Investigating the Function of a Transport Protein: Where is ABCB6 Located in Human Cells?
Published online:
Abstract
One of the challenges of teaching Cell Biology is helping students understand important research methods used to study cells. The goal of understanding cell biology methods is especially challenging in courses without a laboratory component. When studying cells, determining the location of a protein is important for understanding the protein's cellular function. In this lesson, upper-division Cell Biology students will learn about two commonly used methods for protein localization: 1) immunoblotting after differential centrifugation, and 2) immunofluorescence microscopy. They will work in small groups to answer questions in a problem set written in the spirit of Process-Oriented Guided Inquiry Learning (POGIL). Students will explore key points of the two methods and then apply their knowledge to the analysis of protein localization data from the primary literature. Analyzing protein localization data will help students develop the ability to “apply the process of science”, which is one of the Vision and Change core competencies.
Citation
Stanton, J.D., and Dye, K.M. 2017. Investigating the Function of a Transport Protein: Where is ABCB6 Located in Human Cells? CourseSource. https://doi.org/10.24918/cs.2017.19Society Learning Goals
Cell Biology
- Methods & Tools of Cell Biology
- How do the methods and tools of cell biology enable and limit our understanding of the cell?
Lesson Learning Goals
How do protein localization methods enable and limit our understanding of the cell?Lesson Learning Objectives
At the end of this activity students will be able to:- describe the use of two common research techniques for studying proteins: SDS-PAGE and immunoblot analysis.
- determine a protein’s subcellular location based on results from: 1) immunoblotting after differential centrifugation, and 2) immunofluorescence microscopy.
- analyze protein localization data based on the limitations of differential centrifugation and immunofluorescence microscopy.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Students of Cell Biology need to understand how new knowledge is obtained in the field. For example, proteins carry out critical functions for cells. How do cell biologists begin to characterize a protein's function in a cellular context? Through genomics and bioinformatics efforts, scientists have identified an impressive number of protein-coding genes, but our understanding of the functions of their products is less advanced. A first step towards understanding a protein's function is to identify the cellular location of the protein. In our 300-level Cell Biology course, we wanted to help students understand two major methods for localizing proteins within cells: 1) immunoblotting after differential centrifugation, and 2) immunofluorescence microscopy. Then, we wanted them to be able to interpret data from these methods. We realized that to do so, we first had to help them review common research techniques covered in their pre-requisite courses, such as SDS-PAGE and immunoblot analysis (or Western blotting). To meet these goals, we created a student-centered lesson in the style of Process Oriented Guided Inquiry Learning (POGIL) (1, 2).
Background on protein localization methods
Protein localization is an important biological process and its study spans research across the life sciences including fundamental biology research and medical research. It even has application in therapeutic treatments of human disease (3). One way to determine a protein's cellular location is by using differential centrifugation followed by immunoblot analysis (4, 5). In differential centrifugation, cell components are separated based on size and density. Hypotonic solutions and sonication or homogenization are used to break open or “lyse” cells, resulting in a “whole cell lysate”. The lysate is centrifuged at a low speed to pellet nuclei, which are the largest cell component. The supernatant of the low-speed spin contains all other cell components besides nuclei. The low-speed spin supernatant is then centrifuged at a higher speed to pellet other cell components. This process is repeated with progressively higher centrifuge spins. The higher the centrifuge speed, the smaller the cell component that can be obtained in a pellet. At the end of differential centrifugation, the pellets from the different centrifuge steps represent different “fractions” of the cells. Proteins from each pellet can be separated based on size and charge using SDS-PAGE. Separated proteins are then transferred to a membrane for immunoblot analysis. Primary antibodies that recognize marker proteins are used to identify fractions that contain a particular organelle. For example, an immunoblot probed for the mitochondrial marker protein, porin, will indicate which pellets contain mitochondria. If a protein of interest is present in the same fractions as porin, the protein of interest is likely to be located in mitochondria. If a protein is localized to the mitochondria, this suggests that the protein may have a role in mitochondrial structure or function.
