Follow the Sulfur: Using Yeast Mutants to Study a Metabolic Pathway
Published online:
Abstract
Students are frequently overwhelmed by the complexity of metabolic pathways and they think they have "learned" the pathway when they have memorized the individual reactions. This laboratory lesson helps students to understand the significance of individual reactions in the pathways leading to methionine synthesis in the budding yeast, Saccharomyces cerevisiae. Students appreciate that methionine is one of only two sulfur-containing amino acids, and students do not find it difficult to follow the "yellow" sulfur atom in the pathway. In the lesson, students use three different yeast met strains, each of which lacks a single gene involved in methionine synthesis. Working in groups of three, students identify the missing MET gene in each of the three deletion strains by analyzing the abilities of the deletion strains to grow on several defined media in which methionine has been replaced with alternative sulfur sources. Students also determine the position of mutant genes in the pathway relative to sulfite reductase, using indicator media that reacts with sulfide, the product of the reaction catalyzed by sulfite reductase. For the analysis, students prepare serial dilutions of yeast cultures and spot the dilution series on agar plates. This lesson is part of a semester-long research investigation into the evolutionary conservation of the genes involved in methionine synthesis. The lesson can also be used as a stand-alone exercise that teaches students about biochemical pathways, while reinforcing basic microbiological techniques.
Citation
O'Connor CM. 2016. Follow the sulfur: Using yeast mutants to study a metabolic pathway. CourseSource. https://doi.org/10.24918/cs.2016.23
Society Learning Goals
Biochemistry and Molecular Biology
- Energy is required and transformed in biological systems
- How is energy of chemical processes coupled in metabolic pathways?
Genetics
- Genetic variation
- How do different types of mutations affect genes and the corresponding mRNAs and proteins?
Science Process Skills
- Process of Science
- Pose testable questions and hypotheses to address gaps in knowledge
- Plan, evaluate, and implement scientific investigations
- Interpret, evaluate, and draw conclusions from data
- Construct explanations and make evidence-based arguments about the natural world
- Address novel questions through authentic research experiences
- Modeling/ Developing and Using Models
- Recognize the important roles that scientific models, of many different types (conceptual, mathematical, physical, etc.), play in predicting and communicating biological phenomena
- Make inferences and solve problems using models and simulations
- Build and evaluate models of biological systems
- Quantitative Reasoning/ Using Mathematics and Computational Thinking
- Apply the tools of graphing, statistics, and data science to analyze biological data
- Communication and Collaboration
- Share ideas, data, and findings with others clearly and accurately
Lesson Learning Goals
Students will:
- understand the metabolic pathways involved in yeast methionine and cysteine biosynthesis.
- be able to predict how mutations in specific genes affect the ability of yeast to grow on various sulfur sources.
Lesson Learning Objectives
At the end of this lesson, students will be able to:
- use spot plating techniques to compare the growth of yeast strains on solid culture media.
- predict the ability of specific met deletion strains to grow on media containing various sulfur sources.
- predict how mutations in specific genes will affect the concentrations of metabolites in the pathways involved in methionine biosynthesis.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
The introduction of research experiences into undergraduate laboratory courses has been shown to positively affect student learning, particularly in introductory classes (1-3). Course-based undergraduate research experiences (CUREs) vary significantly in the degree to which students participate in an authentic research investigation (1, 3-5), ranging from guided inquiry to an independent research project. This lesson is part of an introductory laboratory course, Investigations in Molecular Cell Biology that engages ~180 students in a semester-long research project in functional genomics. In the course project, students study the evolutionary conservation of genes involved in methionine synthesis, ultimately using complementation of Saccharomyces cerevisiae met deletion strains to test gene function. The course design makes use of the information and resources generated by whole genome sequencing (WGS) projects. WGS projects are highly automated projects that use computational methods to identify potential genes and to predict their functions, based on their similarities to known proteins (6). Since functional tests of a gene product's predicted activity are beyond their scope, WGS projects for many species provide a wealth of potential research questions for undergraduate investigation.
