Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon
Published online:
Abstract
The trp operon exemplifies important biological concepts, including the central dogma of molecular biology, feedback inhibition, and gene regulation. For this reason, regulation of the trp operon is a common theme in introductory biology, genetics, and molecular biology curricula. The concepts associated with prokaryotic gene regulation, especially the function of operons, are challenging for many students to comprehend. To help students understand the trp operon and its associated concepts, we developed a Lesson that uses simulations of a computational model through an online modeling platform, Cell Collective Learn (https://learn.cellcollective.org). In general, computational models and simulations enable students to observe and perturb complex biological systems in order to discover relationships not readily observable in static textbook diagrams. In our Lesson, students explore the important structural and regulatory aspects of the trp operon by manipulating a computational model of the system. As students interact with the model, they are encouraged to think critically about the underlying mechanisms that govern system function. Students are asked to predict the simulation output and the underlying mechanism, test their prediction with simulation, report their observations, and provide an accurate cellular mechanism to support their simulation results. The Lesson requires little guidance from the instructor as all of the needed resources and information are provided to students within the Lesson. It can be implemented in a variety of educational settings without the need for significant changes to existing curricula.
Citation
Crowther, A., Bergan-Roller, H.E., Galt, N.J., Booth, C., Dauer, J.T., and Helikar, T. 2018. Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon. CourseSource. https://doi.org/10.24918/cs.2018.9Society Learning Goals
Genetics
- Genetic variation
- How do different types of mutations affect genes and the corresponding mRNAs and proteins?
Microbiology
- Information Flow and Genetics
- How can cell genomes be manipulated to alter cell function?
Lesson Learning Goals
Students will:- Discover how computational modeling and simulations can be used to observe and semi-quantitatively measure the dynamics of gene expression and regulation.
- Recognize the biological significance of the trp operon.
- Be able to think mechanistically about a system.
- Discover how prokaryotic cells maintain homeostasis in the context of the trp operon.
- Evaluate how mutations influence gene expression.
Lesson Learning Objectives
Students will be able to:- Perturb and interpret simulations of the trp operon.
- Define how simulation results relate to cellular events.
- Describe the biological role of the trp operon.
- Describe cellular mechanisms regulating the trp operon.
- Explain mechanistically how changes in the extracellular environment affect the trp operon.
- Define the impact of mutations on trp operon expression and regulation.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Understanding biological processes from a systems perspective is an important skill to foster in undergraduate biology students (1). The regulated expression of the prokaryotic trp operon is a dynamic system that is well-suited to develop students' system thinking skills. Understanding regulation of the trp operon requires an understanding of the connection between external stimuli and changes to the intracellular environment and how those changes affect transcription of the trp operon genes. The trp operon is commonly taught in undergraduate biology courses ranging from introductory biology and genetics to upper-level molecular biology. The trp operon is a classic example of prokaryotic gene regulation and showcases core biological concepts such as the central dogma of molecular biology and feedback regulation (2-4). However, mastering the trp operon and gene regulation in general is challenging for many students (5-8).
Students have considerable misunderstandings about basic genetic principles that do not appear to be rectified by traditional teaching strategies (7,8). Students struggle with concepts related to the central dogma of molecular biology, including the relationship between genes and their protein products as well as the relationship between changes to DNA and gene function (8-11). With such a gap in core genetics concepts, it is no surprise that students struggle with more complex genetic issues and processes (8). Traditional teaching strategies often do not significantly improve understanding of introductory genetics (7). Computer simulations can have a positive impact on student understanding (12-17) by providing an inherently interactive learning experience (12,15,16). Students can directly interact with complex biological systems that are otherwise difficult to visualize; they can systematically manipulate key features of these systems in order to understand overall biological implications. Computer simulations have been successfully implemented in a variety of science courses, including physics, chemistry, and biology (12,15-17). Classrooms utilizing computational simulations in science education have observed improvements in student comprehension and problem-solving skills (12-17).
Biological processes and systems at almost every scale of biological organization are governed by many components (e.g., genes, proteins, etc.) that form complex networks that give rise to non-linear dynamics. To teach students about biological systems, specifically to help students overcome conceptual misunderstandings and develop systems thinking, we developed a teaching approach that uses interactive simulations of computational models in the publicly available Cell Collective Learn platform (https://learn.cellcollective.org, 18-20). In lessons using this approach, students investigate biological systems by predicting simulation outputs and the underlying cellular mechanisms, testing their prediction with simulations, reporting their observations, and providing updated cellular mechanism based on their simulation results. The lessons developed with this approach are grounded in theories of learning and implement evidence-based best practices specific to using simulations to support science learning (12,21,22). We have previously shown that a lesson using this approach, in the context of cellular respiration, helped students learn both content and skills (13,14,23).
Here, we describe a lesson on gene regulation called Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon. This Lesson aims to enhance students' system thinking skills and understanding of the trp operon system, as well as to experience the use of modeling and simulations (1). The Lesson includes both pre-class and in-class activities. During the in-class portion of the Lesson, students simulate a given computational model of the trp operon system using the web-based modeling platform, Cell Collective Learn. The in-class portion is versatile; students can complete it individually or within small groups. Students receive all the needed instructions and prompts within the Lesson itself and can progress through the Lesson without additional help.
INTENDED AUDIENCE
This Lesson was designed for undergraduate biology students at the introductory level. The Lesson could also be provided to students in higher-level courses, such as genetics and biochemistry, to remind them of the basic concepts of gene regulation before they learn about more complicated gene regulatory systems.
REQUIRED LEARNING TIME
The pre-class assignment should take students about 30 minutes to complete and is done outside of class. The in-class portion of the Lesson takes students about 60 minutes to complete.
