Antibiotic Resistance Genes Detection in Environmental Samples
Published online:
Abstract
Our aim is to provide an authentic research experience for undergraduate students in a variety of biology courses through collaboration between a four-year university and a community college. This lab series teaches transferable and universal skills using antibiotic resistance as the focus. Antibiotics have been vital to the treatment of infectious diseases since the late 1940s. Antibiotic resistance has become an increasing concern in the battle against infectious diseases. We focus specifically on detecting a group of ampicillin-resistance genes. In this curricular research experience, students use PCR and gel electrophoresis to detect genes (Bla-1, Bla-SHV, and Bla-TEM) encoding for different β-lactamases that confer resistance to ampicillin. Through a series of experiments students obtain an understanding of core biological principles including scientific process, cell structure, genetics, the role of the environment, and application of molecular biology techniques while contributing to ongoing primary research.
Citation
Bell, J.H., Thrun, L., LeBeau, M., Makarevitch, I., Goldberg, J., and Martin, P. 2016. Antibiotic Resistance Genes Detection in Environmental Samples. CourseSource. https://doi.org/10.24918/cs.2016.3Society Learning Goals
Microbiology
- Evolution
- Why is the traditional concept of species not readily applicable to microbes due to asexual reproduction and the frequent occurrence of horizontal gene transfer?
- How do human impacts on the environment influence the evolution of microorganisms (e.g., emerging diseases and the selection of antibiotic resistance)?
- Cell Structure and Function
- Even though microscopic eukaryotes (e.g., fungi, protozoa, and algae) carry out some of the same processes as bacteria, how do many of the cellular properties fundamentally differ?
- Information Flow and Genetics
- How can cell genomes be manipulated to alter cell function?
Science Process Skills
- Process of Science
- Locate, interpret, and evaluate scientific information and primary literature
- Pose testable questions and hypotheses to address gaps in knowledge
- Plan, evaluate, and implement scientific investigations
- Interpret, evaluate, and draw conclusions from data
- Construct explanations and make evidence-based arguments about the natural world
- Address novel questions through authentic research experiences
- Modeling/ Developing and Using Models
- Recognize the important roles that scientific models, of many different types (conceptual, mathematical, physical, etc.), play in predicting and communicating biological phenomena
- Make inferences and solve problems using models and simulations
- Build and evaluate models of biological systems
- Quantitative Reasoning/ Using Mathematics and Computational Thinking
- Apply the tools of graphing, statistics, and data science to analyze biological data
Lesson Learning Goals
Students will gain an authentic primary research experience in the classroom that stems from and contributes to an ongoing faculty research project focused on the prevalence and implications of spreading antibiotic-resistance genes in the environment.Lesson Learning Objectives
After completing this laboratory series, students will be able to:- apply the scientific method in formulating a hypothesis, designing a controlled experiment using appropriate molecular biology techniques, and analyzing experimental results;
- conduct a molecular biology experiment and explain the principles behind methodologies, such as accurate use of micropipettes, PCR (polymerase chain reaction), and gel electrophoresis;
- determine the presence of antibiotic-resistance genes in environmental samples by analyzing PCR products using gel electrophoresis;
- explain mechanisms of microbial antibiotic resistance;
- contribute data to the Antibiotic Resistance Genes Network;
- define and apply key concepts of antibiotic resistance and gene identification via PCR fragment size.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Within a few decades of introduction of the first antibiotics, the number of antibiotics used in medicine had expanded rapidly, and medicine has become highly dependent on antibiotics to treat and prevent bacterial infections. Unfortunately, the development of bacterial resistance to antibiotics was first observed shortly after the introduction of antibiotics in the 1940s (1,2). Over the years, the frequency and types of clinically-relevant antibiotic resistance has increased dramatically, so that the problem of antibiotic resistant infections is currently recognized as a serious and growing threat to human health and modern medical treatments (3,4,5,6,7,8). In 2013, the Centers for Disease Control [CDC] released a report identifying antibiotic resistance as a major threat to public health in the United States (3). The report estimated that over 200,000 people in the U.S. suffer from antibiotic-resistant bacterial infections each year, and that approximately 30,000 of those people die as a result of their infection (3). In 2014, both the World Health Organization and the Council of Scientific Advisors to the President of the United States produced reports that emphasized the seriousness of the problem of antibiotic resistance and urged that effective measures be developed and implemented as soon as possible to reduce the rate of increase of antibiotic resistant infections (4,5,6). As result of these reports, the United States government has announced a National Strategy for Combating Antibiotic-Resistant Bacteria (7) and a National Action Plan for Combating Antibiotic-Resistant Bacteria (8).
The lesson described here evolved through collaboration between a four-year university (Hamline University) and a community college (Century College). It allows students to investigate the distribution and frequency of specific antibiotic-resistance genes in the environment. This lesson was specifically designed to be completed and produce results that could be utilized independently at an institution or as part of a larger collaborative. It was also designed to allow introductory level college students (first and second year) to participate in an authentic research experience that was embedded as a laboratory lesson in one of their introductory science courses. This laboratory lesson uses antibiotic resistance as the focus to teach transferable and universal science skills, including experimental design, pipetting, sample plating, growing bacterial cultures, PCR, and gel electrophoresis. Students use these techniques to screen environmental soil samples for the presence of ampicillin-resistant bacteria that carry one or more β-lactamase genes.
Course-based inquiry and research experiences are becoming a popular tool in the undergraduate classrooms in response to national calls for improving science education (9,10). A growing body of evidence demonstrates that undergraduates benefit from early and consistent engagement in research (reviewed in 11,12,13). Students engaged in course-based undergraduate research experiences (CUREs), which involve students in addressing research problems or questions in the context of a class, report cognitive gains such as the development of knowledge and skills, affective gains, such as satisfaction with their research experience, and psychosocial gains, such as feeling like a scientist (reviewed in 13). These research experiences are available to students early in their undergraduate careers when the experiences have greater potential to influence a student's academic trajectory (14,15,16). In addition, CUREs aimed at non-science majors and freshmen have the potential to involve students who might not otherwise have access to science research opportunities (16,17). Authentic undergraduate research experiences have been repeatedly shown to benefit students in a variety of ways, leading them to learn to think like a scientist, find research exciting, and pursue graduate education or careers in science (15,18,19). Several national projects working to advance course-based research experiences have been successful in providing students with high-impact learning experiences in biology research (11,20,21,22,23,24). The Course-based Undergraduate Research Network (CUREnet) provides support to and helps foster collaboration among faculty interested in incorporating research experiences into their classrooms (25). In addition, tools have been developed that allow assessment of student learning as a result of research experiences in individual courses and comparison of individual courses with other courses with embedded research experiences (15,18).