S. cerevisiae is an ideal choice for undergraduate research, since it is non-pathogenic, inexpensive, easy to culture, and it has a long history as a model organism (7). Because of its commercial importance in fermentation, yeast have been actively studied by microbiologists and genetics since the beginning of the 20th century. By the time that the yeast genome sequence was published in 1996 (6), geneticists had already elucidated many metabolic pathways, including the genes involved in the synthesis of sulfur-containing amino acids, cysteine and methionine (8). After the S. cerevisiae genome sequence became available, the Saccharomyces community produced well-designed and experimentally validated collections of genome-wide mutant strains (9) and clones (10). These collections were made available to researchers at low cost, thereby enabling a wide range of experiments.
This lesson, which can be described as a guided inquiry (1,4), typically occurs in the third week of the course, after students have learned to use micropipettes and have some experience with sterile culture techniques. The lesson uses a set of yeast deletion strains (9,11), each of which possesses a targeted deletion in one of the genes involved in methionine (Met) biosynthesis (8). Student groups are given a set of three deletion strains and charged with identifying which of three MET genes has been deleted in each strain. Students are reminded that professional scientists will likewise verify their strains prior to beginning an experiment. In this lesson, students identify the strains by their ability to grow on a variety of defined and indicator media. Some media contain other sulfur sources in the place of Met. Mutant strains that are able to grow on the alternative sulfur sources carry deletions in genes that are upstream of the particular sulfur source in the pathway. The Met/Cys pathway (Figure 1) is particularly easy for students to follow because of the sulfur atom in their precursors. Students can visualize the "yellow" atom that stands out in molecular structures and they have probably learned about the Hershey and Chase's classic "blender experiment" in which the presence of sulfur was used to distinguish proteins from DNA (12).
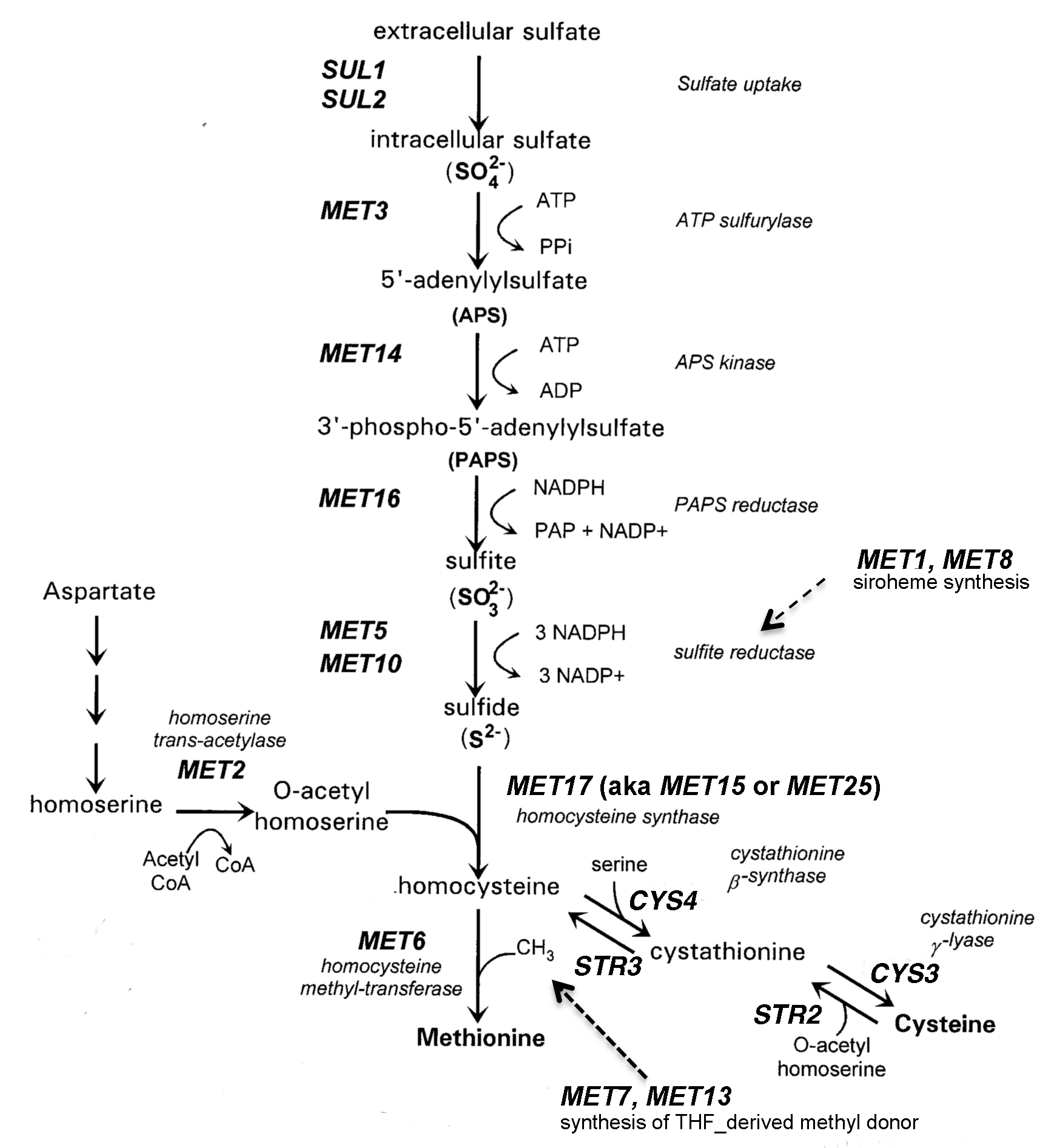