PREREQUISITE STUDENT KNOWLEDGE
Students should understand the basics of genetics and genetic material (i.e. DNA contains protein-coding regions called genes, the definition of mutation, principles of transcription and translation). Students should also be able to define and distinguish between prokaryotes and eukaryotes. These topics are commonly discussed in introductory biology courses prior to the gene expression and regulation unit. Also, students should possess basic computer and internet skills such as navigating a web browser, typing in numeric and text responses when prompted, and being able to find and maneuver "play" and "pause" symbols.
PREREQUISITE TEACHER KNOWLEDGE
Before providing the Lesson to students, instructors should have a thorough understanding of the same content and skills described above for students. In addition, instructors should have an understanding of operons and specifically the trp operon, including the biological role of operons (i.e. to coordinate the expression of related genes), the biological role of the trp operon (i.e. to make the amino acid tryptophan when environmental tryptophan is low), and the mechanism of trp operon regulation via repression (i.e. intracellular tryptophan binds to and activates the trp repressor, leading to the inactivation of the trp operon) and transcription attenuation (i.e. when tryptophan levels are high, ribosomes rapidly translate the upstream attenuator sequence of the trp mRNA, yielding an RNA structure that terminates transcription). Most of this information is provided in the background reading and insight sections of the Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon activity packet (Supporting File S1: Prokaryotic Gene Regulation - Activity Packet). In addition to the biological background, instructors should be familiar with the Cell Collective Learn online learning platform (https://learn.cellcollective.org). Familiarity with the platform will allow instructors to facilitate student learning during the Lesson. Instructors can gain knowledge about the biology and the software by working through the activities in the Lesson prior to implementing the Lesson with students. Additionally, we have created a training lesson designed to familiarize new users to the Cell Collective Learn platform, Cell Collective Training Module: Factors Influencing Exam Scores, which is publicly available on Cell Collective Learn (https://learn.cellcollective.org).
SCIENTIFIC TEACHING THEMES
ACTIVE LEARNING
Students are active participants in all parts of the Lesson. Students read about prokaryotic gene regulation and the trp operon in the provided background material and answer questions to facilitate reading comprehension. Students investigate the trp operon system by predicting, observing, reporting, and reflecting upon the biological mechanisms dictating system behavior. Students are responsible for completing their own work (e.g., reading the background material, running simulations, recording results) and are encouraged to discuss in small groups.
ASSESSMENT
Instructors can use components of this Lesson as an assessment tool in several ways. In the pre-class assignment activity, students answer reading comprehension questions based on background reading which can be used to evaluate student understanding. During the Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon Lesson, students predict the dynamics of the trp operon model, test their predictions with the simulation, report on their observations, and interpret the results. Instructors can use collected student responses as formative assessment of student knowledge. Students self-assess the correctness of their prediction. During our implementation of the Lesson, we formatively assessed student responses to the pre-class assignment questions, their observations, and their interpretation of simulation results.
INCLUSIVE TEACHING
Students can work in small groups, ranging from two to four students, promoting the exchange of ideas among themselves, and encouraging feedback from peers when individual instructor feedback is not readily available. This type of classroom dynamic has been shown to be beneficial for fostering student gains in underrepresented student groups, such as first-generation college and black students (24). Additionally, the Lesson utilizes computational modeling, a tool not typically used at the introductory level. The computer-based learning medium may appeal to a variety of students, including some who struggle to succeed with traditional teaching techniques.
LESSON PLAN
An overview and timeline for the Lesson plan are provided in Table 1. The learning objectives for this Lesson are informed, in part, by the key concepts for undergraduate biology (1) and specifically introductory biology defined by Khodor, et al. 2004 (25). The Lesson is modular, providing flexibility to adjust Lesson timing for different classroom needs. The Lesson modules include i) background information about operons, focusing on the trp operon, ii) reading comprehension questions, iii) an introduction to the web-based modeling platform, Cell Collective Learn, iv) guided simulation experiments of the trp operon, and v) a data analysis activity. All modules are grouped together in the Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon activity packet (Supporting File S1: Prokaryotic Gene Regulation - Activity Packet).
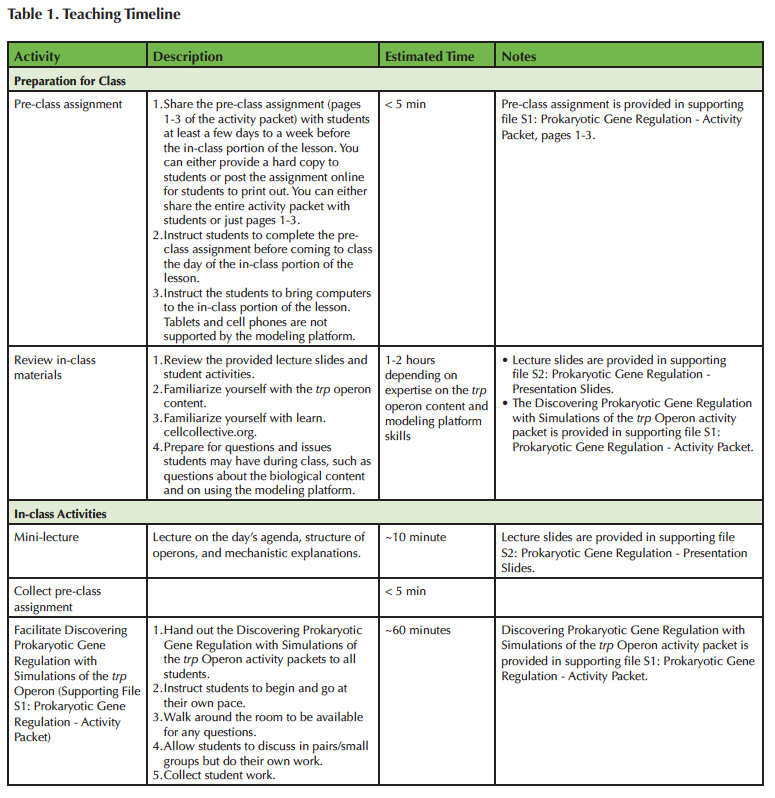
Table 1. Teaching Timeline.