Another approach commonly used by cell biologists to determine protein localization is immunofluorescence microscopy (6, 7). Similar to immunoblotting, immunofluorescence microscopy relies on the use of primary antibodies specific for a protein of interest and marker proteins. Cells are “fixed” to slides by chemically treating the cells to kill and preserve them. Cell fixation is also important for permeabilizing cell membranes to allow antibodies to enter and bind to their target proteins. Then cells are viewed using a fluorescence microscope with filters for each fluorophore, compounds that emit light of a specific wavelength when excited by another wavelength of light. If the signal for a lysosomal marker protein, such as Lamp1, overlaps with the signal for a protein of interest, the protein of interest is likely to be in the lysosomes. Thus, it can be hypothesized that the protein has a role in the structure or function of lysosomes. For a more detailed explanation of these techniques we recommend instructors consult Alberts' Molecular Biology of the Cell (8), or Lodish's Molecular Cell Biology (9).
Background on Process-Oriented Guided Inquiry Learning (POGIL)
POGIL is an active-learning approach centered on small-group work. One of the principles behind the POGIL approach is the idea that a learner must construct knowledge in their own mind in order to learn (10). In addition, POGIL acknowledges that students working in groups can learn more, understand more, and remember more than when they work alone (11, 12). POGIL encourages students to explore content, outline concepts, and apply concepts (1), while focusing on the use of process skills such as teamwork, communication, and problem solving (13). We use POGIL in our Cell Biology class because it allows our students to actively learn and use cell biology concepts while developing process skills that they can transfer to a variety of settings.
POGIL involves small groups of three to six students with each student playing a defined role. Because we teach our Cell Biology class in a SCALE-UP room with tables designed for groups of three (14), we limit our groups to three students playing the following roles: Manager, Recorder, or Presenter (1). The Manager keeps the group focused and makes sure all members contribute. The Recorder writes down the group’s answers on the official copy of the problem set, which is the only one from the group that will be graded. The Presenter speaks on behalf of the group in class discussion and shares answers on the white boards. All students contribute their ideas to help complete the problem set. The problems sets are designed to guide students from knowledge-based questions to analysis and evaluation questions (1). We do not call our problem sets “worksheets” because our students say this reminds them of being in grade school.
Our protein localization lesson
An education in the life sciences involves both the acquisition of knowledge and the development of skills to apply that knowledge. In some circumstances, it is not possible to provide students with hands-on experience in the laboratory. We know from cognitive learning theory that providing our students with the opportunity to construct their own knowledge during an engaging activity leads to improved learning (15, 16). Using primary literature to provide students with data to analyze is one way to involve them in investigating commonly used research methods. Applying the process of science in this way gives students the opportunity to “discover” the way a method works and then to analyze real data without the constraints of having to successfully perform the method in the laboratory.
We provide a lesson in the spirit of POGIL to allow students to explore protein localization methods. This activity is completed in a single class period and is completely student-driven, meaning that it involves no lecture time. When students encounter unfamiliar terms or concepts they work together to develop their understanding. Students work in small groups on a problem set designed to guide them from knowledge-based questions to analysis and evaluation questions. At the end of the lesson, students engage in a class discussion. Students volunteer to share their answers to the problem set and the instructors address misunderstandings that may have emerged during the activity. For example, some students may imagine that the four panels shown in question 3 are from one immunoblot. To address this, we remind students that four different immunoblots were performed and the researchers cropped the immunoblot images so readers can compare the results from each fraction more easily.
We centered our POGIL activity around a paper by Kiss et al. because the paper’s results provided students with the opportunity to analyze cell biology localization data (17). The authors of the paper studied a type of transport protein from the adenosine triphosphate–binding cassette (ABC) family called ABCB6. ABCB6 was predicted to be responsible for porphyrin uptake into mitochondria. The authors wanted to characterize the function of ABCB6, and they started by investigating the protein’s location. One of the major research questions they asked was: “Where is ABCB6 found in mammalian cells?” The authors used multiple cell biology methods to answer this question, and they were surprised to conclude that ABCB6 does not localize to mitochondria. One reason we chose this paper is because it underscores the importance of documenting the location of a protein rather than just predicting its location. Another reason we chose this paper is because the data are not as clear as they are in textbook cartoons. We wanted our students to appreciate that real data can sometimes be challenging to interpret and that using additional methods can help strengthen conclusions.