Figure 1. Synthesis of sulfur-containing amino acids in S. cerevisiae.
The experiment reinforces core concepts of biology associated with information transfer, metabolic pathways and systems biology (13). Students also acquire some core competencies of professional scientists as they perform the experiment, analyze the results and communicate their findings in an oral presentation and written report. This lesson encompasses the entirety of one three-hour class period and lesser amounts of class time in two following class sessions. The lesson uses a "flipped classroom" approach in which students are required to read background information and complete an online quiz about methionine synthesis prior to the first class. At the beginning of the second class, students record their data and discuss it with their team members. In the third session, teams present their data to the entire section. They use feedback from their TA and classmates as they prepare a short written report. In the Boston College class, this lesson is followed by one in which students use PCR to verify their strain identifications (14).
Intended Audience
This lesson is part of a large introductory laboratory course in molecular cell biology at a research university. The course meets twice each week in three-hour sessions. Most of the students are sophomore biology or biochemistry majors. Other students are interested in attending medical school after graduation, although they are not majoring in the life sciences. For almost all the students, this is their first college biology laboratory.
Required Learning Time
The module typically spans three class periods at Boston College. (See the teaching timeline in Table 1) Most of the work is done during the first session, requiring two to three hours. A period of five to seven days separates sessions 1 and 2, which allows sufficient time for cells to grow on the solid media. Students record their data during the second session, which requires ~ 30 minutes. Students present their results during ~30 minutes of the third lab period.
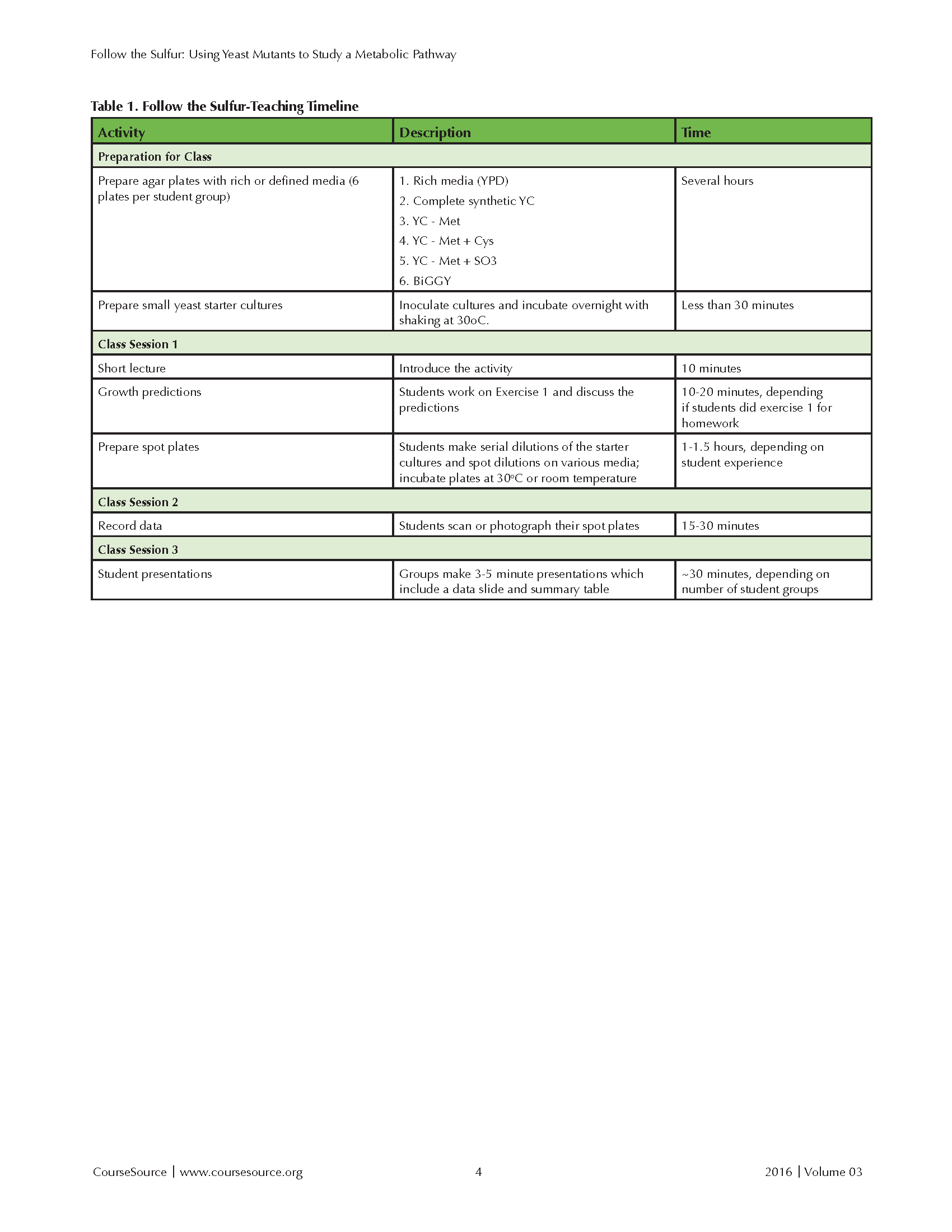
Table 1. Follow the Sulfur - Teaching Timeline
Pre-requisite Student Knowledge