CUREs vary in the degree of autonomy and independence experienced by the students in the classroom. While some CURE projects allow students to ask their own independent questions, many projects rely on the same set of questions and a specific lab protocol to investigate various genes or samples that are picked by students based on their hypotheses (27,36,37,38,39,40). In this laboratory lesson, students ask independent questions and test independent hypotheses using the soil samples of their choice, while implementing a standard research protocol. Such an approach is typical for many research labs, as well as other CUREs, including the SEA-phages project (26), UC-Riverdale Transposon project (27), Ciliate Genome Consortium (28), and many others, and does not contradict the research nature of the project.
Intended Audience
By adjusting its focus, this laboratory lesson can be directed to different populations. We have incorporated it into non-majors courses as well as courses intended for Biology majors. For first-year students and students not majoring in biology, the focus of the laboratory lesson is on basic biological principles, such as the scientific method, antibiotic resistance, DNA replication, and evolution by natural selection. In the courses intended for Biology majors, the focus is on molecular biology techniques while reinforcing the basic biology principles. For microbiology students, who are mainly allied health majors, the focus is on antibiotic resistance mechanisms.
Required Learning Time
This laboratory lesson is divided over five lab periods, individually requiring 30 to 150 minutes of time. This lesson has been incorporated into courses that have a required lecture component. The lecture portion has been delivered in both hybrid and face-to-face formats and discusses relevant biological concepts. The lecture portion includes between 100 and 150 minutes per week of instructional time. In a typical semester, six to ten lab sections of 24 students incorporated this laboratory lesson during their 170 minute, weekly lab period. The Teaching Timeline (Table 1) includes the time required for both presentation and discussion of background material, as well as completion of the lab protocol. With the exception of Lab 4, the lab lessons also allow ample time to complete other laboratory exercises during a standard 170 minute lab session. If lab time is limited, the activities in Labs 1 and 5 can be moved to the lecture component of the course.
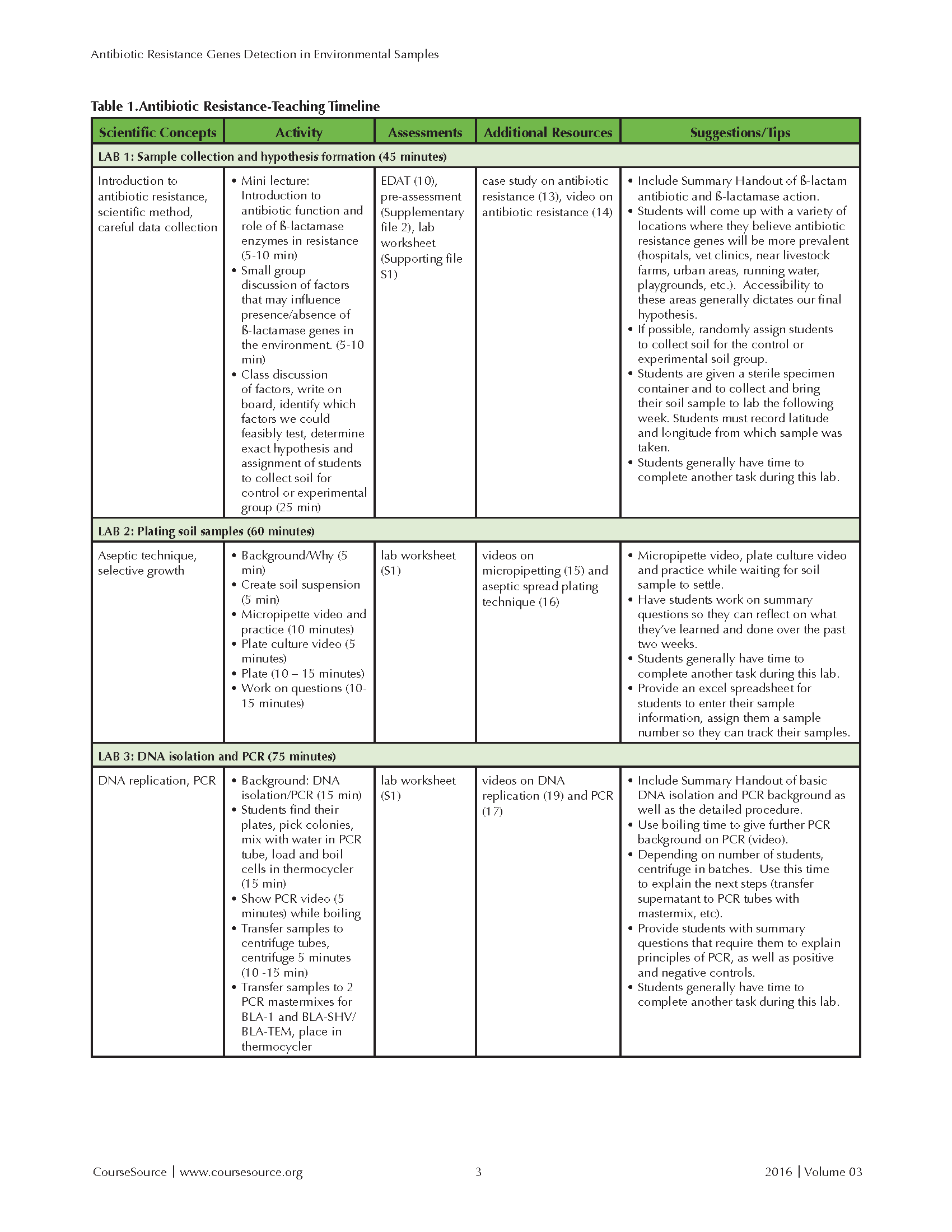
Table 1-Antibiotic Resistance-Teaching Timeline-Page 1
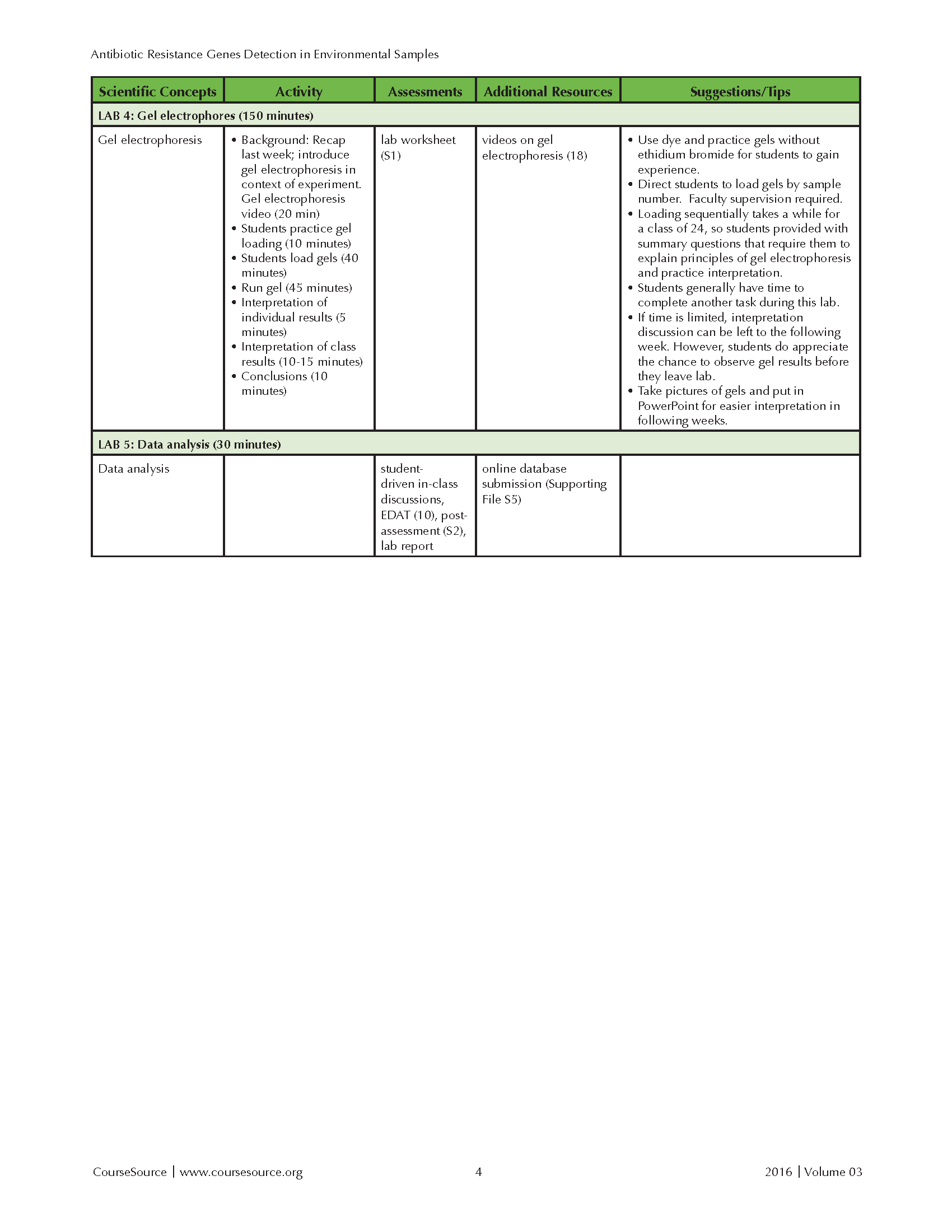
Table 1-Antibiotic Resistance-Teaching Timeline-Page 2
Pre-requisite Student Knowledge
The pre-requisite knowledge depends on the course level and focus. For courses directed at first-year, non-biology majors, completion of a high school biology course is recommended, although no prerequisite knowledge is required. In courses for biology majors, students are assumed to have basic knowledge of the scientific method and basic biological concepts, including the process of DNA replication as well as the structure and function of enzymes. In the microbiology courses, students should have an understanding of bacterial cell structure, antibiotic mechanisms of action, the scientific method, and basic biological concepts.
SCIENTIFIC TEACHING THEMES
ACTIVE LEARNING
This laboratory lesson uses multiple approaches to engage students in active learning. After brainstorming ideas in groups of four to six, students work together as a lab section to develop a unified hypothesis regarding the relative frequency of antibiotic-resistance genes in local soils. The students are first asked to examine the hypothesis based on feasibility and then vote on one hypothesis for that lab section. This approach engages students in a discussion of experimental design and hypothesis development. During the laboratory periods, students complete worksheets with questions that highlight concepts and techniques used in that laboratory. Students start analyzing the data on their own and then discuss their ideas and interpretations in lab, with peers and an instructor. We also discuss, as a lab section, what the absence of a band or an unexplained band means and how primer specificity allows us to detect specific genes.