We implemented the Lesson with the background information and reading comprehension questions assigned as a pre-class assignment (i.e. students read the background material and answer the reading comprehension questions before coming to class). The background information, introduction to Cell Collective Learn, Investigations of the trp operon, and data analysis activity modules are grouped together as the in-class activity (i.e. students have access to the background material while completing the trp Investigations and data analysis activities). The background reading is included in both the pre-class and in-class portions of the Lesson. In the pre-class assignment, the background reading provides a basic introduction to the trp operon, and covers all of the required material for Lesson completion, so that the Lesson does not rely on a textbook or lectures. During class, the background reading is provided as a reference during the activity.
Below is a list of materials that are necessary and/or useful for using the lesson, all of which are provided as supporting materials or links to website:
- Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon activity packet (Supporting File S1: Prokaryotic Gene Regulation - Activity Packet)
- In-class presentation slides (Supporting File S2: Prokaryotic Gene Regulation - Presentation Slides)
- Cell Collective Training Module: Factors Influencing Exam Scores (accessible at https://learn.cellcollective.org)
- Prokaryotic Gene Regulation: the trp operon model, accessible at Cell Collective Learn (https://learn.cellcollective.org)
The Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon activity packet answer key is available by request from the authors.
OVERVIEW OF COMPUTATIONAL MODELS AND SIMULATIONS IN CELL COLLECTIVE LEARN
Computational models in Cell Collective Learn are network models consisting of nodes connected with directed edges. The network nodes represent various components of the modeled biological process, such as genes, proteins, cells, other generalized biological processes (e.g., transcription), etc. The edges represent causal relationships among the components, such as activation/inhibition of one protein by another one. They can also represent more complex interactions and regulatory mechanisms that, for example, incorporate conditional relationships such as co-factors, localization, priming, etc. In the background of the software, regulatory mechanisms of the components and their interactions are mathematically described with logical expressions that determine the activity level of each component and enable users to explore the dynamics of the model over time (26-28). In Cell Collective Learn, the activity level of each model component is discrete (active or inactive) at any specific time point. The activity of the components can also be represented as semi-continuous (ranging from 0 to 100%), representing the (percent) probability of the component being active at a given time. It is important to note that the activity levels of individual model components do not directly correspond to biological concentrations or any other measurable property; rather they provide a semi-quantitative measure to describe the relative activity level of a particular model component. The data representation as discrete or continuous is defined by the Sliding Window control dial or directly by setting the activity levels of the model's external signals in Cell Collective Learn (19,29-31). Both aspects are leveraged in each lesson as needed in order to ensure the output representation is useful for students to understand the appropriate dynamical concepts.
BEFORE CLASS
ASSIGN PRE-CLASS ASSIGNMENT
Prior to the in-class portion of the Lesson, assign reading of the background material (pages 1-2 of Supporting File S1: Prokaryotic Gene Regulation - Activity Packet), and ask students to answer the reading comprehension questions (page 3 of Supporting File S1: Prokaryotic Gene Regulation - Activity Packet). By reading the background information, students will get a concise introduction to the structure and function of operons in general, structure and function of the trp operon specifically, and the basic mechanisms of gene regulation. The questions ensure that students complete the reading and are prepared for the in-class portion of the Lesson. Students should be alerted that their answers to these questions will be collected at the beginning of the class period. The pre-class assignment takes students around 30 minutes to complete. Instructors can share this assignment with students by either providing a print out of the assignment or by posting it to a learning management system at the instructor's institution, with the expectation that students will print out the assignment. Instructors also have the option to provide the entire Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon activity packet (Supporting File S1: Prokaryotic Gene Regulation - Activity Packet) with students before the in-class portion of the Lesson or just pages 1-3 of the activity packet.
When assigning the pre-class assignment, inform students that they will need a computer with Internet connection and a web browser for the upcoming class, and encourage students to bring their own. Tablets and cell phones are not currently supported by the Cell Collective Learn platform. It is ideal for every student to have his or her own computer. However, the activity can be done in groups of up to four students with one or more computer per group. Alternatively, instructors could schedule the activity in a computer lab, if appropriate.
REVIEW IN-CLASS MATERIALS
To prepare to lead the in-class portion, review the in-class presentation slides (Supporting File S2: Prokaryotic Gene Regulation - Presentation Slides); this resource provides an introduction to the content that will be covered during class. Additionally, go through the Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon activity packet (Supporting File S1: Prokaryotic Gene Regulation - Activity Packet). This experience will give instructors a deeper understanding of the content, develop skills for navigating the Cell Collective Learn platform, and help prepare for potential issues and questions from students. All of the necessary materials (S1-2) are provided in this article. Additionally, the Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon activity packet (Supporting File S1: Prokaryotic Gene Regulation - Activity Packet) is available within the Prokaryotic Gene Regulation: the trp operon model at https://learn.cellcollective.org. The Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon activity packet answer key is available upon request by validated instructors, to prevent easy access by students.
PRINT IN-CLASS MATERIALS
Each student will also need a copy of the Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon activity packet (Supporting File S1:Prokaryotic Gene Regulation - Activity Packet), which includes all background, instructions, and question prompts. Instructors can provide copies for each student or require that students print their own copy and bring it to class.
IN-CLASS
COLLECT PRE-CLASS ASSIGNMENT
First, collect students' pre-class assignments (specifically the reading comprehension questions). The pre-class assignment primarily serves to ensure that students read the accompanying background material. Students will investigate the content further during the in-class portion of the Lesson.
