Using QIIME to Interpret Environmental Microbial Communities in an Upper Level Metagenomics Course
Published online:
Abstract
Metagenomics is rapidly evolving due to advances in sequencing technologies and bioinformatics, but without proper training to interpret these datasets, student understanding remains limited. Aimed at undergraduate juniors and seniors and graduate students, our eight-week metagenomics course introduces students to different approaches for producing and analyzing metagenomics data to test hypotheses. Here we present our approach for introducing students to the powerful command-line-based sequence analysis pipeline: Quantitative Insights into Microbial Ecology (QIIME, pronounced "chime"). Used extensively in metagenomics research, QIIME performs quality control analysis of raw data from high-throughput sequencers, clusters the data by taxonomic relatedness, and produces a data table of taxonomy by sample. This data table can be further analyzed with QIIME, or used in statistical analysis programs such as R or Primer. In our lesson, students first familiarize themselves with the format of QIIME commands with a worksheet that deconstructs a command to help students understand the input, output, and arguments. Our students have the most trouble with getting a functional version of QIIME running without error, so we have created instructional videos for the setup and running of basic QIIME commands. During one lab period, we lead the class through processing of a sample subset to identify issues and discuss limitations of this approach. Finally, student pairs analyze their own samples and present their findings to the class. This process builds student confidence in their ability to analyze high-throughput sequencing results of metagenomic datasets with the open-source QIIME pipeline.
Citation
Stevens, J.L., DeHority, R., and Goller, C.C. 2017. Using QIIME to Interpret Environmental Microbial Communities in an Upper Level Metagenomics Course. CourseSource. https://doi.org/10.24918/cs.2017.3Society Learning Goals
Bioinformatics
- DNA - Information Storage [GENOMICS]
- Where are data about the genome found (e.g., nucleotide sequence, epigenomics) and how are they stored and accessed?
- How can bioinformatics tools be employed to analyze genetic information?
- Ecology and Evolution [METAGENOMICS]
- How can bioinformatics tools be employed to examine ecological niches?
Microbiology
- Systems
- Why do most bacteria in nature live in biofilm communities?
Science Process Skills
- Process of Science
- Interpret, evaluate, and draw conclusions from data
- Construct explanations and make evidence-based arguments about the natural world
- Modeling/ Developing and Using Models
- Build and evaluate models of biological systems
- Quantitative Reasoning/ Using Mathematics and Computational Thinking
- Apply the tools of graphing, statistics, and data science to analyze biological data
- Communication and Collaboration
- Share ideas, data, and findings with others clearly and accurately
Lesson Learning Goals
Students will:- be able to discuss the steps of data processing of metagenomic amplicon sequences.
- value the importance of interpreting only high-quality sequences.
- know the limitations of metagenomic amplicon surveys.
Lesson Learning Objectives
Students will be able to:- list and perform the steps of sequence processing and taxonomic inference.
- interpret microbial community diversity from metagenomic sequence datasets.
- compare microbial diversity within and between samples or treatments.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Metagenomic studies are becoming increasingly common, and new, less expensive tools allow a wide diversity of researchers to participate in and contribute to this field. However, the large datasets, number of tools, and availability of numerous analytical approaches can be a barrier to novice scientists entering this field. Our challenge in the Biotechnology Program (BIT) at North Carolina State University was to create an eight-week course for juniors, seniors, and graduate students that provides an overview of key tools using evidence-based pedagogical approaches. This course is taken by biologists, chemical engineers, and entomologists, among other majors and graduate programs. Thus, lessons within this course had to: a) be accessible to participants with diverse educational backgrounds; b) use established tools without requiring extensive computer programming training; and c) highlight important concepts and misconceptions in the field.