ASSESSMENT
During lab periods, students completed worksheets and lab quizzes that focus on data analysis and critical thinking (Supporting File S1). To assess overall knowledge gain and measure the degree of change, a pre-test, given at least one week prior to beginning the project, and a post-test, given one week after the last laboratory period, were used to assess student learning of the relevant biological concepts (see Supporting File S2 for sample pre- and post-test). We used the CURE Survey (29) to evaluate students' perception of the lab experience in relation to developing critical thinking, data analysis, and communication. Students' ability to design experiments and interpret data was evaluated using the EDAT survey (30). The alignment of the lesson's learning objectives with lesson activities and assessment instruments is described in Supporting File S3.
INCLUSIVE TEACHING
The CURE developed through this laboratory lesson is inherently inclusive because of how well it serves the community college population. Courses implementing this laboratory lesson are designed for first- and second-year undergraduate students majoring in biology and/or allied health, as well as students majoring in non-science fields. Implementation of this laboratory lesson was primarily at Century College, a community college. Importantly, this laboratory lesson provides a research experience for community college and introductory level students, who ordinarily do not have the chance to participate in primary research. When designing the project, we were mindful of the inherent cultural and socioeconomic diversity, particularly of our community college student population.
Engaging community college students in research has been shown to benefit students in a variety of ways, including their transfer to four-year STEM programs, and several research programs targeting community college students are successfully implemented (31,32). Despite the focus on competency-based and experiential learning in higher education, it is still relatively uncommon for community college students to conduct primary, original research on the large scale that this lesson provides. According to data from the American Association of Community Colleges in 2012, an estimated 12.8 million students enrolled at a community college, accounting for 45% of all undergraduates in the United States. 56% of STEM undergraduate students attended a community college at some point of their studies (31). These large enrollment numbers underscore the increasing impact that community colleges are having on the undergraduate education of in the United States. Community college students vary greatly in age and other demographic variables such as income and ethnicity. According to the American Association of Community Colleges, the average age of the community college student population is 28 with approximately 36% enrolled as first-generation college students (33). Moreover, 36% of community college students receive Pell grants. In 2015, at Century College, approximately 55% of the students at the community college were first generation college students with little or no previous exposure to science courses (34).
While the lesson is inherently inclusive, there are also varieties of specific activities that will accommodate the needs of students with diverse learning styles. From development of the group hypotheses to analysis of results, the laboratory lesson is collaborative, "easing the way" to scientific experimentation for students. Students learn how to balance an 'ideal' experimental design with feasibility, specifically of obtaining soil samples from locations to which students have access. Discussion throughout the laboratory lesson emphasizes the prevalence and timely importance of spread of antibiotic resistance in the environment as it relates to the greater community. In addition, pedagogical approaches implemented in this laboratory lesson accommodate a variety of learning styles through the use of demonstrations, videos, hands on lab activities, mini-lectures, worksheets, and case studies.