Intended Audience
We teach this lesson in a 300-level Cell Biology course at a public research university. The lesson we share has been taught in more than ten sections of Cell Biology, including by the authors and other instructors. Our Cell Biology students are typically junior or senior life science majors.
Required Learning Time
We teach this lesson in a single 75-minute class period. We have also taught it in a 50-minute class period (see Adaptations in Teaching Discussion).
Pre-requisite Student Knowledge
Our Cell Biology students learn about SDS-PAGE and immunoblotting in their pre-requisite courses, which include Genetics and Biochemistry. If your students are not familiar with SDS-PAGE and immunoblotting, you can introduce them to these techniques in class or through a homework assignment.
Pre-requisite Teacher Knowledge
You should read the Kiss, et al., paper on the ABCB6 transporter, which contains the original data used in the lesson (17). You should be familiar with all of the methods covered in the problem set: SDS-PAGE, immunoblotting, differential centrifugation, and immunofluorescence assay. Alberts' Molecular Biology of the Cell (8) and Lodish's Molecular Cell Biology (9) have excellent coverage of these topics. Older versions of both books are available at no cost via the National Center for Biotechnology Information Bookshelf (www.ncbi.nlm.nih.gov/books).
SCIENTIFIC TEACHING THEMES
Active Learning
Active learning occurs when instructors step away from the traditional lecturer role and allow students to take control of developing their own conceptual understanding (18). This student-driven lesson involves three major activities.
- Students work collaboratively in groups to answer the problem set questions. These peer-to-peer interactions allow the students to share their prior knowledge and discover new understanding together.
- Students work together to interpret data from the primary literature. Working together to decipher the data models the collaborative nature of the scientific process.
- Students present their findings during class discussion. These brief presentations help students build their communication skills while also keeping the responsibility for learning focused on the students rather than to the instructor.
Assessment
Formative Assessment
Each group of three students turns in one official copy of the problem set for grading. This is the Recorder’s problem set, which will be returned to the Recorder after grading. The instructor or a teaching assistant provides detailed written feedback. We have taught this lesson in class sizes ranging from 40 to over 200 students. We cut grading down to a third by only providing feedback on the Recorder’s official copy. We often have teaching assistant (TA) support for grading, but we have also given feedback ourselves when TA support was not available. When the official copies of the problem set are returned we encourage group members to photograph the written feedback if they did not serve as the Recorder. We discuss common points of confusion at the beginning of the next class session.
Summative Assessment
We include “matched-pair” questions on our unit exam and/or final exam that are related to those in the problem set (Supporting Materials File S4). The exam questions are similar to, but not the same, as the ones on the problem set. Matched-pair exam questions require students to apply the knowledge and skills they gained in the lesson to new situations (19).
Inclusive Teaching
This lesson uses groups of three with roles that were defined by the POGIL method. Each group member has a specific role: Manager, Recorder, and Presenter; explained below. The roles rotate weekly to ensure that students have the opportunity to experience each role. This structure encourages students with different backgrounds to share their knowledge. For example, students from underrepresented groups will have the chance to play each type of role, rather than only the roles that students from the predominant group might unconsciously restrict them to.
LESSON PLAN
We designed this lesson for a single 75-minute breakout period, but we have also adjusted it for a 50-minute class period. We teach the lesson in a SCALE-UP classroom equipped with round tables and computers that students can use in groups of three. The lesson can also be taught in classrooms with moveable chairs or even lecture halls with fixed seats (see Adaptations). Students will need internet access and an electronic device to answer some of the questions. A suggested timeline is provided in Table 1.