With growing interest in microbiome research in several scientific fields, the skills acquired through this course are increasingly important for our undergraduate and graduate life science students. This field has led to discoveries on the ubiquity of microbes and their roles in both medically and ecologically significant processes. Prokaryotes have been implicated in human health disorders (e.g., 1,2), personalized medicine (3), agriculture (4), and even forensics (5,6). Sequencing of the 16S rRNA gene to identify bacteria from environmental samples has been the accepted approach since the phylogeny of prokaryotes based on this gene was published in 1980 (7). Since then countless studies have built on our knowledge of prokaryotic diversity and contributed to extensive, publicly available databases to which researchers can compare their samples for identification (e.g., 8). With the advancement of next-generation sequencing technologies, the ability to conduct high-throughput sample processing for high quality, meaningful sequences from large multi-gigabit datasets has become a necessary skill. In addition to high quality sample input, extensive quality filtering of sequence output drastically improves diversity analyses of complex community samples (9). To this end, our lesson seeks to provide students with the power to use open source software to analyze metagenomic amplicon datasets from their own research projects and understand the limitations of this approach. Rather than allowing our diverse students to simply submit their samples to "black-box" automatic filtering software, we guide them through each step of filtering, using the command-line based QIIME pipeline (10). Although other free software packages are available to process large metagenomic datasets (e.g., UPARSE, (12); mothur, (13)), QIIME is a relatively user friendly introduction to working in command line. We have found that our introduction to QIIME often leads to increased confidence in working in the command line; several students have taken it upon themselves to run further analyses in R (14). We are unaware of similar lessons published for this audience.
INTENDED AUDIENCE
This lesson was designed for juniors, seniors, and early graduate students from diverse majors and programs. We have taught this lesson to students in chemical engineering, biological sciences, soil science, entomology, functional genomics, biomanufacturing, and physiology concentrations. This lesson could also be useful for students in computer science, agriculture, and food science.
REQUIRED LEARNING TIME
In-class time required for this lesson is approximately 3-5 hours, a typical laboratory period. Out-of-class time to complete worksheets and follow download instructions requires approximately 2-3 hours.
This lesson is part of an eight-week, lab-based metagenomics course in which students prepare and sequence their own samples. This lesson prepares participants to use QIIME tool to analyze their own sequences by first learning about QIIME with a small but authentic data set. During the remainder of the course, students learn how to extract DNA from microbial communities, prepare libraries for high-throughput sequencing, and use a series of bioinformatics software to analyze their sequences. Student pairs typically spend approximately 6-10 hours analyzing their data and preparing presentations. Instructors and teaching assistants are available to support student analyses during office hours in preparation for their presentations.
PRE-REQUISITE STUDENT KNOWLEDGE
Students in our metagenomics course have successfully completed a semester-long molecular biology course where they learn key molecular biology techniques, including primer design, PCR, Western blotting, protein purification, plasmid purification, and restriction mapping. Previous computer programming experience is helpful but not required; the lesson presented here includes a brief introduction to working in a command line environment. Knowledge of basic biochemistry, genetics, and microbiology is required.
PRE-REQUISITE TEACHER KNOWLEDGE
Teachers should have a working knowledge of molecular biology, genetics, and be comfortable using command line programs. By completing our lesson prior to instruction, teachers will be able to test their level of comfort with the techniques. Teachers should also have familiarity with metagenomic amplicon sequencing.
SCIENTIFIC TEACHING THEMES
ACTIVE LEARNING
Throughout this lesson, students perform analysis tasks along with the teacher. Students remain engaged with each step of sequence processing, assuring that they are able to successfully run each QIIME command. With each step, students tend to ask the teacher conceptual questions about decisions being made for filtering. When students arrive at technical hang-ups, they most often help each other figure out how to move forward and, by the end of lesson, are able to self-troubleshoot. This lesson has the goal of preparing students in the course to use QIIME to analyze the sequences from their own student-processed samples. To summarize results of their sequencing efforts, students work in pairs to process and analyze samples following the methods of this QIIME lesson, then present their analysis to the class in a conference-style scientific presentation.