Students are expected to have been introduced to molecular cell biology and genetics concepts in an introductory biology course. Students are expected to have learned basic laboratory skills during a semester of freshman chemistry laboratory.
Pre-requisite Teacher Knowledge
Instructors with a graduate degree in biochemistry or biology will be able to teach this lesson. Instructors should have a basic knowledge of metabolic pathways as well as an understanding of how selective media can be used to identify which steps in a pathway are affected in mutant strains. Instructors should also be familiar with basic microbiological methods used for culturing microorganisms, such as the preparation of culture media, sterile transfer techniques, serial dilutions and plating.
SCIENTIFIC TEACHING THEMES
Active Learning
Before class, students read background material on methionine synthesis and yeast strains, prepare lab notebooks, and complete an online quiz. During class, students predict the growth properties of mutant strains on various media, prepare spot plates of yeast strains on with various sulfur sources, and analyze the growth of mutant strains on selective and indicator media.
Assessment
Before class, students take an online quiz and prepare a lab notebook that is checked for completion at the beginning of the first class session. Students analyze their experimental results and present their findings in both an oral presentation and a written report. The focus of the report is a data figure prepared in the format of a scientific publication and a summary table. Reports are limited to two pages, excluding the figure and summary table.
Inclusive Teaching
Students work in teams that are organized with balance (major, gender, graduation year) in mind. At the beginning of the semester, students self-describe their talents with the following prompt: “ If you were working in a widget factory, rate yourself in the following roles: CEO, widget designer, marketing guy, or widget fabricator." These responses are used to help compose balanced teams. Students typically work in teams of three. The activities in the module accommodate a variety of learning styles. Pre-class materials include readings and an online quiz. Class activities are hands-on, and groups can organize their work to build on individual strengths.
