyle, M.D. (2010). “Shovel-ready” Sequences as a Stimulus for the Next Generation of Life Scientists. In Journal of Microbiology & Biology Education: JMBE 11, p. 38.
- Davey, J.L., and Blaxter, M.W. (2010). RADSeq: next-generation population genetics. In Briefings in Functional Genomics 9, p. 416-423.
- McCormack, J.E., Maley, J.M., Hird, S.M., Derryberry, E.P., Graves, G.R., and Brumfield, R.T. (2012). Next-generation sequencing reveals phylogeographic structure and a species tree for recent bird divergences. In Molecular Phylogenetics and Evolution 62, p. 397-406.
- Vera, J.C., Wheat, C.W., Fescemyer, H.W., Frilander, M.J., Crawford, D.L., Hanski, I., and Marden, J.H. (2008). Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. In Molecular Ecology 17, p. 1636-1647.
- Meyer, E., Aglyamova, G.V., Wang, S., Buchanan-Carter, J., Abrego, D., Colbourne, J.K., Willis, B.L., and Matz, M.V. (2009). Sequencing and de novo analysis of a coral larval transcriptome using 454 GSFlx. In BMC Genomics 10, p. Article No.: 219.
- Chan, Y.F., Marks, M.E., Jones, F.C., Villarreal, G., Shapiro, M.D., Brady, S.D., Southwick, A.M., Absher, D.M., Grimwood, J., Schmutz, J., Myers, R.M., Petrov, D., Jonsson, B., Schluter, D., Bell, M.A., and Kingsley, D.M. (2010). Adaptive Evolution of Pelvic Reduction in Sticklebacks by Recurrent Deletion of a Pitx1 Enhancer. In Science 327, p. 302-305.
- Peterson, M., Whittaker, D., Ambreth, S., Sureshchandra, S., Mockatis, K., Buechlein, A., Choi, J., Lai, Z., Colbourne, J., Tang, H., and Ketterson, E. (2012). De novo transcriptome sequencing in a songbird, the dark-eyed junco (Junco hyemalis): Genomic tools for an ecological model system. In BMC Genomics 13.
- Peterson, M.P., Rosvall, K.A., Choi, J.-H., Ziegenfus, C., Tang, H., Colbourne, J.K., and Ketterson, E.D. (2013). Testosterone affects neural gene expression differently in male and female juncos: A role for hormones in mediating sexual dimorphism and conflict. In PLOS ONE 8, p. e61784.
- Peterson, M.P., Rosvall, K.A., Taylor, C.A., Lopez, J.A., Choi, J.-H., Ziegenfus, C., Tang, H., Colbourne, J.K., and Ketterson, E.D. (2014). Potential for sexual conflict assessed via testosterone-mediated transcriptional changes in liver and muscle of a songbird. In The Journal of experimental biology 217, p. 507-517.
- Kodama, Y., Shumway, M., and Leinonen, R. (2012). The Sequence Read Archive: explosive growth of sequencing data. In Nucleic acids research 40, p. D54-D56.
- Meyer, F., Paarmann, D., D'Souza, M., Olson, R., Glass, E.M., Kubal, M., Paczian, T., Rodriguez, A., Stevens, R., Wilke, A., Wilkening, J., and Edwards, R. (2008). The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. In BMC Bioinformatics 9, p. 386.
- Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. In Nature Reviews Genetics 10, p. 57-63.
- Robles, J.A., Qureshi, S.E., Stephen, S.J., Wilson, S.R., Burden, C.J., and Taylor, J.M. (2012). Efficient experimental design and analysis strategies for the detection of differential expression using RNA-Sequencing. In BMC Genomics 13, p. 184.
- Wit, P., Pespeni, M.H., Ladner, J.T., Barshis, D.J., Seneca, F., Jaris, H., Therkildsen, N.O., Morikawa, M., and Palumbi, S.R. (2012). The simple fool's guide to population genomics via RNA-Seq: an introduction to high-throughput sequencing data analysis. In Molecular ecology resources 12, p. 1058-1067.