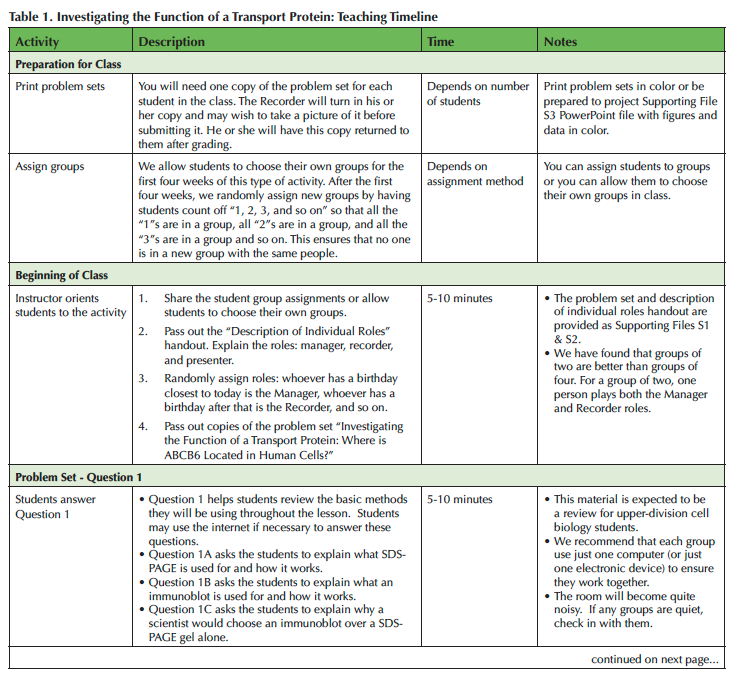
Table 1. Investigating the Function of a Transport Protein: Teaching Timeline, page 1
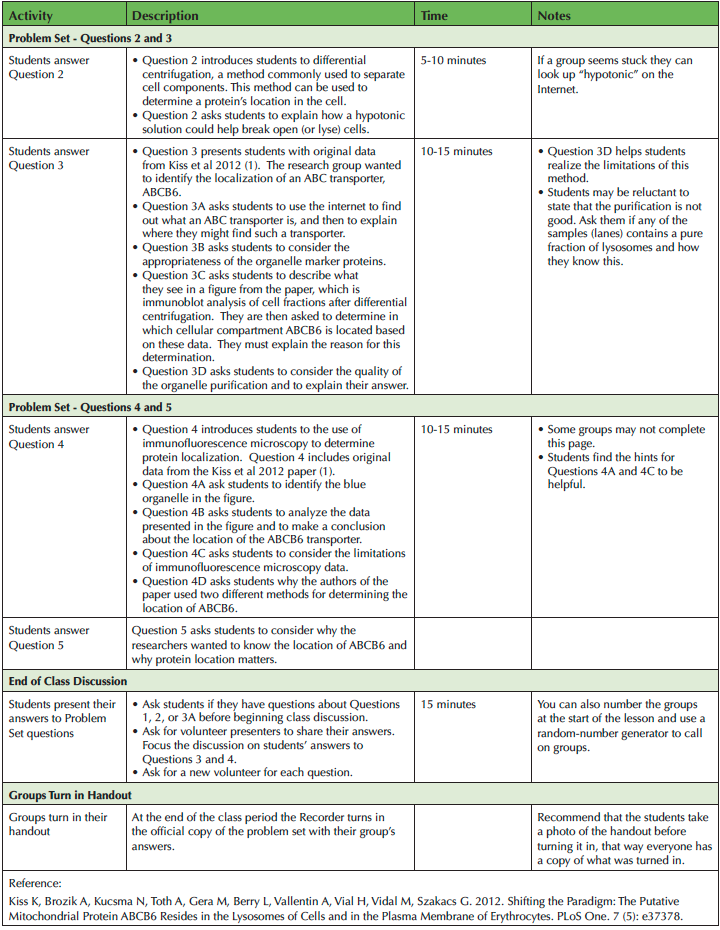
Table 1. Investigating the Function of a Transport Protein: Teaching Timeline continued
Pre-class Preparation
Before class you will need to assign students to groups (if you choose to) and print copies of the problem sets (see Supporting Materials File S1). The problem set is five pages long and requires color printing. If you are unable to print in color you can project the figures that require color for interpretation using an LCD projector (see Supporting Materials File S3).
Beginning of Class
We use this as the first activity of the semester; if your students are already used to working in groups you may be able to spend less time preparing them for the activity. You will begin by explaining and assigning roles: Manager, Recorder and Presenter (see Supporting Materials File S2). The Manager keeps the group on task, the Recorder writes the group's answers on the official copy of the problem set, and the Presenter presents the groups answers during the class discussion at the end of class. The group roles are randomly assigned. The student with the birthday closest to that day is the Manager, the student with the birthday next closest is the Recorder, and so on. Give the students a few minutes to meet each other and determine who has what role. Once the roles are determined, the groups answer the questions provided in the problem set in the order in which they appear. We ask our students to use just one computer per group so that all three members of a group work together rather than working on their own in parallel.