LESSON PLAN
This lesson involves one full laboratory period and small segments of two following lab periods. A sample timeline is shown in Table 1. Students prepare the spot plates during the first laboratory period, collect the data during the second lab period, and present their data during the third lab period. The first lab can be completed in 2-3 hours, depending on the experience of the students with sterile culture techniques. The time required for the first class can be shortened by having students complete the prediction exercise in the student reading before the first lab class.
BEFORE CLASS
Several weeks in advance
Teachers should order the yeast deletion strains (9,11) needed for the experiment. (See Supporting File S1 for ordering information.). The major work required for this lesson is the preparation of agar plates containing various growth media (see Supporting File S2). The culture techniques used in this lesson are standard ones that are familiar to most instructors. Before class, instructors will probably find it helpful to read the background material provided to students on yeast methionine synthesis (see Supporting File S3) and preparing spot plates (see Supporting File S4). We generally follow the timeline below to prepare for class.
Several days in advance
The microbiological techniques required for the lesson are standard procedures that should not require practice. Instructors will need to prepare or arrange to have prepared one set of 6 plates for each team of students: YPD, YC (complete synthetic media), YC-Met, YC-Met+Cys, YC-Met+SO3 and BiGGY indicator agar (see Supporting File S2). Plates should be prepared several days in advance so that the media has sufficiently dried so that the "spots" with yeast will dry quickly on the plates and not run into each other.
Day before class
Instructors should prepare or arrange to have prepared liquid cultures of deletion mutants and the parental BY4742 strain in YPD medium. It is convenient to start these cultures the previous day so that the cultures have reached stationary phase by the time of the experiment. Stationary cells will remain viable for at least several days if the cultures are stored at 4 ˚C.
DURING CLASS
Class 1 - Spot plating of strains on selective and indicator media
Each group of students should be given small (<1 mL) cultures of three different deletion strains in rich media for the experiments. The cultures are labeled with the strain names but not the genotypes of the strains. Students are provided with the names of the genes that are deleted in the three strains, but they do not know which gene is defective in each strain. Students will use their experimental data to associate each strain with a particular deletion. We usually give teams strains carrying mutations in different parts of the pathway so that the students will observe three distinct growth patterns. For example, a team might be given a met3 strain that is able to use either sulfite or Cys as its sulfur source, a met5 strain that is able to use Cys as a sulfur source, and a met6 strain that grows only on plates containing Met.
Predict the growth properties of mutant strains (10-20 minutes)
If the students have not already done so, have the students complete the table in Exercise 1 of the student reading. Students should discuss their individual predictions with their teams for a few minutes to develop a consensus, after which the instructor leads a class discussion. I find it helpful to draw the grid from Exercise 1 on the board and to record the class predictions during the class discussion. (Answers to Exercise I are included in Supporting File S5.)
Spot plate the diluted strains (1-1.5 hr, depending on student proficiency)
Students typically work in groups of three. Each student in the group should assume responsibility for preparing the dilutions and spotting ONE of the three deletion strains. This ensures that all students will develop proficiency with the procedures. Teams can decide how to divide responsibilities for the parent strain.
Cultures will be spotted on six different media. Stress to students the importance of (1) labeling the plates with their initials and strain numbers and (2) using the same pattern of strains/rows on all six plates. Students should not invert the plates for ~30 minutes after spotting them, which allows time for the yeast cells to settle and attach to the media.
Plates can be incubated either at 30 ˚C or room temperature, but should not be allowed to over-grow. S. cerevisiae should NOT be grown at or above 37 ˚C, which induces a stress response. The growth rate of the cells varies with the media. Cultures grow most rapidly on YPD and slowest on media containing SO3 as the sulfur source. Plates are ready when individual colonies are evident in the most dilute spots.