- Woodin, T., Carter, V.C., and Fletcher, L. (2010). Vision and change in biology undergraduate education, a call for action—initial responses. In CBE-Life Sciences Education 9, p. 71-73.
- Buonaccorsi, V.P., Boyle, M.D., Grove, D., Praul, C., Sakk, E., Stuart, A., Tobin, T., Hosler, J., Carney, S.L., Engle, M.J., and others (2011). GCAT-SEEKquence: genome consortium for active teaching of undergraduates through increased faculty access to next-generation sequencing data. In CBE-Life Sciences Education 10, p. 342-345.
- Buonaccorsi, V., Peterson, M., Lamendella, G., Newman, J., Trun, N., Tobin, T., Aguilar, A., Hunt, A., Praul, C., Grove, D., and others (2014). Vision and Change through the Genome Consortium for Active Teaching Using Next-Generation Sequencing (GCAT-SEEK). In CBE-Life Sciences Education 13, p. 1-2.
- Campbell, A.M., Ledbetter, M.L.S., Hoopes, L.L., Eckdahl, T.T., Heyer, L.J., Rosenwald, A., Fowlks, E., Tonidandel, S., Bucholtz, B., and Gottfried, G. (2007). Genome Consortium for Active Teaching: meeting the goals of BIO2010. In CBE-Life Sciences Education 6, p. 109-118.
- Anders, S., and Huber, W. (2010). Differential expression analysis for sequence count data. In Genome Biology 11, p. R106.
- Alexa, A., and Rahnenfuhrer, J. (2010). topGO: Enrichment analysis for Gene Ontology. In .
- Charif, D., and Lobry, J. (2007). SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis.. In Bastolla, U., Porto, M., Roman, H., and Vendruscolo, M. (Ed.), Structural approaches to sequence evolution: Molecules, networks, populations, Springer Verlag.
- Warnes, G.R., Bolker, B., Bonebakker, L., Gentleman, R., Liaw, W.H.A., Lumley, T., Maechler, M., Magnusson, A., Moeller, S., Schwartz, M., and Venables, B. (2014). gplots: Various R programming tools for plotting data. In .
- Goslee, S.C., and Urban, D.L. (2007). The ecodist package for dissimilarity-based analysis of ecological data. In Journal of Statistical Software 22, p. 1-19.
- Auchincloss, L.C., Laursen, S.L., Branchaw, J.L., Eagan, K., Graham, M., Hanauer, D.I., Lawrie, G., McLinn, C.M., Pelaez, N., Rowland, S., Towns, M., Trautmann, N.M., Varma-Nelson, P., Weston, T.J., and Dolan, E.L. (2014). Assessment of Course-Based Undergraduate Research Experiences: A Meeting Report. In CBE Life Sci Educ 13, p. 29:40.
- Aronson, E., Blaney, N., Stephin, C., Sikes, J., and Snapp, M. (1978). The jigsaw classroom.. Sage.
Article Files
Login to access supporting documents
Teaching RNAseq at Undergraduate Institutions: A tutorial and R package from the Genome Consortium for Active Teaching(PDF | 143 KB)
Supplemental Material S1 Part1. RNAseq-sampleData.zip(ZIP | 59 MB)
Supplemental Material S1 Part2. RNAseq-sampleData.zip(ZIP | 58 MB)
Supplemental Material S2. RNAseq-dataForStartingAtR.zip(ZIP | 344 KB)
Supplemental Material S3. RNAseq-learningR_2014Nov03.pdf(PDF | 384 KB)
Supplemental Material S4. RNAseq-rnaseqTutorial_2014Nov03.pdf(PDF | 761 KB)
Supplemental Material S5. RNAseq-tutorialSourceFiles.zip(ZIP | 1 MB)
Supplemental Material S6. RNAseq-data.zip(ZIP | 4 KB)
Supplemental Material S7. RNAseq-parallelScripts.zip(ZIP | 4 KB)
Supplemental Material S8. RNAseq-sampleTeachingMaterials.zip(ZIP | 28 MB)
- License terms

Comments
Comments
There are no comments on this resource.