Problem Set Page One
Question 1
Question 1 of the problem set guides students through a review of SDS-PAGE and immunoblotting. It includes a cartoon depiction of both methods (S1-Figure 1). Students explain the purpose of SDS-PAGE and how it works (S1-Question 1A). This information is prior knowledge for our Cell Biology students, but we encourage them to use an online search engine to look up any information they cannot remember. The process of looking up forgotten information keeps the responsibility for learning focused on the students rather than on the instructor. Students then explain the purpose of an immunoblot and how it works (S1-Question 1B). Finally, students explain why a scientist would choose immunoblotting instead of an SDS-PAGE gel alone (S1-Question 1C). These questions set students up for the next section of the problem set where SDS-PAGE and immunoblotting are both used for protein localization.
Problem Set Page Two
Question 2
Question 2 introduces students to the use of differential centrifugation to purify cell components. The students are asked to read about differential centrifugation and examine Figure 2. Then they are asked about how a hypotonic solution can help break open or lyse cells. Differential centrifugation is used to obtain the data in Question 3.
Question 3
Question 3 introduces students to data from Kiss, et al., 2012 (17), where the authors report their investigation of the cellular location of ABCB6, a member of the adenosine triphosphate-binding cassette (ABC) transporter family. The researchers used differential centrifugation, SDS-PAGE, and four immunoblots. Figure 3 is the data from immunoblots of organelle pellets obtained from human K562 leukemia cells.
Question 3A asks students to find out what an ABC transporter is, using an online search engine if necessary, and to explain where they might find such a transporter. Question 3B asks about the suitability of the organelle marker proteins. Their answer prepares them for Question 3C, which asks them to describe the data they see in Figure 3. They are asked to determine where ABCB6 is located based on these data. They are then asked to provide a justification for their analysis.
Question 3D asks students whether or not the researchers were able to achieve a high level of organelle purity based on the data in Figure 3. Students are also asked to provide a rationale for their conclusion. This question helps students learn to analyze data critically.
Problem Set Page Three
Question 4
Question 4 introduces students to immunofluorescence assay, an additional method for identifying the localization of a protein. The description explains how immunofluorescence labeling of proteins uses similar principles as immunoblotting. The description then introduces the use of fluorescent probe-conjugated antibodies to visualize protein locations.
Question 4A asks students to identify the “blue” organelle in the micrographs. Most students will correctly identify the blue organelle as the nucleus, especially with the provided hint to consider the shape and size of the blue organelle. If some students say "plasma membrane", encourage them to think about where the lysosomes and mitochondria are located. These organelles are shown outside the "blue" organelle. Thus, the "blue" organelle cannot be the plasma membrane.
Question 4B asks students to interpret the data in the figure and make a conclusion about where the ABCB6 transporter is located in the cell. The three-dimensional nature of the cell versus the two-dimensional nature of a micrograph may confuse students. Some will say the mitochondria should not be inside the nucleus. Encourage them to consider how a micrograph is different from an actual cell, and whether or not the image indicates that one organelle is inside the other.
Question 4C asks students whether co-localization data can definitively tell you the location of an organelle. Similar to Question 3D, students must consider the limitations of using immunofluorescence assay for determining protein location.
Question 4D asks students to explain why the researchers used two approaches to test the location of ABCB6. This question will prompt them to consider the limitations of the methods.
Question 5
Question 5 asks students to think about why it is useful to know the location of a protein in a cell. This brings a greater context to the lesson.
Completion of Problem Set/End of Class
When about 15 minutes of class remain, ask students to stop working. Then ask for volunteers to come to the front of the room to share their answers. A document camera can be used to project students’ answers. Begin the discussion with Question 3, unless students have questions about the earlier problem set questions. Ask for one of the Presenters to volunteer to answer question 3C. In discussing Question 3C, students should mention that the patterns of Lamp1 (lysosomes) and the ABC transporter are similar, but that the results are not conclusive since there are no samples with pure lysosomes.
