Sequence Similarity: An inquiry based and "under the hood" approach for incorporating molecular sequence alignment in introductory undergraduate biology courses
Published online:
Abstract
Introductory bioinformatics exercises often walk students through the use of computational tools, but often provide little understanding of what a computational tool does "under the hood." A solid understanding of how a bioinformatics computational algorithm functions, including its limitations, is key for interpreting the output in a biologically relevant context. This introductory bioinformatics exercise integrates an introduction to web-based sequence alignment algorithms with models to facilitate student reflection and appreciation for how computational tools provide similarity output data. The exercise concludes with a set of inquiry-based questions in which students may apply computational tools to solve a real biological problem.
In the module, students first define sequence similarity and then investigate how similarity can be quantitatively compared between two similar length proteins using a Blocks Substitution Matrix (BLOSUM) scoring matrix. Students then look for local regions of similarity between a sequence query and subjects within a large database using Basic Local Alignment Search Tool (BLAST). Lastly, students access text-based FASTA-formatted sequence information via National Center for Biotechnology Information (NCBI) databases as they collect sequences for a multiple sequence alignment using Clustal Omega to generate a phylogram and evaluate evolutionary relationships. The combination of diverse, inquiry-based questions, paper models, and web-based computational resources provides students with a solid basis for more advanced bioinformatics topics and an appreciation for the importance of bioinformatics tools across the discipline of biology.
Citation
Kleinschmit, A., Brink, B., Roof, S., Goller, C., and Robertson, S.D. 2019. Sequence Similarity: An inquiry based and “under the hood” approach for incorporating molecular sequence alignment in introductory undergraduate biology courses. CourseSource. https://doi.org/10.24918/cs.2019.5Society Learning Goals
Bioinformatics
- Computation in the life sciences
- What computational concepts are important in bioinformatics?
- DNA - Information Storage [GENOMICS]
- How can bioinformatics tools be employed to analyze genetic information?
Lesson Learning Goals
Students will be able to:- Explain computational concepts used in bioinformatics (e.g., meaning of algorithm, bioinformatics file formats).
- Use bioinformatics tools (e.g., BLAST, NCBI) to examine phylogenetic relationships.
- Explain the basic theory behind common bioinformatics algorithms.
- Use bioinformatics tools to approach biological questions.
Lesson Learning Objectives
At the end of this lesson, students will be able to:- Define similarity in a non-biological and biological sense when provided with two strings of letters.
- Quantify the similarity between two gene/protein sequences.
- Explain how a substitution matrix is used to quantify similarity.
- Calculate amino acid similarity scores using a scoring matrix.
- Demonstrate how to access genomic data (e.g., from NCBI nucleotide and protein databases).
- Demonstrate how to use bioinformatics tools to analyze genomic data (e.g., BLASTP), explain a simplified BLAST search algorithm including how similarity is used to perform a BLAST search, and how to evaluate the results of a BLAST search.
- Create a nearest-neighbor distance matrix.
- Create a multiple sequence alignment using a nearest-neighbor distance matrix and a phylogram based on similarity of amino acid sequences.
- Use appropriate bioinformatics sequence alignment tools to investigate a biological question.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Biology has entered the era of 'big data' (1), in which the generation of large data sets is common and economically feasible for most investigators. There is also a growing niche of biologists pursuing research projects that lack a wet-lab component, and instead utilize rich publicly available data sets to mine data in silico. This massive amount of data generated from high-throughput techniques is changing the biological research landscape and bringing with it a bottleneck of data analysis (2-6). This bottleneck partially stems from insufficient knowledge of basic bioinformatics tools and a poor understanding of their underlying computational algorithms and limitations (2). Individuals trained within the interdisciplinary field of bioinformatics are therefore in high demand in the workforce (7,8). Integration of bioinformatics concepts in undergraduate curricula is key, yet the breadth of knowledge and rapid evolution in this field makes it challenging for instructors (9,10). There is a gap between the quickly expanding bioinformatics tools and techniques used by the biology and computer science research communities, and readily available resources for educators to integrate bioinformatics into the biology classroom. By introducing bioinformatics in the classroom, we are preparing the next generation of life scientists to handle the tidal wave of big data.
Computational tools have changed the way we practice biology; it is essential for biology programs to reflect this shift through the integration of bioinformatics early within the undergraduate curriculum (11). Curricular efforts to integrate bioinformatics into the classroom have steadily increased over the past decade, with varying approaches from dedicated courses to learning modules within a course (12-14). A majority of these resources either focus on more advanced students (e.g., a course focused on bioinformatics) or introduce students to introductory computational tools and their output, but largely ignore the algorithm behind the tool and treat it as a magical black box (15-19). This exercise was designed to provide an introduction to computational tools to introductory biology students with no previous bioinformatics exposure and also, to pique their interest in future, more intensive bioinformatics courses. To facilitate student learning of the underlying bioinformatics principles on which the computational tools are based, we focused on the algorithms "under the hood" while addressing biological questions.