LESSON PLAN
BACKGROUND: HOW BETA-LACTAMASES WORK
Antibiotics have been vital to the treatment of bacterial diseases since the late 1940s (1,2). Specific classes of antibiotics include tetracyclines, aminoglycosides, carbapenems, and beta-lactams (β-lactams). β-lactam antibiotics, commonly used to treat bacterial infections, are characterized by the presence of a nitrogen-containing β-lactam ring. β-lactams bind to the active site of penicillin-binding proteins, which are bacterial enzymes needed to synthesize a properly cross-linked cell wall. This inhibition results in a weakened cell wall that cannot resist the high internal osmotic pressure of the bacterial cell, leading to cell wall rupture and lysis of the bacterial cell (35).
Bacteria are continuously developing new mechanisms of resistance. Identification and mapping of these resistance mechanisms may provide insight into the sources of resistance and help in the design of new antimicrobial agents (36). One large group of enzymes responsible for resistance are the β-lactamases. β-lactamases can break the β-lactam ring, thus destroying the ability of β-lactam antibiotics to act as a competitive inhibitor and thereby destroying the antibacterial function of the antibiotic. β-lactamase-producing bacteria are important contributors to resistant infections that have been of increasing concern in the battle against infectious diseases.
β-lactamases may be classified in different ways based on sequence homology or on the enzymes' functional properties (37,38). This lesson focuses on TEM- and SHV-type β-lactamases: enabling us to examine different two families of β-lactamases. Although the SHV family thought to be originally derived from Klebsiella spp. and TEM was initially found in E. coli isolates, these gene families can now be found in multiple enterobacteria species creating a significant threat to treatment (36). The SHV and TEM families have substrate and inhibition profiles normally inhibited by active site-directed β-lactamase inhibitors (38). This profile confers resistance to broad-spectrum penicillins such as ampicillin and first generation cephalosporins (36,38). Derivatives of these genes continue to form and evolve to confer an even broader resistance profiles.
LESSON
Lab Safety
All laboratories working with microorganism should follow the American Society for Microbiology Laboratory Safety Guidelines (39). The foremost concerns in this laboratory lesson are the bacterial growth plates (Lab 2) and the use of ethidium bromide in the gels (S1 and S4: Lab 3 and 4). Special concern and safety precautions are provided for students with immunosuppressive conditions.
Laboratory Lesson Overview
In this laboratory lesson, we focus on detecting the presence of ampicillin-resistance genes in soil samples using two sets of primers for the TEM-type gene (Bla-1 and Bla-TEM) one set for the SHV-type gene (Bla-SHV). The activities of the lab and lecture are intentionally coordinated so that the laboratory reinforces material that students are learning in the lecture portion of the course. Students begin each lab period after they have received instruction during lecture about relevant content, e.g., scientific method, DNA replication, PCR, gel electrophoresis, antibiotics, and antibiotic resistance mechanisms. The laboratory lesson is divided into five lab periods: hypothesis generation, culturing soil microbes, DNA isolation and PCR, gel electrophoresis, and data analysis (Table 1). Step-by-step student instructions for each lab can be found in the Supporting File 1. See Supporting File 4 for a list of required materials for each lab and instructor preparation guidelines.
LAB 1: Introduction to Antibiotic Resistance and Hypothesis Formation (45 MINUTES)
During lecture, we introduce hypothesis-based science, if necessary, as well as the function of antibiotics, the mechanism(s) of antibiotic resistance, and the increasing public health concerns related to antibiotic-resistant bacteria. A description including the importance and global relevance of β-lactamases is provided in the student instructions (Supporting File S1). We then emphasize that, during lab, students will be able to contribute new data to the Antibiotic Resistance Genes Network database (see Supporting File S1, Lab #1), while at the same time testing a hypothesis about expected occurrence of genes providing resistance to the antibiotic ampicillin in the environment (10 minutes).
During lab, students work in groups of four that are consistently maintained during the series. The group beings by identifying places they think ampicillin-resistance genes may be more prevalent (5-10 minutes). For example, students may hypothesize that ampicillin-resistance genes will be more commonly found in soils from urban areas or near hospitals, vet clinics, livestock farms, playgrounds, water sources, etc. Students then write their ideas on the board and share them with the entire lab section during the laboratory period. We then discuss which factors we could most feasibly test, with accessibility to particular areas often dictating our final hypothesis. Once a location has been chosen (e.g., urban areas), we discuss the need for a control group appropriate for the hypothesis being tested. The control group will depend on the hypothesis of that lab section during that semester. For example, if the group proposes that there will a greater prevalence of ampicillin resistance genes in urban areas, samples from rural areas will be needed as a control. After discussion of variables that need to be considered for soil collection, students are randomly assigned to collect either the control or experimental samples. The group then finalizes their hypothesis and records all of this information on their Lab #1 handout (Supporting File S1).
Students then receive sterile sample containers provided by the instructor (a sterile 50 ml plastic test tube or a sterile urine specimen cup) and instructions about aseptic collection of the soil samples (See Supporting File 4). Students record their name, date, and the latitude and longitude in decimal form of the sample collection site. We encourage them to also take a photo of the site. They store the containers in a cool area before bringing the soil sample to the next lab period. All of these activities (group discussion, final hypothesis selection, random assignment to control/experimental sample collection, and sample container distribution) requires approximately 25-30 minutes. Pre-assessment of students' baseline content knowledge may also be conducted at this point (10-15 minutes).
In the microbiology courses, students work in groups and complete a case study on antibiotic mechanisms as a preparatory worksheet. The case study focuses on mechanisms of penicillin and vancomycin (β-lactam antibiotics) (40). All students watch a documentary by Frontline entitled 'Hunting the Nightmare Bacteria' (41). This activity requires that students complete an instructor-created worksheet and can be assigned to be completed as an out-of-class activity. This activity humanizes the growing concern of antibiotic resistance in allied health professions.
LAB 2: Plate and Incubate Environmental Soil Samples (60 minutes)
Students receive Lab #2 Handout a week in advance (Supporting File 1). Lab #2 begins with a five-minute mini-lecture describing the overall goal of this lab, which is to extract bacteria from their soil samples and culture them in nutrient media containing a bactericidal concentration (100-200 ug/mL) of β-lactam antibiotic, ampicillin (100ug/mL). The inclusion of low-dose bactericidal ampicillin restricts growth of ampicillin-sensitive bacteria, selecting for ampicillin-resistant bacteria. This enrichment makes it more likely that we detect the ampicillin-resistance genes of interest. It may be important to note that, although the growth of bacterial colonies indicates resistance to ampicillin, it does not determine the specific mechanism of that resistance. Students are assigned a sample number, and then record information about their sample in the Excel spreadsheet shared by all of the lab members. (Information includes sample number, the latitude/longitude, and a description of the location from which they collected their sample.)
After brief instruction, students suspend their soils in an approximately equal volume of sterile water and shake (10 minutes). While the soil is settling, students watch a short video on basic pipetting techniques (42), with the instructor stopping to emphasize important aspects of proper technique including the difference between the first and second stops and when they should be used. Students are then given time to practice pipetting with a container of water.