ASSESSMENT
To meet the learning objectives, two worksheets are assigned and graded (complete/incomplete), and a final data analysis project/presentation is evaluated by reviewers using a rubric (Supporting files S1, S2, and S3). The worksheets are treated as low-stakes assignments intended to encourage students to research the tools and explain them in writing. Worksheets are returned with feedback to students and contributed to their in-class assignments grade (5% of final grade). The final presentations are graded by averaging the rubrics from four independent reviewers (JS, CG, the graduate teaching assistant, and an invited postdoctoral fellow). If reviewers are not available, the instructor can grade final presentations based on the rubric. Final presentation/data analyses scores contributed to 15% and 10% of the course grade for undergraduate and graduate students, respectively.
INCLUSIVE TEACHING
The lesson uses a scaffolded approach by providing students with pre-activity worksheets and online video tutorials. We assign the worksheets as homework to minimize stress and to allow students time to research the tools and formulate their answers. We provide the worksheets both as physical handouts and as editable electronic documents. The ongoing video tutorial series can be shown in the classroom or used as a stand-alone resource on the basics of QIIME and the command line interface. The videos we developed are hosted on YouTube (see Supporting File S4 for a list and links), and therefore completely public and free to use. Using QIIME, a free-to-download software, rather than expensive bioinformatics tools increases the likelihood that those who do not have access to commercial tools are still able to learn. Also, with inclusion in mind, the videos do not assume the viewer has extensive technical knowledge and they do not skip steps. Every video is closed-captioned to increase accessibility. Finally, during the in-class QIIME activities, we check student progress at each step to ensure nobody falls behind. Often, these check-ups encourage students to help each other, promoting peer discussion and learning.
LESSON PLAN
Teacher Preparation
The best preparation for our QIIME lesson is for the teacher to run through the entire activity in advance of beginning the lesson in class. To do this, the annotated sequencing pipeline (Supporting File S5, link to folder: https://drive.google.com/open?id=0B33BU08B2owHbzI0YURiMHNIQnM) should be used as a guideline. This pipeline is kept up to date with QIIME updates at (https://github.com/JuliaLStevens/qiime_workshop_pipeline). Teachers should also familiarize themselves with the "QIIME Scripts" page maintained by developers (http://QIIME.org/scripts/). Additionally, teachers should have computers with proper QIIME installs ready for students who are unable to use their own laptops. For loading QIIME onto a PC, see our instructional video for running QIIME through VirtualBox. Mac users can use MacQIIME. Detailed download and install instructions are provided by SUNY Cortland's Werner Lab (http://www.wernerlab.org/software/macqiime/macqiime-installation).
Student Pre-Lesson Activity
Worksheets
We developed two worksheets to prepare students prior to the QIIME in-class lesson.The first worksheet, assigned at least a week before the in-class activities, focuses on fundamental QIIME commands and mapping files (see Supporting File S1). Completed worksheets were reviewed by the teacher, returned with feedback to the student, and discussed in class. During class, we gave students a second worksheet covering two important mothur (http://www.mothur.org/) tools and assigned the worksheet as homework (see Supporting File 2). This completed worksheet was reviewed by the teacher, discussed in class, and returned to students prior to the QIIME activities. Depending on your preference, both worksheets can be assigned simultaneously.
Video tutorials
To prepare students for the QIIME lesson, we developed a series of three tutorials to help students install the software on their personal computers (using VirtualBox on PCs). In addition, we provided students with two basic command line tutorials focusing on essential commands for working with files. Students were encouraged to watch the videos in an email with a link to the NCSU BIT YouTube channel and a reminder to complete their second worksheet. In one offering of the course, the command line video was played in class immediately before using QIIME, to remind students how to navigate in the command line environment. Additionally, we produced two videos to help students create and validate QIIME mapping files using a Google Sheets add-on called Keemei.
In-Class QIIME Lesson
After checking that students had access to a working version of QIIME and that the version was up to date, we provided students with a compressed file containing raw sequence files from a previous experiment. The sequences were produced from samples taken from warming chambers in Duke Forest for a study that assessed the microbial diversity as part of a multi-year climate change experiment (see: http://robdunnlab.com/projects/warming-chambers/). The compressed file also contains: a) QIIME-formatted mapping file with corresponding environmental metadata, b) output files of long-running commands to streamline the lesson, c) the annotated sequence processing pipeline. This entire file is available as a compressed file in supporting material (Supporting File S5, link to folder: https://drive.google.com/open?id=0B33BU08B2owHbzI0YURiMHNIQnM).