PRESENT MINI-LECTURE
Second, present the slides (Supporting File S2: Prokaryotic Gene Regulation - Presentation Slides) to inform students of the day's tasks and guide them through the activity. The presentation helps to prepare students for the expected content and length of the activity. The presentation slides include an itinerary, an overview of operon structure, an outline of the Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon activity, and instructions on creating mechanistic explanations. We provide commentary in the notes section of the slides to guide instructors on what to do with each slide.
FACILITATE THE DISCOVERING PROKARYOTIC GENE REGULATION WITH SIMULATIONS OF THE TRP OPERON ACTIVITY
Third, start the Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon activity by handing out the activity packet (Supporting File S1: Prokaryotic Gene Regulation - Activity Packet). This packet (Supporting File S1: Prokaryotic Gene Regulation - Activity Packet) provides students with all of the background information, directions, and question prompts they need to complete the activity successfully.
Exercise 1. Describe the interactions of the trp operon
In the first part of the activity, students are presented with a diagram of the trp operon computational model (Figure 5 in the activity packet). This model represents the molecules and processes that impact trp operon activity, either directly or indirectly, including tryptophan, the trp operon, trp mRNA, trp enzymes, trp synthesis, and the trp repressor. In the figure, these components are connected by either black arrows that represent a positive relationship between components (i.e. activation) or red, blunted lines that represent a negative relationship between components (i.e. inhibition). In Table 1, students describe each relationship depicted in the model.
Exercise 2. Setup computational simulations
In the second part of the activity, students use their computers to access Cell Collective Learn (https://learn.cellcollective.org) and either create or log into their free Cell Collective Learn account. Students that need to register for a Cell Collective Learn account will need to select the "Create Account" icon and provide their full name along with their email and their institution of study. After logging into their Cell Collective Learn account, students are then prompted to open the Prokaryotic Gene Regulation: the trp operon model. Students set up simulations to monitor the outputs and regulatory elements of the trp operon computational model. Simulation settings have been optimized to provide clear visual outputs of the system. Students may need to refer back to these instructions during later parts of the activity.
Students are able to simulate the Prokaryotic Gene Regulation: the trp operon model without a Cell Collective Learn account by selecting the "Explore All Learning Modules" icon. However, students will not be able to utilize all of the features of Cell Collective Learn, including the ability to create and save new models as well as any other Lesson-related work that requires the saving of data on the Cell Collective Learn servers. For this reason, we strongly recommend that instructors have students create a free Cell Collective Learn account before interacting with any Cell Collective Learn models.
Exercise 3. Investigate the impact of changes to the trp operon
In the third part of the activity, students learn how the trp operon is regulated by investigating the impacts of perturbations on the operon. We call these exercises Investigations. Because the computational model of the trp operon is based on probabilistic logical modeling (described above), students should focus on whether the activity of a given component is affected positively or negatively (as opposed to specific values) in response to mechanistic changes in the modeled system (this is similar to the interpretation of the output of many biological experiments, including Western Blots, differential gene expression analysis, etc.).
The content of each Investigation is unique in that it closely examines the regulation of the trp operon under different environmental and intracellular conditions. In Investigation 1, students examine how the trp operon is regulated when environmental tryptophan is present. In Investigation 2, students examine how the trp operon is regulated when environmental tryptophan is absent. In Investigation 3, students explore how a mutation in the trp repressor affects the regulation of the trp operon. In Investigation 4, students explore how a mutation of a trp operon gene affects trp operon regulation. Investigations 1 and 2 are designed to have students observe how regulation of the trp operon maintains homeostasis of intracellular tryptophan levels. The objective is for students to observe how changes in extracellular conditions influences internal gene activity and the mechanism behind that influence. Investigations 3 and 4 are designed to emphasize the connection between changes in DNA and changes in protein function. Our goal for these two Investigations is to have students recognize how DNA mutations influence gene regulation by influencing protein function, particularly transcription factors and biosynthetic enzymes.
Each Investigation follows specific steps of A) making predictions, B) developing a mechanistic explanation to support the prediction, C) testing the prediction using simulations, D) recording the simulation results, E) evaluating the prediction based on simulation results and retesting as necessary, and F) providing an updated mechanistic explanation supported by simulation results. For example, in Investigation 1 Step A, students are asked to predict how the presence of environmental tryptophan will affect the activity of the trp operon. In Step B, students support their prediction in Step A with a mechanism based on the provided network diagram of the computational model. In Step C, students set up and run a simulation to test their prediction. In Step D, students report their simulation results. In Step E, students use the simulation results to evaluate the correctness of their prediction. If their simulation results do not align with their prediction, students are prompted to repeat Steps A-E until both their prediction and their results match. Finally, in Step F, students attempt to describe the mechanism responsible for the biological phenomena represented by the simulation results.
We designed Steps A - F based on the recommended best practices of using simulations to facilitate science learning (12,21,22), specifically to have students
- make explicit their conceptions as predictions (Step A) (22),
- provide reasons for their predictions (Step B) (22),
- confront alternative conceptions with simulation results (Step D) (12,21),
- evaluate the consistency of their predictions with their results (Step E) (22), and
- use evidence (from simulation results) to support findings (Step F).
In Investigation 1, Step B, students are asked to provide their mechanistic explanation through a fill-in-the-blank question format. This format provides scaffolding (32) to help students build a high quality, accurate mechanistic explanation in the context of the trp operon. This scaffolding is removed for Investigations 2-4 where Step B uses an open-ended question format.