Each student labels an LB-AMP plate with their name and sample number. Students then watch a short video on aseptic spread plating methods (43), with the instructor stopping to emphasize important aspects of aseptic technique. Students are asked to sample from 10 different colonies and then aseptically spread 100μl of their soil "supernatant" on the LB-AMP plate. After plating their soil samples, students complete the remaining handout questions, which requires them to reflect on what they accomplished during the lab period and relate that work to what was learned in the previous lab session. Incubate the plates in an incubator with the appropriate biohazard label. Plates are incubated for 24 hours at 37°C and then refrigerated until the following week's lab. The samples are grown at 37°C to enrich for bacteria that would be more relevant to human health and disease.
LAB 3: Bacterial DNA Isolation and Amplification of Antibiotic Resistance Genes by PCR (75 minutes)
Students receive Lab #3 Handout (Supporting File 1), and the instructor provides ~10 minute mini-lecture on the overall goal of this lab, which is to extract DNA by heating a bacterial cell suspension to 95°C, which causes the bacterial cell to rupture. A brief explanation of PCR is included. Students then use the bacterial genomic DNA as the template in a PCR reaction to amplify the antibiotic-resistance genes of interest.
Students should don appropriate personal protective equipment, remove the plates to a biohazard safety cabinet, and collect the bacterial colonies for DNA isolation. Once the DNA is isolated, the rest of the laboratory work can be performed on the benchtop. To isolate the bacterial DNA, students collect a small amount of bacterial cells by gently touching a sterile pipette tip to ten different bacterial colonies and shaking the tip in 100μl of sterile distilled water in a PCR tube that has been labeled with their sample number. The tubes are placed in the thermocycler to heat for five minutes. During the boiling cycle, students watch a short video on PCR (44) and receive further instruction on the principles of PCR, including the roles of temperature cycles, Taq polymerase, and primers. Two different colored PCR tubes containing mastermixes for amplification of Bla-1 or Bla-TEM/Bla-SHV are distributed to each student (see instructor guide, Supporting File S4 for relevant details). Students must label each tube with their sample number and keep these tubes on ice. Students then retrieve their bacterial samples from the thermocycler and transfer 3 ul of the lysed bacterial solution into each PCR tube. The instructor can make positive and negative controls for both PCR reactions during this time using an E. coli cloning vector containing Bla-1 ampicillin resistance gene for the positive and sterile nuclease free water for the negative. Emphasize sterile technique (wear gloves, dispose of pipette tips after each use to avoid contamination). Instruct students to flick PCR tube to mix contents and settle all liquid at bottom of tube.
In order of their sample numbers, students then load their Bla-1 samples into one thermocycler and Bla-SHV/Bla-TEM samples into another thermocycler, and the PCR protocols are run. While PCR is running, students must complete summary questions from the handout, requiring them to reflect on the day's procedure and to explain that principles of PCR and positive and negative controls. Students may also have time to complete another lab exercise, such as a DNA replication simulation.
LAB 4: Gel Electrophoresis (at least 90 minutes for introduction and gel loading, another 60 minutes to run and view gels)
Students receive Lab #4 Handout (Supporting File S1) and use gel electrophoresis to analyze the PCR products obtained from their samples in Lab 3. The instructor uses 15 minutes to provide a brief explanation of the principles of gel electrophoresis and shows a related video (45). Another 10 minutes is spent demonstrating proper loading of gel wells and giving students a chance to practice loading (i.e. pipetting methylene blue in 1X loading buffer into gels without ethidium bromide). Students must use gloves when handling the gels containing ethidium bromide. After practicing, students take turns loading their PCR samples, with the instructor closely supervising and directing where students should load each sample (45 minutes). Students complete Lab #4 summary questions on the handout while waiting for the rest of the students to load their samples. The instructor or a volunteer student loads the positive and negative control samples during this time. The gels are then run for 45 minutes. The time required for the samples to run can be used to further discuss the principles of gel electrophoresis and show a video that provides context about why antibiotic resistance should be of concern to students. Students will typically have just enough time to get a look at the finished gels under UV light, but not complete their analysis. The instructor photographs the finished gels and the saves the images for evaluation in the next lab or class period. Properly dispose to gels and used running buffer in appropriately labeled disposal containers.
LAB 5: Data analysis (30 minutes)
Students view the photographs of their gels, looking for DNA bands that match the sizes of the expected PCR amplicons. Interpretation goes more smoothly if instructor creates an organized PowerPoint file of the gel images. As a group, students examine the data for every sample. In a single lab section of 24 students, typically this includes two to three gels with 20 samples per gel. Students are asked to interpret the results in the positive and negative control lanes and determine if the bands in their experimental lanes are true positives. Students examine the samples and record the number and size of the bands in their lanes. The number of positive results for each ampicillin resistance gene (Bla-1, Bla-TEM, Bla-SHV) is tallied for both the control and experimental groups. Based on the 2015-2016 data collected with 268 students, approximately 32% of the samples are positive for Bla-1, 3% for Bla-SHV and 5% for Bla-TEM. The data lead to a discussion of whether the data support the group hypothesis, how confident we are in our conclusions given our sample size, and other factors that could have affected our results. Students use a Google Form to submit their results to the online database (Supporting File 5). Students are then given a week to study their Lab #1-5 handouts (Supporting File 1) to prepare for the post-assessment questions, which can be administered during a subsequent laboratory, lecture, or exam.
TEACHING DISCUSSION
This lesson provides a platform for students to explore the scientific method in practice and increases student understanding of molecular biology and microbiology concepts. Students design their own experiments, collect data from soil samples of their choice, conduct analyze their results in light of their original hypotheses. Students contribute their original data to the student-driven research program at Hamline University. This research project investigates the spread of β-lactamase genes in the environment and is driven by Hamline undergraduate researchers in and outside of the classroom. Faculty who use this lesson and would like to participate in this project are welcome to contact Jessica Bell or Irina Makarevitch so that the resulting data can be added to the database.
Role of Hypothesis Development
The process of hypothesis development as a group is critical for contextualizing the complex nature of antibiotic resistance. During the first semester that this research was implemented, some instructors had students collect soil samples from places that were convenient to students without first developing a hypothesis. One course, however, created a hypothesis as a group, which then guided the sample collection process. After soliciting feedback from students during this semester, students expressed a substantially higher level of interest in and overall satisfaction with the project when unified by a group hypothesis. Not only did hypothesis development guide students where to collect samples, but it provided a natural opportunity to discuss experimental design along with all the potential difficulties that exist in 'real life' experiments compared to those conducted from a lab manual.
Diverse Teaching Pedagogies
A number of instructors have implemented this laboratory lesson in classes with a variety of teaching formats. Some use 'traditional' lecture with hands-on laboratories whereas others are implementing elements of a flipped classroom and whole-class discussion approaches. The basics of the lesson successfully translate across a variety of biological disciplines and teaching styles. The topic as well as the format allows the lesson to be modified and adapted to whatever style an instructor may use. To provide context and help prepare students for the lesson, instructors used supplemental assignments such as worksheets to complete while viewing the Frontline episode 'Hunting the Nightmare Bacteria' (41), videos (46) and the accompanying Science article (47). The supplemental assignments provided current and real cases of antibiotic resistance in action, bridging the gap between antibiotic resistant microbes and the presence of antibiotic-resistance genes.