Physical models have commonly been used to effectively facilitate learning of dynamic abstract molecular mechanisms such as DNA replication and transcription (20-22). Here we take a similar approach to clarify the abstract processes happening under the algorithm's "computational hood" by using paper models. Electronic models are frequently used to teach bioinformatics (23), but we opted to provide students with a kinesthetic activity in contrast to the static computer screen.
There have been repeated calls for the integration of inquiry-based, investigation-rich exercises into the introductory biology laboratory (24,25). For a short introductory bioinformatics exercise, we present here a guided inquiry activity to provide context and applicability for introductory bioinformatics tools (26). Exercises have been previously published which provide students with abstract, treasure hunt style questions to solve with bioinformatics tools (27). Our approach differs by having students apply similar bioinformatics tools to address engaging scientific questions that are likely to resonate with first-year students (investigating the evolution of alcohol metabolism, a Zika outbreak, and a veterinary medicine case study). While other inquiry-style bioinformatics exercise modules are available that take students through a depth of concepts over a series of laboratory sessions (28), this exercise focuses on the core concept of quantifying sequence similarity via alignment in a single three-hour stand-alone module (or two two-hour modules) within a broader introductory biology survey course. Additionally, our exercise comes with three guided inquiry scenarios that the instructor can choose from, based on interest or appropriateness for the course. The inquiry scenarios can also be used directly in class with students in a "choose your own adventure" style approach to appeal to a wide variety of students. The scenarios range from micro- to macro-level biological principles and involve asking how humans evolved alcohol metabolism, seeking the origin of the Zika virus strain that led to the 2015-2016 Brazilian outbreak, and diagnosing the type of pathogen responsible for corneal ulcers in horses.
This activity also addresses barriers that interfere with student learning and the integration of bioinformatics into the curriculum (10). The exercise relies solely on web-based bioinformatics applications (as opposed to command line-based computational programs), which reduces the learning curve for students and instructors (29). Although there is a wide spectrum of individuals (basic user to software engineer) in need of training for bioinformatics competencies (30,31), here we focus on biologists who access data resources and use computational tools as part of a larger research program, but are most comfortable with tools using a graphical user interface (32).
This laboratory module provides introductory biology students with an exploration of a basic set of bioinformatics concepts and tools. The exercise utilizes simple paper models to help students understand matrices and algorithms and then uses web-based computational tools for more robust alignments. Students first define sequence similarity and investigate how similarity can be quantitatively compared between two similar length proteins using a Blocks Substitution Matrix (BLOSUM) scoring matrix. Students then find local regions of similarity between a sequence query and a database of subject sequences using the Basic Local Alignment Search Tool (BLAST) algorithm. Lastly, students practice accessing text-based FASTA-formatted sequence information via National Center for Biotechnology Information (NCBI) databases as they collect data for a multiple sequence alignment for the generation of phylogenetic trees. Students wrap up the activity by applying the concepts to a set of inquiry-based biological questions that can be investigated through use of the bioinformatics tools learned within the activity.
INTENDED AUDIENCE
This activity has been implemented in an undergraduate introductory-level life science majors course at a primarily undergraduate institution. The activity is integrated within the semester that focuses on cellular/molecular biology with the aim of exposing students to principles of bioinformatics early within the biology program. Although the activity has been implemented as a laboratory exercise using laptops in a wet-laboratory, it could be given in a computer laboratory if access to laptops is logistically challenging. The short modules, which introduce the core concepts behind aligning and scoring sequence similarity, could be used at any level of life-science education as an introduction to bioinformatics.
REQUIRED LEARNING TIME
A single three-hour laboratory session is appropriate, although the authors have implemented the activity in a course with two-hour laboratory sessions by splitting the modules across two consecutive sessions. "Similarity and Sequence Alignment" and "Sequence Alignment to a Database of Sequences" modules were implemented together during the first week, and "Phylogenetic Analysis of Homologous Sequences" and the "Inquiry-based Investigation" were implemented during the second week.
PRE-REQUISITE STUDENT KNOWLEDGE
In a two-semester major's introductory biology course, the module is best used in the semester that focuses on cellular and molecular biology and includes a brief unit on an introduction to genomes. As background for the module, students should have been exposed to the following concepts: evolution including natural selection, mutation and phylogenetic trees, nucleic acid structure, Mendelian inheritance, DNA replication, and the Central Dogma of molecular biology. Our curriculum includes macro-level biology principles including evolution in a first semester course that precedes the cellular/molecular course. As stated above, the activity could be adapted as an introduction to bioinformatics in more advanced biology courses.
PRE-REQUISITE TEACHER KNOWLEDGE
There are no teacher prerequisites for instructors with a general background in biology. Teachers without prior experience with bioinformatics tools should run through the activity in its entirety, and refer to the suggested reflection question solutions before implementing it in the classroom. The resource provides enough background information and step-by-step instructions to be used as a training piece for instructors. The activity focuses on introductory bioinformatics tools that instructors can easily learn prior to implementing the module even if they have not used the specific computational tools presented here. Teachers who are familiar with accessing and utilizing the following web-based tools (NCBI web portal, FASTA format, alignment algorithms, and BLOSUM scoring matrices) should be able to implement the activity with minimal preparation time.