The lesson began with a brief introduction to important Unix commands used to navigate within the shell (Supporting File S6) and discussion of terminology (Supporting File S7). Once students were comfortable moving in and out of directories and keeping track of where they were working, we started QIIME. The workflow begins with joining the paired end reads of six samples and one laboratory control. After joining, fastq files must be converted to fasta files; here we taught students how to automate this process using the custom python command in our pipeline. For ease, this conversion could also be done one sample at a time. Once converted, all sequences were combined into one fasta file. Our students now began to count sequences after each step to keep track of how their sequence processing was cleaning the dataset.
At this point, we used QIIME's ability to switch between software programs to start mothur (5). Within mothur, we ran summary statistics on our combined dataset. Take this opportunity to discuss with students the importance of knowing details of your target sequence. In our case, using primer pair 515F/806R, we expect a 291 base pair sequence, but summary statistics show that more than half of the sequences in this dataset are only 35 base pairs. This amount of short sequence reads indicates a lot of primer dimer, so now we discussed what we could have done in the lab prior to sequencing to reduce the sequencing of extraneous, false sequences. Additionally, a region of repeating base pairs (i.e., homopolymers) longer than eight base pairs is statistically improbable and indicative of sequencing errors, so we also wanted to remove samples with homopolymers longer than eight base pairs and any sequences with ambiguous base pairs.
After returning to QIIME, students were now ready to pick operational taxonomic units (OTUs). In our pipeline, we discussed "chimera checking" with our students and showed them vsearch, freely available software that can handle the computational load of a large Illumina dataset. To cut down on lesson time and complications of downloading another software program, we chose to give our students the output file. Using our provided chimera-free file, students moved forward with OTU picking. There are several options for OTU picking that can be explored on the QIIME commands webpage, but we chose to remain basic in our commands: we picked OTUs based solely on their similarity to other sequences within the dataset; anything with similarity 97% and above grouped into one OTU. Using these groupings, a representative fasta file was constructed using only one sequence per OTU to reduce computational load.
The representative sequence set needs to be aligned against a reference database. For this purpose, we used the default Greengenes database (2). Sequences that successfully aligned were now filtered to remove spurious alignments and gaps. Both alignment and filtering are computationally expensive and can take multiple hours to complete, so here again we provided students with the output files to move forward in the lesson. Using default QIIME arguments, taxonomy was assigned for the filtered, aligned representative sequence fasta file against the Greengenes database with the RDP algorithm (6). Students then made a phylogenetic tree using the fasttree algorithm so that they could use the Unifrac phylogeny-based community similarity method downstream. Finally, an OTU table was created based on per sample OTU abundances excluding sequences that did not successfully align. The OTU table was filtered to remove singleton sequences, unclassified sequences, and any OTUs present in the laboratory control sample. This sample is processed through all laboratory steps but without sample DNA added serving as a negative reagent control.
With this filtered OTU table, phylogenetic tree, and mapping file, students ran the core diversity analysis command, choosing the rarefaction number based on the sample with the lowest sequence representation. This command runs a variety of standard alpha and beta diversity tests based on given metadata variables. The lesson ended with exploring the extensive output of the core diversity analysis command and conversion of the BIOM-formatted OTU table to a text file. This text file could now be further analyzed in our students' choice of statistical analysis platforms.