In addition to our own CURE, other projects have examined the worldwide health threat of antibiotic resistance by incorporating laboratory exercises into the undergraduate science courses. The Small World Initiative is an international collaboration funded by crowd-sourcing (48). This initiative uses introductory biology students to perform hands-on field and laboratory research on soil samples in the hunt for new antibiotics. Another national program is the Prevalence of Antibiotic-Resistant microbes in the Environment (PARE) program (49). The PARE program is designed to be implemented in two traditional undergraduate laboratory class periods where students evaluate soil samples for antibiotic resistant bacteria. This program extends to high school classrooms through partnerships with undergraduate institutions. While our project and similar projects are designed to be incorporated into an existing scientific curriculum, others have designed entire courses or programs around antibiotic resistance and drug discovery (50,51). These course based experiences teach biological concepts in the context of an ongoing antibiotic discovery project carried out by the students. While these programs are well designed and effective, some of the CURE programs may be cost prohibitive to community colleges and primarily undergraduate teaching institutions. We have consciously developed an economical alternative (less than $8.00/student) to be utilized in a wide variety of courses.
This laboratory lesson could be easily adapted to teaching upper level courses in biology. Advanced students could start this project by investigating primary literature to propose possible antibiotic-resistance genes they could screen for and designing the PCR primers for the screening. Students could be asked to present their ideas and data in a variety of formats, from short chalk talks to their peers and lab notebooks to posters and presentations. Students could also independently design follow up experiments, for example, identifying antibiotic resistant bacteria and resistance alleles using microbiology staining and culturing techniques, DNA sequence analysis and antibiotic sensitivity profiles.
Assessment of Student Learning
We used a combination of subjective and objective assessment approaches to assess student learning as the result of this lab series. First, students were asked to complete a multiple choice test containing seventeen questions assigned to four categories: DNA, molecular biology techniques, microbiology, and data interpretation and analysis. Learning gains were estimated by comparing student answers prior to the lab lesson and after the lab lesson. On average, the proportion of correct answers increased from 37% to ~72% (normalized learning gain of 0.56). Although there are no established criteria for what constitutes acceptable learning gains on these tests, a normalized gain of >=0.50 probably represents a substantial achievement. Students in the microbiology course demonstrated significantly stronger learning gains (0.68 on average) compared to students in genetics and general biology courses on questions targeting microbiology concepts. We were specifically interested in assessing students' skills in the experimental design. The experimental design assessment test (EDAT) survey (30) implemented to assess learning gains in students' ability to list the elements of experimental design also showed strong learning gains as the result of the lab lesson (an increase from 4.3 out of 10 to 8.2 out of 10, on average). To assess students' success in formulating and testing hypotheses and analyzing data, group lab reports were scored using a detailed rubric (see Supporting File S3). Student's perception of the lab lesson and the learning gains was assessed using a CURE survey (29). 384 of our students participated in the CURE survey before and after the course integrated research experience. Students reported perceived learning gains higher or comparable with learning gains reported by all CURE participants in 16 of 21 categories, with the largest gains in learning to work independently and understanding science as part of the larger grouping of understanding scientific process. On 10 of those criteria, students scored higher than the 4800 CURE participants in the database. In general, our assessment data strongly support the effectiveness of the curricular research experience on antibiotic resistance in teaching and learning the elements of experimental design and understanding concepts in molecular biology and microbiology. In addition, students perceived the lab lesson as highly effective in developing data analysis and communication skills as well as increasing their understanding of the scientific process.
Challenges in Implementing Primary Research
Students at Century College, a community college, were unfamiliar with equipment and molecular biological techniques that may seem routine to students at other colleges and universities. To address this challenge, students viewed a series of videos from a variety of sources including JoVE, BioRad, and YouTube (Table 1). Reviewing the basics of proper micropipette use as well as DNA replication and PCR were invaluable and helped students feel more confident when they began to use these tools in their research project.
Another challenge can be collecting enough samples for students to conduct meaningful data interpretation. Because most of our courses have about 24 - 48 students, sample sizes are relatively small. Although it is possible for completion of some statistical analysis, it can be difficult for students to formulate a cohesive story at the end of the four-week project. Coordination between multiple sections of the same course and access to the online database, however, increases sample size and provides a larger set of data for analysis at the end of the project. To achieve this goal, students from each school have been contributing their data to a large collection to produce an Antibiotic Resistance Genes Network maintained at Hamline University and used by collaborators at Hamline University, Century College, and North Hennepin Community College.
Determining which antibiotic-resistance genes to study was a crucial and important first step in designing the lesson. The genes utilized for the project described were part of a screen that was the focus of summer research by four students at Hamline University. The genes of interest were chosen based on: 1) the high use of beta-lactam antibiotics such as penicillin and related antibiotics; 2) the PCR products produced clearly distinguishable bands when using a quick, simple DNA isolation technique [boiling colonies]; and 3) the frequency of occurrence of these particular genes (it was important that the specific genes not be found in every sample, but also not detected too infrequently so as to yield only negative results). The last of these criteria may be the most important when considering other antibiotic-resistance genes to augment this lesson plan. Given the surfeit of published primers and PCR protocols for thousands of antibiotic-resistance genes, instructors could easily change the focus to other mechanisms of resistance or classes of antibiotics, such as tetracyclines or aminoglycosides among others. However, this adaptation would require preliminary testing prior to implementation in lab.
Benefits of the Lesson
The learning outcomes illustrate the effectiveness of this lesson. First, students gain an understanding of the ubiquity of antibiotic resistant microbes in the soils of their communities. The sheer number of microbes able to grow on the plates containing ampicillin amazes many students and students are unaware of the role that soil microbes have on human health (52). Moreover, students recognize the disparity between drug resistant microbes (e.g. number of colonies growing) and those samples positive for the β-lactamase genes we investigated (e.g. samples with PCR amplicons). After first being puzzled by the fact that not all samples were positive for the β-lactamase genes tested, students discover the vast number of β-lactamase genes (36), as well as other antibiotic-resistance genes currently identified (53).
This lesson also served as a platform to discuss the variety of ways that bacteria can acquire drug resistance genes. Without lengthy literature reviews on the topic, we were able to discuss ideas related to metagenomics and bacterial genetics, both of which would normally be beyond the scope of our community college courses for students seeking degrees in allied health. Students gained a better understanding of the potential risks of the mere presence of antibiotic-resistance genes and the importance of horizontal gene transfer, relating our project to the rise of antibiotic resistance throughout the world.