Class 2 - Students record their data and being working on their presentation slide (~30 minutes)
Students record the growth of their strains using either a scanner or their cell phone cameras. Cameras give a wide range of variability, so we prefer to have students use the scanner. Students arrange the plates on the scanner in the same orientation, so results obtained on the various media can be easily compared. Depending on the resolution of your scanner, you can have students remove the lids from the plates and invert the plates on the scanner platen or leave the plates upright - whatever works best. It is important that the four variants of YC media be recorded together. Results from the BiGGY plates should be taken in color, since the colonies appear in various shades of brown, but color is not important for YPD and YC media. Figure 2 shows an example of a student spot plate of strains growing on BiGGY media. The parental strain (P) does not carry a met mutation, and the colonies appear light brown. Strain YMP16 appears white, since it carries a deletion in the MET5 gene, which codes for one of the subunits of sulfite reductase, which reduces sulfite to sulfate. Colonies of strain YMP14 colonies are slightly lighter than the parental strain, because YMP14 has a deletion in the MET16 gene, which occurs upstream of sulfite reductase in the pathway (Fig. 1). Conversely, colonies of strain YMP24 colonies are darker than those of the parental strain, because YMP24 carries a deletion in MET2 gene. Sulfide accumulates in the strain, because it does not produce the O-acetyl serine required for the conversion of sulfide to homocysteine.
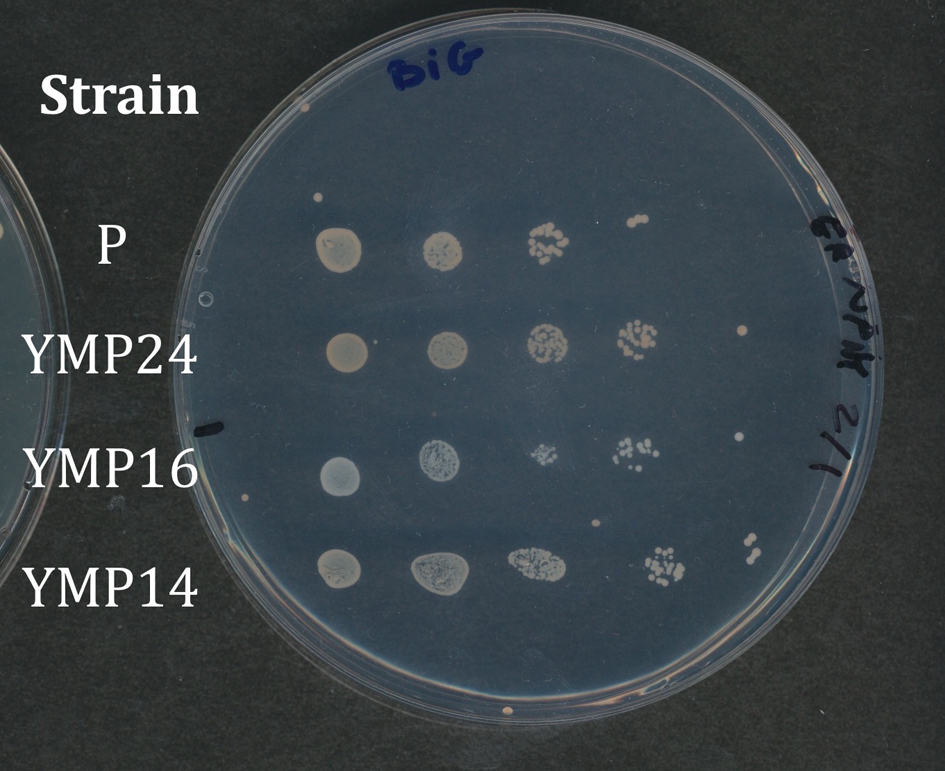
Figure 2. Student plate showing strain growth on indicator media
Class 3 - Students present and discuss their data (~30 minutes)
Students make short group presentations of their data. The heart of each group's presentation is the scanned image comparing growth of the strains on defined media containing different sulfur sources. Groups are also encouraged to include a summary table in their presentation that contains their predictions as well as the observed results. We usually limit presentations to 3-5 minutes, because the groups have been working in parallel and students are already familiar with the methods.
Before the presentations begin, I draw a master grid on the board with the names of the various strains listed in the first column and blank columns for each sulfur source, and a blank column for the names of the MET genes. After each presentation, student presenters fill in the blank columns with their growth results and the name of the MET gene that they hypothesize is deleted in each strain. This table helps to keep the other students engaged during the presentations, which can seem repetitive to them. In this experiment, multiple groups may have tested the same strains on the various media. Students find it interesting to see if other groups have reached the same or different conclusions. When the data table is complete, have the students group construct a Venn diagram of strains that are able to grow on similar sulfur sources. Students may be able to see that strains with mutations in the same portion of the pathway group together. They may also see that the strains able to grow on the largest number of sulfur sources have mutations in the early part of the pathway (see Supporting File S5).