Student Pair Project
After the QIIME tutorial, students were challenged to analyze their own metagenomic data using QIIME and other tools such as the MG-RAST web server, Geneious software, or the CyVerse Discovery Environment web portal. Students worked in pairs on their presentations for two weeks. During that time, teacher and teaching assistant office hours were used to help support students with their analyses. Presentations were graded by four reviewers using a rubric (Supporting File S3), although reviews can be conducted by the teacher alone if no other appropriate reviewers can be found. This conference-style oral presentation encouraged student groups to analyze their data, make connections to other datasets (publicly available or class data from other groups), summarize their methods and results, and, importantly, consider limitations and future directions. We encouraged students to discuss the limitations of amplicon-based metagenomic approaches after experiencing some of the challenges of preparation of 16S libraries in the laboratory and how the QIIME pipeline filters and classifies reads to create OTU tables. These limitations include lack a taxonomic resolution assessed by short read sequencing, difficulty in sequence analysis, and lack of information on bacterial function within microbial communities. The student pair project also provides students with the opportunity to practice oral communication skills and evaluate their own analyses and those of their peers.
TEACHING DISCUSSION
This lesson was a successful practice for our students in building computer coding confidence and informed decision making during sequence data processing. To this end, our lesson purposefully avoided the batch commands offered by QIIME that would otherwise streamline many of the steps our students performed. A possible extension on this lesson would be to include instructions for writing batch commands, which can then be run on a high performance computing (HPC) cluster such as the Virtual Computing Lab at NCSU (https://vcl.ncsu.edu/). As each computer core has specific instructions for using their facility, we recommend contacting your cluster's point person to write these extension instructions.Localized processing would allow participants to access the software from their own devices.
In this lesson, we focus on processing 16S rRNA gene amplicon sequences from environmental samples. Many of the same processing steps can be used for processing of other amplicons, but we stress the importance of making informed decisions for size filtering and database selection. Our laboratory section is five hours long, so we conduct this lesson in one class which typically takes approximately four hours. It is possible for teachers to break this lesson into two sessions. If breaking up into multiple days, we recommend having students attempt to run the alignment and filtering steps overnight themselves. This step will make a good stopping point for session one, and session two can then begin with building the OTU table leaving plenty of time for fully exploring the core diversity analyses output.
To ensure students watch the videos and successfully install QIIME on their computers, a formative assessment in the form of an in-class quiz with questions about the videos and QIIME would be helpful. To better assess the skills of participants after the lesson, a practical quiz on QIIME tools can be used. Students would be provided with a challenging task and sample dataset to process using QIIME. This task could be performed either individually or in groups. Teachers could tailor the assignment to address aspects such as familiarity with command line, processing of raw sequences, OTU picking, and/or diversity analyses. Participants would complete the task during a (computer) lab period, following a worksheet describing the nature of the dataset and objectives and recording their results after indicated steps. This assessment would allow teachers to gauge how participants are using QIIME after the lesson. In addition, a practical quiz on QIIME tools would emphasize learning with real data, retrieval practice, and provide participants with an additional opportunity to receive feedback and gain familiarity with QIIME. The practical quiz could be graded or simply be an in-class activity. Worksheets would be collected by teachers and evaluated to improve the QIIME activity and provide additional clarification, if needed. Worksheets could be submitted either as hard copies or as part of electronic lab notebooks. For example, in the fall of 2015 and 2016, and spring of 2016 we used LabArchives (http://www.labarchives.com/) electronic lab notebooks in this course.
Our experience conducting this QIIME lesson in three offerings of the metagenomics course highlighted that coding difficulties often challenge students of all academic levels, both undergraduate and graduate students. In our experience, a handful of students get frustrated and begin to stall. The presence of a knowledgeable and engaged teaching assistant (TA) was critical to keep students on task and moving forward with the analyses. The use of smaller, more manageable but authentic dataset, together with breaks for occasional group discussions to emphasize key points and resolve common issues significantly improved the lesson. Although coding difficulties can be frustrating, helping students troubleshoot and resolve issues on their own often resulted in re-energized participants.