Implementing this lesson produces multiple benefits. Literature in this area is rich with possible resistance genes to investigate outside of a large classroom project as the topic of an independent project. In addition, the topic unifies some themes that cross disciplines: Instructors from immunology, ecology and evolution, geology, chemistry, and even geography could collaborate at one institution to develop capstone or common core projects all related to testing soil characteristics in addition to antibiotic-resistance genes. With respect to biology classes for non-science majors, antibiotic resistance provides one of the best examples of natural selection in action in the real world. A notable and unanticipated result was the high level of intellectual investment and engagement students demonstrated participating in this primary research project. Their enthusiasm was due in part to participating in a lab where results were unknown as well as the idea that students were contributing actual data to a research study. When students are able to participate in hypothesis formation and testing, they are better equipped to put materials and methods used into context. Procedures become more meaningful to students when they use them to answer a specific question of their own design.
Providing them with hands-on experience with molecular biological techniques is essential to keep pace with their peers at other colleges and universities. For many of our allied health students, this laboratory could be their only exposure to research. Based on our initial observations, this lesson seems to foster our allied health students' interest in other areas of science beyond nursing or dental hygiene. For students who transfer to four year colleges, this project may aid in decreasing the opportunity and experience gap with exposure to methods and equipment not available at our community college prior to implementation of this lesson. Through this collaboration and exposing students to scientific research, several of the community college students have fostered a new interest in research and continued their research experiences by conducting summer research projects at Hamline University.
SUPPORTING MATERIALS
- Supporting File S1. Antibiotic Resistance Genes Detection in Environmental Samples-Student lesson and handout
- Supporting File S2. Antibiotic Resistance Genes Detection in Environmental Samples-Pre- and Post-assessment multiple choice questions
- Supporting File S3. Antibiotic Resistance Genes Detection in Environmental Samples-Alignment of learning objectives with lesson activities and assessment instruments.
- Supporting File S4. Antibiotic Resistance Genes Detection in Environmental Samples-Lab Protocol and Instructor Preparation.
- Supporting File S5. Antibiotic Resistance Genes Detection in Environmental Samples-Data Submission Form.
ACKNOWLEDGMENTS
We thank Kathryn J. Malody, John P. DeMay, and Dustin Mielke for providing laboratory support; Ann Kessen for implementing this work in her classroom; Yazmin Cejudo, Michael Cullen, James DeMars, Lara Thao and Pieta Thao for their involvement in obtaining the primary image for this paper; research students in Presley Martin's laboratory for their help in developing the antibiotic resistance project and network. Support was provided by a Howard Hughes Medical Institute grant 52007543 to Hamline University.
References
- Aminov, RI. 2010. A Brief History of the Antibiotic Era: Lessons Learned and Challenges for the Future. Front Microbiol. 2010; 1:134.
- Podolsky SH. 2014. The Antibiotic Era: Reform, Resistance, and the Pursuit of a Rational Therapeutics. Johns Hopkins University Press. Baltimore, MD.
- Centers for Disease Control and Prevention. 2013. Antibiotic Resistance Threats in the United States, 2013. [Press release] http://www.cdc.gov/drugresistance/threat-report-2013/index.html. Accessed February 8, 2016.
- World Health Organization. 2014. Antimicrobial resistance: global report on surveillance 2014. http://www.who.int/drugresistance/documents/surveillancereport/en/. Accessed February 8, 2016.
- World Health Organization. 2014. WHO's first global report on antibiotic resistance reveals serious, worldwide threat to public health. [Press release] http://www.who.int/mediacentre/news/releases/2014/amr-report/en/. Accessed February 8, 2016.
- Reardon S. 2014. WHO warns against 'post-antibiotic' era. Nature News. Nature Publishing Group. http://www.nature.com/news/who-warns-against-post-antibiotic-era-1.15135. Accessed February 8, 2016.
- White House. 2014. National Strategy for Combating Antibiotic-Resistant Bacteria. https://www.whitehouse.gov/sites/default/files/docs/carb_national_strategy.pdf. Accessed February 8, 2016.
- White House. 2015. National Action Plan for Combating Antibiotic-Resistant Bacteria. https://www.whitehouse.gov/sites/default/files/docs/national_action_plan_for_combating_antibotic-resistant_bacteria.pdf
- National Research Council. 2003. BIO2010: Transforming undergraduate education for future research biologists. National Academies Press (Washington, DC U.S.A).
- American Association for Advancement of Science. 2011. Vision and Change: A Call to Action, Washington, DC. http://visionandchange.org/files/2011/03/Revised-Vision-and-Change-Final-Report.pdf. Accessed March 6, 2016.
- Laursen S, Hunter AB, Seymour E, Thiry H, Melton G. 2010. Undergraduate Research in the Sciences: Engaging Students in Real Science, San Francisco: Jossey-Bass.
- Lopatto D. 2010. Science in solution: The impact of undergraduate research on student learning, Tucson, AZ: Council on Undergraduate Research and the Research Corporation for Science Advancement.
- Corwin LA, Graham MJ, Dolan EL. 2015. Modeling course-based undergraduate research experiences: an agenda for future research and evaluation. CBE Life Sci Educ 14:es1.
- Dolan EL, Lally DJ, Brooks E, Tax FE. 2008. PREPping students for authentic science. Sci Teach 75:38-43.
- Lopatto D, Alvarez C, Barnard D, Chandrasekaran C, Chung HM, et al. 2008. Undergraduate research. Genomics Education Partnership. Science 322: 684-685.
- Wei CA, Woodin T. 2011. Undergraduate research experiences in biology: alternatives to the apprenticeship model. CBE Life Sci Educ 10:123-131
- Bangera G, Brownell SE. 2014. Course-based undergraduate research experiences can make scientific research more inclusive. CBE Life Science Education 13:602-606.
- Lopatto D, Hauser C, Jones CJ, Paetkau D, Chandrasekaran V, et al. 2014. A central support system can facilitate implementation and sustainability of a classroom-based undergraduate research experience (CURE) in genomics. CBE Life Sci Educ 13:711-723.
- Thiry H, Laursen SL. 2011. The role of student-advisor interactions in apprenticing undergraduate researchers into a scientific community of practice. J Sci Educ Technol 20:771-784.
- Hanauer DI, Jacobs-Sera D, Pedulla ML, Cresawn SG, Hendrix RW, Hatfull GF. 2006. Inquiry learning. Teaching scientific inquiry. Science 314:1880-1881.
- Campbell AM, Ledbetter MLS, Hoopes LLM, Eckdahl TT, Heyer LJ, Rosenwald A, Fowlks E, Tonidandel S, Bucholtz B, Gottfried G. 2007. Genome Consortium for Active Teaching: Meeting the Goals of BIO2010. CBE Life Sci Educ 6:109-118.
- Campbell AM, Eckdahl T, Cronk B, Andresen C, Frederick P, Huckuntod S, Shinneman C, Wacker A, Yuan J. 2014. pClone: Synthetic Biology Tool Makes Promoter Research Accessible to Beginning Biology Students. CBE Life Sci Educ 13:285-296.