TEACHING DISCUSSION
This experiment usually works very well. Most teams are able to generate spot plates with appropriate dilutions of the cultures. Although not all the plates are publication-quality (!), trends in growth are usually apparent. In a few cases, plates may be contaminated, usually with microorganisms that have been introduced to the water that students used for their serial dilutions. More common problems relate to poor labeling of the plates and an occasional row where the student has spotted the dilution series in the opposite direction from the other rows on the plate.
The greatest difficulty for students is predicting growth properties of the mutant strains, reflecting their limited experiences with metabolic pathways. Analogies can help students to visualize the problem. I find it helpful to describe metabolites as cars moving along a highway (the pathway) that involves a series of bridges (enzyme-catalyzed conversions in the pathway). The color of the car changes as it passes over each bridge. If one of the bridges is broken, cars will pile up before the bridge and cars will be absent on the road downstream of the bridge. The prediction that students find most difficult is that involving met2 mutants. Have students look closely at the pathway (Figure 1, which is reproduced in student reading). Draw their attention to the MET2 gene. The product of the Met2p-catalyzed reaction is O-acetyl homoserine. Next, draw their attention to the Str2p-catalyzed reaction of cysteine with O-acetyl homoserine to form cystathionine. Cells will be unable to catalyze this step if the MET2 gene has been deleted.
This discussion could be brought to a higher level if students have had enough chemistry that they know how to calculate the free energies of individual reactions. Most of the reactions in the pathway are essentially unidirectional because of the hydrolysis of ATP, NADPH, or another high energy donor. The sulfate assimilation process requires considerable expenditure of energy as sulfate is reduced from a +6 oxidation state to a -2 oxidation state in sulfide.
This activity could be modified to substitute another metabolic pathway or a signaling pathway in the place of Met/Cys biosynthesis, as long as selective media are available to discriminate between deletion strains. Any pathway that students analyze should be well-supported by experimental evidence. The Saccharomyces gene deletion project generated single gene deletions for every open reading frame (ORF) in the genome, so it should be straightforward to collect the strains needed to analyze an alternative pathway.
At Boston College, this lesson occurs early in our semester-long research investigation into the evolutionary conservation of MET genes (14). Later in the semester, students will transform their deletion strains with expression plasmids containing homologous genes identified in other organisms. The students will then use complementation analysis to determine if the genes have been functionally conserved during the divergence of S. cerevisiae and the other organism from a common ancestor. This lesson provides students with controls that they can use to interpret those complementation experiments, as well as practice with experimental techniques that they will use throughout the semester.
Other educators have previously reported positive experiences using S. cerevisiae mutants to introduce research experiences into undergraduate research classes. Manney and colleagues developed a collection of yeast strains and developed experiments (15) that can used to introduce students to the principles of Mendelian inheritance. There materials are available to the community through the GENE project (16). Several other groups have used yeast complementation to analyze the phenotypic consequences of mutations involved in human cancers. Gammie and Erdeniz (17) transformed yeast defective in DNA repair with plasmids carrying human homologs of the yeast repair enzymes to see if mutant variants were able to restore DNA repair function. Similarly, Brownell et al. (18) describe a large-enrollment laboratory class in which students use yeast reporter strains to analyze the effects of p53 mutations from human tumors on p53-dependent gene expression. Links to these studies and a variety of other resources are available on the wiki maintained by the Saccharomyces Genome database (19).
SUPPORTING MATERIALS
- S1. Follow the Sulfur-S. cerevisiae deletion strains. Ordering information for S. cerevisiae deletion strains
- S2. Follow the Sulfur-S. cerevisiae growth media. Instructions on preparing agar growth media
- S3. Follow the Sulfur-Sulfur student reading. Background reading and exercises for students
- S4. Follow the Sulfur-Spot plates. Student instructions for preparing and analyzing spot plates
- S5. Follow the Sulfur-Exercise I answers. Growth predictions for strains in exercise 1
- S6. Follow the Sulfur-Micro-report guidelines. Instructions on preparing written micro-reports
ACKNOWLEDGMENTS
It has been a pleasure to work with many Boston College colleagues who helped to develop this lesson. I particularly thank the lab directors, Dr. Douglas Warner and Ms. Holli Rowedder, whose careful planning has consistently ensured the success of this experiment. I would also like to thank dozens of graduate teaching assistants (GTAs) and hundreds of undergraduate students who generously supplied feedback on this lesson.