Exercise 4. Insights about Attenuation
In the last part of the Lesson, students learn about a second mechanism of regulating gene expression from the the trp operon, namely attenuation. During this part of the Lesson, students are provided with a brief overview of attenuation that describes the molecular details of this mechanism in relation to transcriptional control. This information is separate from the background reading section in order to allow students to focus on each regulatory mechanism, repression and attenuation, independently throughout the Lesson. Next, students are asked to perform a thought experiment by placing themselves in the role of a scientist researching the trp operon before attenuation had been discovered. Students are presented with experimental data, adapted from Hiraga, et al. 1967 (33), and expected to analyze the data in order to demonstrate whether they support or refute the hypothesis that a second regulatory mechanism controls trp operon expression. This activity is designed to train students to analyze and interpret experimental data and to draw connections from raw experimental data to biological mechanisms (34).
Ways to Facilitate the Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon Activity
Although outside guidance is not required for this Lesson, we encourage instructors to be engaged with students as they navigate this Lesson. We suggest that instructors be available to address student questions and concerns. To ensure that students are applying mechanistic explanations, read over their answers in Steps B and F of each Investigation. Look for responses that 1) include two or more components, 2) describe the interactions between the components, and 3) provide an explanation of how the phenomena described in the prompt occurs. Be aware that students favor why answers instead of how answers (35). We provide question prompts in the notes section of the presentation slides to help encourage students to provide how answers when writing their mechanistic explanations. Instructors can also use the answer key (available upon request from the authors) to formatively assess student responses. We suggest first checking in with one student within a group and then encouraging group discussion. This strategy provides some feedback to all students in the group, even if indirectly, and promotes peer instruction.
Common areas of difficulty and solutions
The most common issue we encounter when we teach this Lesson is with students incorrectly simulating the perturbations. Students may haphazardly make changes to the simulation settings which frequently leads to incorrect simulation results. While students are welcome to explore the model and the modeling platform, correctly working through the Lesson as designed requires that students run their simulation with the parameters that have been outlined in the instructions. Running the simulation using different parameters, especially when students are not aware of how those parameters influence the simulation output, may prevent them from confronting their alternative conceptions with correct simulation results. Therefore, it is essential for instructors to mediate this activity by verifying that students are adjusting the simulation settings correctly. To do this in a student-driven manner, ask students to focus on smaller predictions about model behavior, such as "if tryptophan is present, should the trp repressor be on or off?" If these simpler predictions do not match their simulation results, guide them to check the simulation settings provided in Step C of the Investigation they are working on. To minimize the need for instructor intervention, remind students to regularly compare and discuss their simulation results with those of their neighbors or group members. This reminder will help students detect and resolve settings errors. Additionally, have students explain to each other why their settings gave the result, to practice argumentation skills.
Many times, students skip the instructions and become confused or frustrated with the simulations. If instructors suspect frustration from a student, have him or her go back to carefully read and apply the instructions. Retracing steps commonly provides clarity and productive engagement with the simulations.
TEACHING DISCUSSION
LESSON IMPLEMENTATION
We implemented this Lesson twice to introductory biology students at a large research university. First, this Lesson was provided to 21 honors students who completed it individually as a part of their honors requirements for the course. During the second implementation, this Lesson was given to 663 students during a single discussion period (i.e. dry laboratory) with about 25 students per discussion period. These discussion periods are intended to reinforce lecture content and last up to three hours. Students worked in groups of three to four. Students were organized into these groups at the beginning of the semester and remained in those groups throughout. While students were encouraged to discuss with their group members, each student was required to complete his or her own in-class activity packet. All data collected were from consenting students only and with institutional review board approval (UNL IRB 20171017504 EX).
Assessment of Student Achievement of the Learning Objectives
The Lesson provides multiple opportunities to assess student achievement of the learning objectives. An alignment of the learning objectives, assessments, and results are provided in Table 2. The data reported in Table 2 were collected from all consenting students (n=87) during implementation in an introductory biology course. Blank and illegible student responses were discarded during analysis of student achievement on a per learning objective basis (see Table 2).
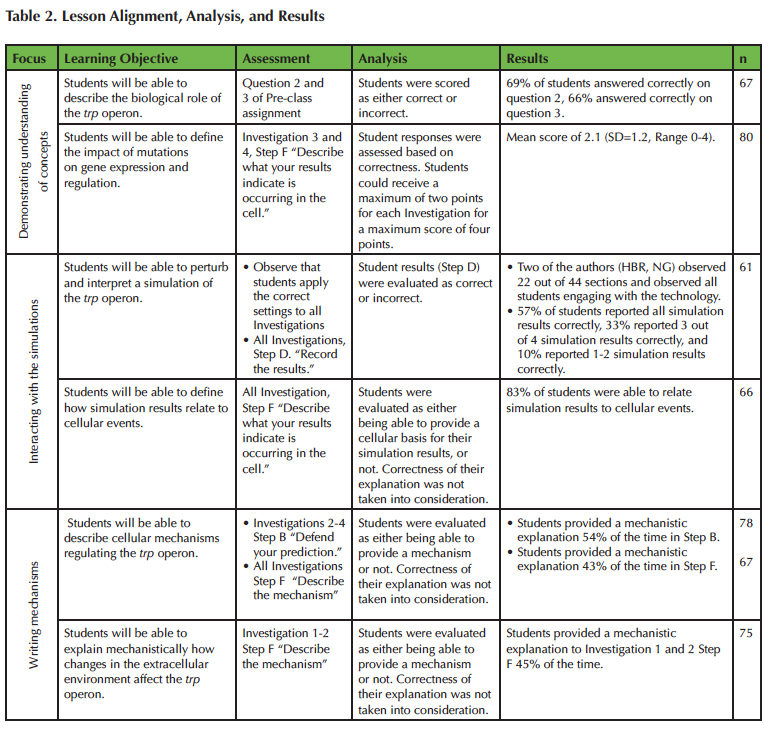
Table 2. Alignment of learning objectives, assessments, analysis, and results.