This Lesson allows participants to work with QIIME and real sequence datasets to familiarize themselves with the workflow and commands for an amplicon-based metagenomics analyses. Importantly, the Lesson engages students with a challenging, authentic task that is scaffolded so that participants can familiarize themselves with key concepts and issues while wrestling with the complexities of the process. Several modalities are used to expose students to the information, including worksheets, a short background lecture on data processing used to introduce background knowledge, hands-on lesson, and videos (Table 1). Summative assessments allow the teacher to gauge student understanding and tailor the pre-lesson mini-lecture/discussion and lesson to the needs of the participants. The student pair project then challenges students to apply the process they learned to their own samples, summarize their methodology and results, and consider future directions and limitations of their approach while promoting practice of oral communication. This Lesson is in alignment with the competencies described by the Vision and Change report (15) and the need to better prepare our students for the challenges of analyzing complex datasets using bioinformatic tools. We hope that the modularity and flexibility of this lesson will encourage other educators to try QIIME in their courses.
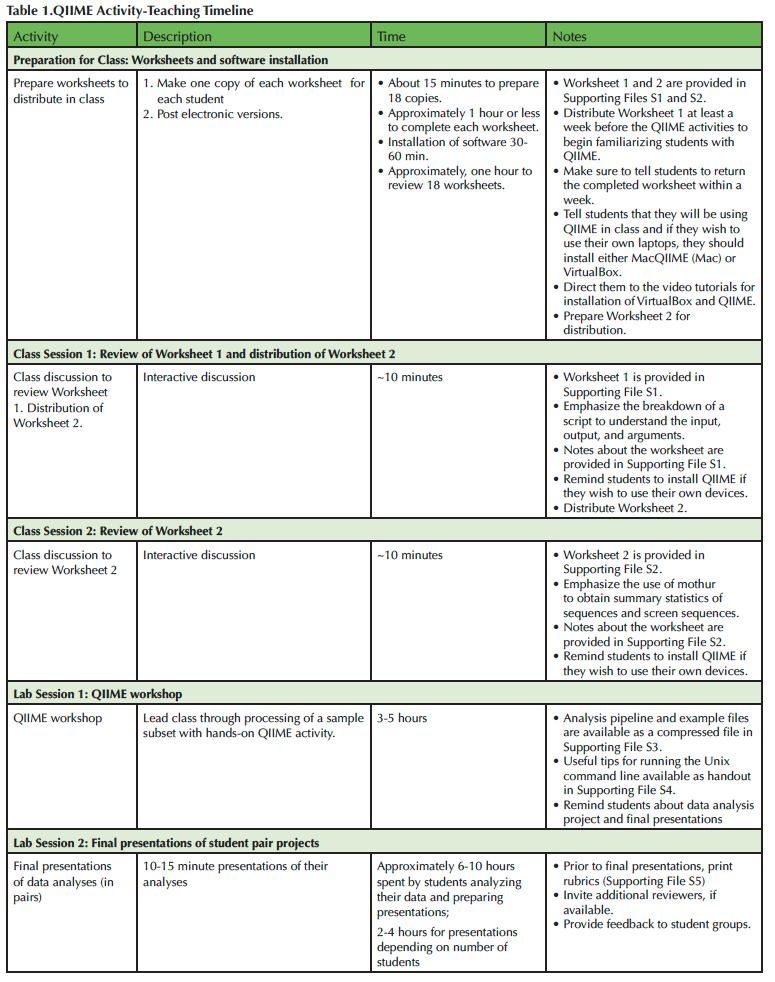
Table 1. QIIME Activity - Teaching Timelline
SUPPORTING MATERIALS
S1. QIIME activity: Assignment 1 Worksheet
S2. QIIME activity: Assignment 2 Worksheet and key
S3. QIIME activity: Pair presentation grading rubric
S4. QIIME activity. List of tutorial videos and links
S5. QIIME activity: Compressed file with raw data, long step output files, annotated pipeline LINK TO FOLDER: https://drive.google.com/open?id=0B33BU08B2owHbzI0YURiMHNIQnM
S6. QIIME activity: Worksheet/handout with Helpful basic commands presentation
S7. QIIME activity: Glossary of commonly used terms in this lesson
ACKNOWLEDGMENTS
The authors wish to thank the NCSU Biotechnology Program (BIT) for support. The NCSU Office of Faculty Development Summer Institute and the Graduate School provided support in learning and applying teaching approaches and tools. Funding was provided by the Biotechnology Program. The authors would like to thank the course participants for their enthusiasm and helpful feedback. The NCSU Office of Information Technology Accessibility program provided funds for captioning the YouTube videos.