Ask for another Presenter to answer 3D. Students should note that all of the fractions have more than one cell component, indicating a lack of purity. You may want to ask them how they could improve the purity (e.g., purification with antibody-conjugated beads or additional gradients). Ask them to predict the immunoblotting results from a fraction of pure lysosomes as a hypothetical 6th fraction.
Request a new Presenter to answer 4A and 4B. For 4A, students will use size and shape to determine that the organelle in question is the nucleus. For 4B, students should discuss the three-dimensional nature of the cell versus the two-dimensional format of a micrograph. If time allows, ask for another Presenter to discuss question 4C and 4D. Students should note that the apparent overlap in a 2D image might represent structure in front of or behind one another, rather than inside one or the other. They should also point out that the use of two methods helps strengthen the authors' conclusion.
When discussion is complete be sure to thank the students for their participation as you collect the Recorder’s completed copy of the problem set.
TEACHING DISCUSSION
Activity’s effectiveness at achieving the stated learning goals and objectives
This activity is effective in helping students understand two protein localization methods. The lesson also helps students interpret data generated by these two methods. The constructive nature of the activity gives the students ownership of the knowledge. Their level of ownership is evident in their confidence while presenting their answers during class discussion. Groups who do not complete the activity have the opportunity to hear how their classmates answered those questions. During the discussion, volunteers also explain and justify their answers. This explanation ensures that those who came to incorrect conclusions or did not complete the questions are exposed to both the correct answer as well the reasoning behind the correct answer. After completing this lesson, our students do well on matched-pair exam questions designed to assess the extent to which they have met the problem set’s learning objectives (see Supporting Materials File S4 for example exam questions).
Reactions to the activity
We have used this activity in at least ten sections of Cell Biology in the past three years and the response from students has been very positive. The discovery nature of the activity often leads to lively discussion and even excitement from the students. Students appreciate the opportunity to learn new methods and apply their understanding to problems.
For students who have little or no experience with small-group work, it is important to allot enough time at the beginning of the lesson to allow them to settle into their groups. Once the students have gotten through this brief “getting to know each other” period, we have found that they will work through the assignment with little or no resistance. Groups that are working very quietly or do not seem to be working together may need encouragement to engage with their classmates.
Adaptations
If you do not have access to a SCALE-UP classroom, this activity can be done in a regular classroom with movable chairs. If necessary, it is possible to do the activity in a room with fixed seating because there are only three people per group. In general, if you are unable to achieve an even distribution of three-member groups, we have found that groups of two stay on task with less social loafing than groups of four.
We have done this activity in both 75-and 50-minute class periods. If you only have a 50-minute class period, you may want to give the first question of the problem set as homework for students to complete before class. Students would then begin collaborating with their groups to answer questions on page 2. Alternately, you can pace the students using a displayed countdown clock (http://timer.onlineclock.net/) to help them progress through the questions.
SUPPORTING MATERIALS
- S1.Transport Proteins-Problem Set: Investigating the Location of a Transport Protein: Where is ABCB6 Functioning in Mammalian Cells? This file contains the problem set that students complete during class. It includes all of the questions they must answer and the figures needed to answer those questions.
- S2.Transport Proteins-Description of Individual Roles: This handout describes the roles (Manager, Recorder, and Presenter) to be used by each group of three. It also includes information about the value of group work.
- S3.Transport Proteins-Figures and Data in PowerPoint Format: This file contains the individual figures from the problem set to be projected in the classroom if the problem set cannot be printed in color.
- S4.Transport Proteins-Matched-Pair Exam Questions: This file contains exam questions that are similar, yet different than the questions in the problem set.
Available for instructors only upon request: (S5) Protein Localization in Cells: Answer Key to Problem Set: A file containing all the answers to the problem set can be requested by emailing the corresponding author, Julie Stanton, at stantonj@uga.edu. (The key is omitted to prevent future students from accessing the answer key online rather than answering the questions themselves.)
IMPORTANT Copyright Information: The authors affirm that they have written permission to use the text, figures, tables, artwork, abstract, summaries and supplementary materials, or they are using these items under a Creative Commons license.
The figures shown in Questions 1 and 2 (SDS-PAGE and immunoblotting figure and differential centrifugation figure) were commissioned from a graphic artist, Ashley Rasys of Athens, GA. Ms. Rasys signed over all rights to use and publish the figures to the authors (Stanton & Dye).