Analysis of the pre-class questions (n=67) indicates that 69% of students were able to determine what cellular signal the trp operon detects (i.e. changes in tryptophan levels), and 66% of students successfully identified the collective function of the proteins encoded in the trp operon (i.e. to produce tryptophan). When asked what cellular signals the trp operon detects (question 2), the most common incorrect student response was that the trp operon detects changes in the internal and external environment, which, while correct, does not address the specific signal detected by the trp operon. When asked about the function of the trp operon (question 3), the majority of students incorrectly described the role of the trp repressor and not the operon as a whole. When examining student responses to Table 1 in the activity packet (Supporting File S1: Prokaryotic Gene Regulation - Activity Packet), 78% of students who answered question 3 incorrectly were able to successfully trace the production of tryptophan from the transcription of the trp operon genes, implying that any confusion or misconception students may have regarding the function of the proteins encoded in the trp operon is corrected later on in the Lesson. We are planning to develop new questions to further probe student understanding and to decipher if incorrect responses to these pre-class assignment questions stem from a lack in biological knowledge or from a misinterpretation of the presented questions.
Analysis of the in-class activities indicates students interact well with the modeling software. Based on student responses to Investigations 1-4, Step D, which asks students to record their simulation results, students are able to conduct the simulation activities and interpret simulation results 86% of the time. Correct simulation results include 1) identifying the constant activity of the trp repressor and absence of trp operon activity when environmental tryptophan is present (Investigation 1), 2) the oscillatory nature of trp operon and trp repressor activity levels when environmental tryptophan is absent (Investigation 2), 3) the constant activity of the trp operon and trp repressor when a mutation prevents repressor binding to the operon (Investigation 3), and 4) the constant activity of the trp operon and trp mRNA and lack of trp repressor, tryptophan synthesis, and tryptophan when there is a loss-of-function mutation in one of the trp operon genes (Investigation 4). Likewise, students successfully translated quantitative output from their simulations into conceptual understanding of component interactions 83% of the time (although the correctness of their interpretation was not taken into consideration).
Most students (75%) were able to at least partially define the impact of mutations on the trp operon system (Investigations 3 and 4, Step F). Students were evaluated on their ability to 1) explain how each mutation impacts protein function and 2) identify how this mutation impacts the overall trp operon expression. In Investigation 3, Step F, 41% of students provided a complete explanation (score of 2) for how a mutation in the trpR gene (that codes for the trp repressor) that prevents repressor binding to the operon impacts regulation of the trp operon (i.e. the absence of the inhibitory effect of the trp repressor binding to the operator causes the trp operon to be constantly expressed), with 78% of students providing at least a partially correct explanation (a score of 1 or 2). In Investigation 4, Step F, 23% of students provided a complete explanation for how a loss-of-function mutation in the trpC gene affects trp operon regulation (i.e. the trp operon is constantly expressed, but the lack of the functional trpC gene product would prevent tryptophan synthesis), with 73% of students providing at least a partially correct explanation.
Many students struggled to identify that a mutation in the trpC gene would cause the trp operon to be continuously expressed, implying that students are struggling with the cyclic nature of the trp operon: that the production of tryptophan, or lack thereof, regulates expression of the trp operon. However, most students that failed to identify this connection (score of 0) between the trpC mutation and trp operon activity omitted any relationship between the trpC mutation and trp operon activity in their explanation rather than providing an incorrect relationship. This pattern of students omitting relationships between components rather than providing incorrect connections is consistent across both Investigations 3 and 4, with only 16% of student who received a score of 0 providing incorrect connections between relevant components. Post-lesson student interviews or additional open-response questions probing these specific connections can help instructors decipher whether students are confused by these relationships or are merely forgetting to include them in their explanations.
Although students were able to identify components of the system 80% of the time in Step B and 71% of the time in Step F, they struggled to describe the connections between components when attempting to write a cellular mechanism. Our definition of mechanistic reasoning is based on work from van Mil and colleagues (2013) (36). We define a cellular mechanism as identifying two or more components connected by at least one relationship. Across all Investigations, students were able to describe a mechanism regulating the trp operon 54% of the time in Step B and 43% of the time in Step F. In Step F, most students were able to provide a mechanistic description for at least one Investigation, but only 24% were able to do so consistently throughout all four Investigations. This result implies that the biggest hurdle for students in this Lesson is describing a system mechanistically. This finding is consistent with other work on students' explanations of biological systems (23,35).
Recent work in other cellular contexts suggests that simulation-based lessons can help improve students' mechanistic reasoning abilities (13,37). Specifically, students improved in their ability to identify relevant components in the system and in the quality of relationships between those components (13,37), which has been identified as an obstacle in student comprehension (14). Additionally, students improved in their ability to predict and explain how changes in other components of the system impact other parts of the system (13). We speculate that this improvement is in part because of repetition: that asking students to predict and observe certain components of the system enhances student understanding (37). Thus, like any skill, we predict that students' ability to write mechanistic explanations will improve with additional practice.
Based on our results, we have identified certain areas in the Lesson where instructors could serve a mentoring role for their students. Specifically, instructors can provide additional guidance in writing mechanistic explanations and probing student understanding of the biological background. How students learn and communicate as well as how students learn with our simulations lessons is an ongoing research focus of our group. Many of the questions brought up in this Lesson are current research questions we are looking to answer in the future. Overall, these results show that students are successfully working through the provided Lesson.
STUDENT AND INSTRUCTOR REACTION TO THE LESSON
Students found our Lesson beneficial to their learning. Honor students who completed the activity during the first implementation took a survey intended to gauge student reaction to the Lesson. Students (n=14) reported feeling more knowledgeable about gene expression and regulation (Figure 1A) and about the trp operon (Figure 1B) after completing our Lesson.