- Ditty JL, Kvaal CA, Goodner B, Freyermuth SK, Bailey C, Britton RA, Gordon SG, Heinhorst S, Reed K, Xu Z, Sanders-Lorenz ER, Axen S, Kim E, Johns M, Scott K, Kerfeld CA. 2010. Incorporating genomics and bioinformatics across the life sciences curriculum. PLoS Biol 8:e1000448.
- Shaffer CD, Alvarez C, Bailey C, Barnard D, Bhalla S, et al. 2010. The genomics education partnership: successful integration of research into laboratory classes at a diverse group of undergraduate institutions. CBE Life Sci Educ 9:55-69.
- Auchincloss LC, Laursen SL, Branchaw JL, Eagan K, Graham M, Hanauer DI, Lawrie G, McLinn CM, Pelaez N, Rowland S, et al. 2014. Assessment of course-based undergraduate research experiences: a meeting report. CBE Life Science Education 13: 29-40.
- Staub NL, Poxleitner M, Braley A, Smith-Flores H, Pribbenow CM, Jaworski L, Lopatto D, and Anders KR. 2016. "Scaling Up: Adapting a Phage-Hunting Course to Increase Participation of First-Year Students in Research. CBE Life Sci Educ. 15(2). pii: ar13. doi: 10.1187/cbe.15-10-0211.
- Burnette JM 3rd and Wessler SR. 2013. Transposing from the laboratory to the classroom to generate authentic research experiences for undergraduates. Genetics. 193(2):367-75. doi: 10.1534/genetics.112.147355. Epub 2012 Nov 19.
- Smith JJ, Wiley EA, and Cassidy-Hanley DM. 2012. Tetrahymena in the classroom. Methods Cell Biol. 109:411-30. doi: 10.1016/B978-0-12-385967-9.00016-5.
- Lopatto D. 2008. Classroom Undergraduate Research Experiences Survey (CURE) 2008 www.grinnell.edu/academics/areas/psychology/assessments/cure-survey. Accessed 15 September 2014.
- Sirum K, Humburg J. The Experimental Design Ability Test (EDAT) Bioscene. 2011; 37:8-16.
- CCURI and CUR. Investing in Impact: The Power of Undergraduate Research (Washington, DC: Council on Undergraduate Research, 2015).
- McCook A. 2011. Two-Year Colleges Are Jumping Into the U.S. Research Pool. Science 333(6049):1572-1573.
- American Association of Community Colleges (AACC). 2016. http://www.aacc.nche.edu/AboutCC/Documents/AACCFactSheetsR2.pdf
- Century College. "About | Century College". 2016. https://www.century.edu/about. Accessed February 8, 2016.
- Kotra LP, Mobashery S. 1999. Mechanistic and clinical aspects of beta-lactam antibiotics and beta-lactamases. Arch Immunol Ther Exp (Warsz). 47(4):211-6.
- Shaikh S, Fatima J, Shakil S, Rizvi SMD, and Kamal MA. (2015). Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment. Saudi Journal of Biological Sciences, 22(1): 90-101. http://doi.org/10.1016/j.sjbs.2014.08.002
- Ambler, R.P. 1980. The structure of ß-lactamases. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 289:321-331.
- Bush K, Jacoby GA, and Medeiros AA. 1995. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233.
- Leonard M. 2012. (30 November 2012, posting date). Antibiotic Resistance: Can We Ever Win? National Center for Case Study Teaching in Science, University at Buffalo. [Online.] http://sciencecases.lib.buffalo.edu/cs/collection/. Accessed February 8, 2016.
- FRONTLINE. 2013 (Air date: 2013, October 21). Hunting the Nightmare Bacteria. United States: PBS Studios. http://www.pbs.org/video/2365104403/. Accessed February 8, 2016.
- BioNetwork. Micropipetting [Video file]. http://www.youtube.com/watch?v=NgosWmRjjAo. Accessed February 8, 2016.
- BioRadLifeScience. Aseptic Technique [Video file]. http://youtu.be/bRadiLXkqoU. Accessed February 8, 2016.
- Howard Hughes Medical Institute. 2015. Polymerase chain reaction [Video file]. http://www.hhmi.org/biointeractive/polymerase-chain-reaction. Accessed February 8, 2016.
- BioRadLifeScience. Agarose Gel Electrophoresis [Video file]. http://youtu.be/vq759wKCCUQ. Accessed February 8, 2016.
- American Society for Microbiology. 2012. Guidelines for Biosafety in Teaching Laboratories http://www.asm.org/images/asm_biosafety_guidelines-FINAL.pdf
- Dolan DNA Learning Center, CSHL in collaboration with Red Green & Blue Company Ltd. (Producer). 2003. DNA Interactive [DVD]. DNA Interactive (DNAi). Cold Spring Harbor Laboratory: James D. Watson.
- Beatson SA, Walker MJ. 2014. Microbiology. Tracking antibiotic resistance. Science. 345(6203):1454-5.
- Small World Initiative. 2015. http://www.smallworldinitiative.org/. Accessed March 6, 2016.
- Prevalence of Antibiotic-Resistant microbes in the Environment (PARE). 2016. http://campuspress.yale.edu/bascomslack/projects/. Yale University. Accessed March 6, 2016.
- Nathan, A. 2014. Course offers hands-on science education. Yale Scientific. http://www.yalescientific.org/2014/07/course-offers-hands-on-science-education/, Accessed March 6, 2016.
- University of Minnesota. 2015. Discover Your Own Drug http://cbs.umn.edu/hhmi-grants/science-education/research-experiences/antibiotic. Accessed March 6, 2016.
- Baumgardner, DJ. 2012. Soil-Related Bacterial and Fungal Infections. J Am Board Fam Med. 25:734-744; doi:10.3122/jabfm.2012.05.110226
- Lin J, Nishino K, Roberts MC, Tolmasky M, Aminov RI, and Zhang L. Mechanisms of antibiotic resistance. Frontiers in Microbiology. 2015;6:34. doi:10.3389/fmicb.2015.00034.
Article Files
Login to access supporting documents
Antibiotic Resistance Genes Detection in Environmental Samples(PDF | 192 KB)
S1.Antibiotic_Resistance-Student_lessonhandout.doc(DOC | 169 KB)
S2.Antibiotic_Resistance-Pre- and Post-assessment multiple choice questions with answer key.docx(DOCX | 200 KB)
S3.Antibiotic_Resistance-Alignment of learning objectives with lesson activities and assessment instruments.docx(DOCX | 18 KB)
S4.Antibiotic_Resistance-Lab protocols and Instructor Preparation Guide.docx(DOCX | 913 KB)
S5.Antibiotic_Resistance-Data Submission Form.pdf(PDF | 318 KB)
- License terms

Comments
Comments
There are no comments on this resource.