The data for Questions 3 and 4 (immunoblot data and immunofluorescence data) can be freely obtained from the Kiss et al., 2012 PLoS One paper, which was published under a Creative Commons license. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0037378
ACKNOWLEDGMENTS
We thank Dr. Marcus Fechheimer for his valuable feedback on an early draft of the problem set. We also gratefully acknowledge Ashley Rasys for her graphic design.
References
- Moog RS, Spencer JN, Straumanis AR. 2006. Process-oriented guided inquiry learning: POGIL and the POGIL project. Metropolitan Universities 17:41-52.
- Moog RS, Spencer JN, Straumanis AR. 2015. Process-oriented guided inquiry learning: POGIL and the POGIL project. Metropolitan Universities 17:41-52.
- Hung M-C, Link W. 2011. Protein localization in disease and therapy. J Cell Sci 124:3381-3392.
- Burnette WN. 1981. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Analytical biochemistry 112:195-203.
- MacPhee DJ. 2010. Methodological considerations for improving Western blot analysis. Journal of pharmacological and toxicological methods 61:171-177.
- Brelje TC, Wessendorf MW, Sorenson RL. 2002. Multicolor laser scanning confocal immunofluorescence microscopy: practical application and limitations. Cell Biological Applications of Confocal Microscopy:165.
- Miller DM, Shakes DC. 1995. Immunofluorescence microscopy. Methods in cell biology 48:365-394.
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. 2002. Molecular biology of the cell. new york: Garland science; 2002. Classic textbook now in its 5th Edition.
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J. 2000. Molecular cell biology 4th edition. National Center for Biotechnology InformationÕs Bookshelf.
- Bransford JD, Brown AL, Cocking RR. 2000. How People Learn: Brain, Mind, Experience, and School., Expanded ed. National Academies Press, Washington DC.
- Hanson DM. 2006. Instructor's guide to process-oriented guided-inquiry learning. Pacific Crest Lisle, IL.
- Chi MT, Wylie R. 2014. The ICAP framework: Linking cognitive engagement to active learning outcomes. Educational Psychologist 49:219-243.
- Bailey CP, Minderhout V, Loertscher J. 2012. Learning transferable skills in large lecture halls: Implementing a POGIL approach in biochemistry. Biochemistry and Molecular Biology Education 40:1-7.
- Beichner RJ, Saul JM, Abbott DS, Morse JJ, Deardorff D, Allain RJ, Bonham SW, Dancy MH, Risley JS. 2007. The student-centered activities for large enrollment undergraduate programs (SCALE-UP) project. Research-based reform of university physics 1:2-39.
- Lawson A. 1995. Science teaching and the development of reasoning. Belmont, CA, Wadsworth.
- Lawson AE. 1999. What should students learn about the nature of science and how should we teach it? Journal of college science teaching 28:401.
- Kiss K, Brozik A, Kucsma N, Toth A, Gera M, Berry L, Vallentin A, Vial H, Vidal M, Szakacs G. 2012. Shifting the paradigm: the putative mitochondrial protein ABCB6 resides in the lysosomes of cells and in the plasma membrane of erythrocytes. PLoS One 7:e37378.
- Andrews T, Leonard M, Colgrove C, Kalinowski S. 2011. Active learning not associated with student learning in a random sample of college biology courses. CBE-Life Sciences Education 10:394-405.
- Simon HA, Hayes JR. 1976. The understanding process: Problem isomorphs. Cognitive psychology 8:165-190.
Article Files
Login to access supporting documents
Investigating the Function of a Transport Protein: Where is ABCB6 Located in Human Cells?(PDF | 141 KB)
S1.Transport Proteins-Where is ABCB6 Problem Set final.docx(DOCX | 3 MB)
S2.Transport Proteins-Where is ABCB6 Description of Individual Roles final.docx(DOCX | 115 KB)
S3.Transport Proteins-Where is ABCB6 Color Figures in PowerPoint Format final.ppt(PPT | 3 MB)
S4.Transport Proteins-Where is ABCB6 Matched Pair Example Exam Question final.docx(DOCX | 121 KB)
- License terms

Comments
Comments
There are no comments on this resource.