References
- Turnbaugh P, Ley R, Mahowald M, Magrini V, Mardis E, Gordon J. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 444.7122: 1027-131.
- Koopen A, Groen A, and Nieuwdorp M. 2016. Human microbiome as therapeutic intervention target to reduce cardiovascular disease risk. Current Opinion in Lipidology. 27(6): 615-622.
- Zmora N, Zeevi D, Korem T, and Segal E. 2016. Taking it personally: personalized utilization of the human microbiome in health and disease. Cell Host & Microbe 19(1): 12-20.
- Zhu S, Vivanco J, and Manter D. 2016. Nitrogen fertilizer rate affects root exudation, the rhizosphere microbiome and nitrogen-use-efficiency of maize. Applied Soil Ecology. 107: 324-333.
- Fierer N, Lauber C, Zhou N, McDonald D, Costello E, and Knight R. 2010. Forensic identification using skin bacterial communities. Proceedings of the National Academy of Sciences. 107(14): 6477-6481.
- Franzosa E, Huang K, Meadow J, Gevers D, Lemon K, Bohannan B, and Huttenhower C. 2015. Identifying personal microbiomes using metagenomic codes. Proceedings of the National Academy of Sciences. 112(22): E2930-E2938.
- Fox G, Stackebrandt E, Hespell R, Gibson J, Maniloff J, Dyer T, Wolfe R, Balch W, Tanner R, Magrum L, Zablen L, Blakemore R, Gupta R, Bonen L, Lewis B, Stahl D, Luehrsen K, Chen K, and Woese C. 1980. The phylogeny of prokaryotes. Science. 209(4455):457-463.
- DeSantis T, Hugenholtz P, Larsen N, Rojas M, Brodie E, Keller K, Huber T, Dalevi D, Hu P, and Andersen G. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology. 72(7):5069-5072.
- Bokulich N, Subramanian S, Faith J, Gevers D, Gordon J, Knight R, Mills D, and Caporaso JG. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nature Methods. 10(1):57-59.
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Gonzalez Pe?a A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widman J, Yatsunenko T, Zaneveld J, and Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 7:335-336.
- Kuczynski J, Stombaugh J, Walters WA, Gonz?lez A, Caporaso JG, and Knight R. 2011. Using QIIME to analyze 16S rRNA gene sequences from Microbial Communities. Current Protocols in Bioinformatics. Ed. Andreas D Baxevanis. Chapter 10.7.
- Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nature Methods 10(10): 996-998.
- Schloss P, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, and Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology. 75(23): 7537-7541.
- Ihaka R and Gentleman R. 1996. R: A language for data analysis and graphics. Journal of Computational and Graphical Statistics. 5(3):299-314.
- American Association for the Advancement of Science, Vision and change in undergraduate biology education: A call to action. (AAAS Press, 2011).
Article Files
Login to access supporting documents
Using QIIME to Interpret Environmental Microbial Communities in an Upper Level Metagenomics Course(PDF | 135 KB)
S1. QIIME activity-Assignment 1 Worksheet.docx(DOCX | 25 KB)
S2. QIIME activity-Assignment 2 Worksheet .docx(DOCX | 20 KB)
S3. QIIME activity-Pair presentation grading rubric .docx(DOCX | 18 KB)
S4. QIIME activity-List of tutorial videos and links.docx(DOCX | 17 KB)
S6. QIIME activity-Worksheethandout with Helpful basic commands presentation.pdf(PDF | 59 KB)
S7.QIIME activity-GlossaryofCommonlyUsedTerms.docx(DOCX | 14 KB)
- License terms

Comments
Comments
There are no comments on this resource.