Figure 1. Student responses from the honors student implementation on a survey that asked students A) how would you rate your knowledge of gene expression before and after the exercise and B) how would you rate your knowledge of the trp operon before and after the exercise. The y-axis represents the number of student responses.
After the second larger implementation, students and teaching assistants (TAs) were asked to reflect on the Lesson. On a survey, students reported that the Lesson enhanced their understanding of the trp operon (Figure 2A). In addition, students found the computer simulations engaging (Figure 2B).
Figure 2. Student responses from the all-class implementation on a survey that asked students to A) rate whether they found the computer simulation experiments enhanced their understanding of the trp operon and B) rate whether they found the computer simulation experiments engaging. The y-axis represents the number of student responses. The percentages on the columns represent the percentage of students that selected each response.
Two TAs were interviewed after implementing the Lesson in their class. Both TAs stated that they liked the Lesson because it fostered student discussion and prompted students to use problem-solving skills. They also reported that the interactive computer simulations fostered a deeper understanding of the system that is difficult to accomplish from textbook diagrams and lectures. Representative comments are presented below.
- "[F]or the other recitations it's more to do with how well the students could retain that knowledge.... This one is different because it challenges the student to how creatively can you think to solve the problem." (TA 1)
- "I love these simulations, I think that they allow you to see the pathways in ways that you don't when they're on paper." (TA 2)
- "I love the [predict and test] process, and I think they [the students] go through it in their heads.... I think making them stop and think about it first was excellent." (TA 2)
- "I love the activity and I like it when they talk. I think having the computers really makes it a nice casual environment, and they're more likely to discuss it with their peers." (TA 2)
- "I think being able sit and play with the levels and manipulate it [the system] really gives you an insight into it that you don't have in a traditional lecture when you're staring at an image of it." (TA 2)
POSSIBLE ADAPTATIONS
Our Lesson is highly flexible and can be adapted to fit a variety of different classroom needs. For example, the pre-class assignment could be implemented in class instead of as homework. However, we recommend that students read the background material before coming to class to prepare them for the in-class activities. The in-class portion of the Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon activity packet can be completed individually outside of class should timing not allow an in-class implementation.
Some instructors and students may prefer teaching this material in a more traditional classroom style but still want to engage students in active learning with this Lesson. Therefore, instructors could add readings and/or lecture on gene regulation to the Lesson. Providing mechanistic explanations is novel for many students. We suggest having students practice creating a simple mechanistic explanation with feedback (38) from the instructor and their peers before they are asked to independently describe the trp operon for the in-class activity. During the Lesson, instructors can prompt students to draw out their mechanisms in Step F of the Investigations, specifying that students need to label the nodes and the relationship between nodes.
Instructors could also add a summative post-assessment after the Lesson to gauge what information students retained from the Lesson. This post-assessment could focus on content related to the trp operon and/or mechanistic descriptions. Instructors could also use the pre-class assignment as a pre- and post-assessment, if desired.
Additionally, while students in this Lesson are only prompted to use the Simulation workspace of Cell Collective Learn, instructors can utilize the other workspaces in the platform to explore different aspects of the trp operon. For example, the Analysis workspace allows users to generate dose-response and titration curves to investigate the input-output relationships between external signals and various components of the model, such as the strength of a trpC mutation (i.e. how effectively the mutation inhibits enzyme function) and tryptophan synthesis. In this Lesson, the Model and Knowledge Base workspaces provide students will additional information about the components. These workspaces can be used if students are curious about the terminology used in the model. A detailed and complete description of the capabilities of Cell Collective Learn can found in Helikar, et al. 2015 (18).
SUPPORTING MATERIALS
- Supporting File S1: Prokaryotic Gene Regulation - Activity Packet
- Supporting File S2: Prokaryotic Gene Regulation - Presentation Slides
ACKNOWLEDGMENTS
We thank Dr. Steven Harris, James Buescher, and the teaching assistants of the LIFE 120 course for their assistance in implementation and their feedback on the lesson. We thank Dr. Trisha Vickrey and Dr. Marilyne Stains for conducting interviews with teaching assistants. We thank Erica Pribil for assisting with the student response analysis. We thank Dr. Lara Appleby for editing the manuscript. This work was supported by the National Science Foundation under Grant No. IUSE #1432001 to T. Helikar and J. Dauer.
References
- AAAS. 2011. Vision and Change in Undergraduate Biology Education: A Call to Action. Washington, D.C.
- Cooper R. 2015. Teaching the Big Ideas of Biology with Operon Models. Am Biol Teach 77: 30-39.
- Santill?n M, Mackey M. 2000. Dynamic regulation of the tryptophan operon: A modeling study and comparison with experimental data. PNAS 98: 1364-1369.
- Stefanski K, Gardner G, Seipelt-Thiemann R. 2016. Development of a Lac Operon Concept Inventory (LOCI). CBE 15: ar24.
- Campbell Biology AP 8th Edition. 2008. Student Misconceptions for Campbell/Reece Biology. Pearson Education, Inc 18-1.
- Campbell Biology 9th Edition - Instructor Resource. 2011. Chapter 18. The Genetics of Viruses and Bacteria. Pearson Education, Inc 124-125.
- Banet E, Ayuso E. 2000. Teaching Genetics at Secondary School: A Strategy for Teaching about the Location of Inheritance Information. Sci Educ 84:313-351.
- Wright K, Fisk N, Newman D. 2014. DNA -> RNA: What Do Students Think the Arrow Means? CBE Life Sci Educ 13: 338-348.
- Briggs AG, Morgan SK, Sanderson SK, Schulting MC, Wieseman LJ. 2016. Tracking the Resolution of Student Misconceptions about the Central Dogma of Molecular Biology. J Microbiol Biol Educ 17:339-350.