References
- Beck C, Butler, A, Burke da Silva K.. 2014. Promoting inquity-based teaching in laboratory courses: Are we meeting the grade? CBE Life Sci. Educ. 13:444-452.
- Lopatto D, Alvarez C, Barnard, D et al. 2008. Genomics education partnership. Science 322:684-685.
- Corwin LA, Graham MJ, Dolan EL. 2015. Modeling course-based undergraduate research experiences: An agenda for future research and evaluation. CBE Life Sci. Educ. 14:es1,1-13.
- Brownell SE, Kloser MJ. 2015. Toward a conceptual framework for measuring the effectiveness of course-based undergraduate research experiences in undergraduate biology. Studies in Higher Education 40:525-544.
- Corwin LA, Runyon C, Robinson A, Dolan EL. 2015. The laboratory course assessment survey: A tool to measure three dimensions of research-course design. CBE Life Sci. Educ. 14:ar37,1-14.
- Goffeau A, Barrell BG, Bussey H et al. 1996. Life with 6000 genes. Science 274:563-567.
- Duina AA, Miller ME, Keeney JB. 2014. Budding yeast for budding geneticists: A primer on the Saccharomyces cerevisiae model system. Genetics 197:33-48.
- Thomas D, Surdin-Kerjan Y. 1997. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 61:503-532.
- Winzeler EA, Shoemaker DD, Astromoff A et al. 1999. Functional characterization of the Saccharomyces cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906.
- Gelperin DM, White MA, Wilkinson ML et al. 2005. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Develop. 19:2816-2826.
- Saccharomyces Genome Deletion Project. http://www-sequence.stanford.edu/group/yeast_deletion_project/deletions3.html Accessed 2/8/16.
- Hershey AD, Chase M. 1952. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J. Gen. Physiol. 36:39-56.
- Bauerle C, DePass A, Lynn D et al. 2011. Vision and Change in Undergraduate Biology Education: A Call to Action. National Science Foundation/American Association for the Advancement of Science, Washington, D.C.
- Investigations in Molecular Cell Biology. http://capricorn.bc.edu/bi204. Accessed 2/8/16.
- Manney TR, Manney ML. 1992. Yeast: A research organism for teaching genetics. Amer. Biol. Teacher 54:426-431.
- GENE Project. https://www.phys.ksu.edu/gene/
- Gammie AE, Erdeniz N. 2004. Characterization of pathogenic human MSH2 missense mutations using yeast as a model system: A laboratory course in molecular biology. Cell Biol. Educ. 3:31-48.
- Brownell SE, Hekmat-Scafe DS, Singla V, Seawell PC Imam JFC, Eddy SL, Stearns T, Cyert MS 2015. A high-enrollment course-based undergraduate research experience improves student conceptions of scientific thinking and ability to interpret data. CBE Life Sci. Educ. 14:ar21,1-14.
- Saccharomyces Genome Database wiki http://wiki.yeastgenome.org/index.php/Main_Page
Article Files
Login to access supporting documents
Follow the Sulfur: Using Yeast Mutants to Study a Metabolic Pathway(PDF | 869 KB)
S1.Follow the Sulfur-Ordering information for S. cerevisiae deletion strains.pdf(PDF | 94 KB)
S2.Follow the Sulfur-Instructions on preparing agar S. cerevisiae growth media.pdf(PDF | 72 KB)
S3. Follow the Sulfur-Background reading and exercises for students.pdf(PDF | 342 KB)
S4.Follow the Sulfur-Student instructions for preparing and analyzing spot plates.pdf(PDF | 343 KB)
S5.Follow the Sulfur-Exercise 1 answers.pdf(PDF | 45 KB)
S6.Follow the Sulfur-Micro-report guidelines.pdf(PDF | 362 KB)
- License terms

Comments
Comments
There are no comments on this resource.