- Queloz AC, Klymkowsky MW, Stern E, Hafen E, K?hler K. 2017. Diagnostic of students' misconceptions using the Biological Concepts Instrument (BCI): A method for conducting an educational needs assessment. PLoS One 12(5): e0176906. https://doi.org/10.1371/journal.pone.0176906
- Lewis J, Kattmann U. 2004. Traits, genes, particles and information: Re-visiting students' understandings of genetics. Int J Sci Educ 26:195-206.
- Smetana LK, Bell RL. 2012. Computer Simulations to Support Science Instruction and Learning: A critical review of the literature. Int J Sci Educ 34:1337-1370.
- Appleby L, Bergan-Roller HE, Galt NJ, Pribil E, Chizinski C, Dauer JT, Helikar T. A computational model-based lesson improves students' systems thinking in the context of cellular respiration. JMBE. Submitted.
- Bergan-Roller HE, Galt NJ, Pribil E, Chizinski C, Helikar T, Dauer JT. Undergraduate Student Knowledge of Cellular Respiration. J Biol Educ. Under Review.
- Bell RL, Smetana LK. 2008. Chapter 3. Using Computer Simulations to Enhance Science Teaching and Learning Technology in the secondary science classroom.
- Rutten N, Van Joolingen WR, Van Der Veen JT. 2012. The learning effects of computer simulations in science education. Comput Educ 58:136-153.
- Kulik CLC, Kulik JA. 1986. Effectiveness of computer-based education in colleges. AEDS J 19:81-108.
- Helikar T, Cutucache CE, Dahlquist LM, Herek TA, Larson JJ, Rogers JA. 2015. Integrating Interactive Computational Modeling in Biology Curricula. PLoS Comput Biol 11:e1004131.
- Helikar T, Kowal B, McClenathan S, Bruckner M, Rowley T, Madrahimov A, Wicks B, Shrestha M, Limbu K, Rogers JA. 2012. The Cell Collective: toward an open and collaborative approach to systems biology. BMC Syst Biol 6:96.
- Cutucache CE, Helikar T. 2014. Cancer biology: an inquiry-based approach. Kendall Hunt, Dubuque.
- Duit R, Treagust DF. 2003. Conceptual change: A powerful framework for improving science teaching and learning. Int J Sci Educ 25:671-688.
- Liew C, Treagust DF. 1998. The Effectiveness of Predict-Observe-Explain Tasks in Diagnosing Students' Understanding of Science and in Identifying Their Levels of Achievement. Paper presented at the annual meeting of the American Educational Research Association, San Diego.
- Bergan-Roller HE, Galt NJ, Dauer JT, Helikar T. 2017. Discovering Cellular Respiration with Computational Modeling and Simulations. CourseSource. https://doi.org/10.24918/cs.2017.10
- Eddy S, Hogan K. 2014. Getting Under the Hood: How and for Whom Does Increasing Course Structure Work? CBE Life Sci Educ 13: 453-468.
- Khodor J, Halme G, Walker C. 2004. A Hierarchical Biology Concept Framework: A Tool for Course Design. CBE 3:111-121.
- Abou-Jaoud? W, Traynard P, Monteiro PT, Saez-Rodriguez J, Helikar T, Thieffry D, Chaouiya C. 2016. Logical Modeling and Dynamical Analysis of Cellular Networks. Frontiers in Genetics 7:94.
- Nov?re NL. 2015. Quantitative and logic modelling of molecular and gene networks. Nature Reviews Genetics 16:146-158.
- Helikar T, Kochi N, Konvalina J, Rogers JA. 2011. Boolean Modeling of Biochemical Networks. The Open Bioinformatics Journal 5:16-25.
- Helikar T, Rogers JA. 2009. ChemChains: a platform for simulation and analysis of biochemical networks aimed to laboratory scientists. BMC Systems Biology 3:58.
- Helikar T, Kowal B, Rogers JA. 2013. A Cell Simulator Platform: The Cell Collective. Clinical Pharmacology & Therapeutics 93:393-395.
- Helikar T, Kowal B, Madrahimov A, Shrestha M, Pedersen J, Limbu K, Thapa I, Rowley T, Satalkar R, Kochi N, Konvalina J, Rogers JA. 2012. Bio-Logic Builder: A Non-Technical Tool for Building Dynamical, Qualitative Models. PLoS ONE 7.
- Pea RD. 2004. The Social and Technological Dimensions of Scaffolding and Related Theoretical Concepts for Learning, Education, and Human Activity. J Learn Sci 13:423-451.
- Hiraga S, Ito K, Hamada K, Yuta T. 1967. A new regulatory gene for the tryptophan operon of Escherichia coli. Biochem Biophys Res Commun 26:522-527.
- Brodland GW. 2015. How computational models can help unlock biological systems. Semin Cell Dev Biol 47-48:62-73.
- Abrams E, Southerland S. 2001. The how's and why's of biological change: How learners neglect physical mechanisms in their search for meaning. Int J Sci Educ 23:1271-1281.
- van Mil MHW, Boerwinkel DJ, Waarlo AJ. 2013. Modelling molecular mechanisms: a framework of scientific reasoning to construct molecular-level explanations for cellular behaviour. Sci and Educ 22:93-118.
- Bergan-Roller HE, Galt NJ, Chizinski C, Helikar T, Dauer JT. 2018. Simulated computational model lesson improves systems thinking in biology students. BioScience, biy054, https://doi.org/10.1093/biosci/biy054
- Bell B, Cowie B. 2001. The characteristics of formative assessment in science education. Sci Educ 85:536-553.
Article Files
Login to access supporting documents
Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon(PDF | 207 KB)
S1_Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon.pdf(PDF | 582 KB)
S2. Prokaryotic Gene Regulation_ Presentation Slides.pptx(PPTX | 196 KB)
- License terms

Comments
Comments
There are no comments on this resource.