SCIENTIFIC TEACHING THEMES
ACTIVE LEARNING
Students will actively engage in learning concepts through a combination of paper-based models and web-based computational algorithms. Using paper-based alignment models, students explore how the BLAST algorithm aligns local sequence "words" to a collection of sequences within a database. Students also manually calculate values for a distance matrix in order to create a phylogram with a group of sequences. The distance matrix exercise guides students to align two sequences and calculate the ratio of mismatches to total positions aligned, thus requiring students to understand the algorithm by performing the steps sequentially. These hands-on exercises are critical for students to engage with and understand how computational algorithms score and present sequence alignments. Without these physical exercises, the computational algorithms are essentially a black box. Student pairs work through the exercise and are encouraged to collaborate with a second pair of students to facilitate peer learning. In a classroom or computer lab with a traditional layout, student pairs can be instructed to collaborate with a second pair of students directly in front or behind them. The lesson finishes with a set of guided inquiry-based investigative questions to pique student interest and engage them to address one of the following real biological issues using bioinformatics tools: propose how humans evolved to metabolize alcohol, determine the origin of a strain of Zika virus, or identify a microbe that causes ocular pathology.
ASSESSMENT
Students work through a series of reflection questions integrated within the exercise, to be turned in before the following laboratory session and graded for summative assessment. Students are encouraged to discuss the questions with their peers during the laboratory investigation and seek out guidance from the instructor when appropriate. The instructor should make classroom rounds to help stimulate laboratory group/table discussion and answer questions or help students when they are off track. During the classroom rounds, the instructor should also encourage students to verbally explain solutions for the thought questions, integrated within the activity, to the group as a metacognitive exercise and to check for understanding. The instructor can use these formative assessment data to determine when it is appropriate to bring the class back together for follow-up demonstrations in a just-in-time teaching style.
The reflection questions are also compiled into an informal/short answer-based laboratory report that is attached to the last page of each exercise as part of the collective activity (Supporting Files S1-S6: Sequence Similarity - Exercise 1, 2, 3, 4.1, 4.2, and 4.3). Students are instructed to write down solutions in their own words after informally discussing the questions with their peers. The informal laboratory report is turned in before the next laboratory for summative assessment. The instructor can provide teaching assistants with a suggested solution key integrated at the end of each exercise (Supporting File S1-S6 Sequence Similarity - Exercise 1, 2, 3, 4.1, 4.2, and 4.3) and then verify grading quality while entering grades into the course gradebook.
INCLUSIVE TEACHING
In this activity, students work collaboratively in pairs and are directed to further interact with a second pair during the allotted laboratory time. The instructor should encourage teamwork and peer-instruction to complement the instructor's explanations, which present the content in verbal, visual, and physical contexts. The instructor may assign pairs and pairs-of-pairs to facilitate interaction with peers with diverse views and approaches to learning. The instructor can create heterogeneous groups by drawing on differences in student academic performance in the class, discipline, or differences in writing and/or organizational skills. Peer-to-peer teaching may also be effective in instructor-assigned groups in which students have heterogeneous levels of familiarity with bioinformatics tools and databases. The materials are accessible by complying with universal design principles (33,34). Information is presented in different ways including paper models, computational algorithms, teacher demonstrations, and peer-to-peer interaction. Students can express their level of understanding through verbal interactions with the instructor, individual exploration and reflection, group discussion and reflection, and individual summative assessment. The activity also stimulates student interest and motivation to learn by having students kinesthetically interact with physical paper models, visually observe demonstrations, verbally communicate understanding to peers, and promote higher order thinking through the investigation of application questions.
LESSON PLAN
OVERVIEW
As stated above, the exercise can be implemented in a single three-hour session or split into two separate two-hour laboratory sessions within a general biology laboratory. The activity could easily be adapted for a classroom where students bring in laptops or relocated to a computer lab. It is highly recommended, if possible, that the instructor uses a collaborative environment with square or round tables for the activity to promote an interactive group effort for discussing and working through the exercises. At Adams State, this environment was the laboratory space, which also promoted the idea that bioinformatics tools are used within the biology laboratory to complement wet-lab based experiments. In the lecture session before the laboratory activity, the students were introduced to some introductory concepts related to studying genomes including: DNA sequencing, biological databases, and comparative and functional genomic approaches to genome annotation and analysis.
PRE-CLASS PREPARATION
The instructor should work through the exercises prior to classroom implementation to become familiar with the activity and bioinformatics tools utilized (Table 1). Walking through the activity is necessary to confirm the web-based computational tools and/or the web interfaces have not changed; new versions and updates to database records happen continually. In the activity handouts (Supporting Files S1-S3: Sequence Similarity - Exercise 1, 2, and 3), instructors should especially pay attention to the web-based tool protocols that are found within grey boxes since this is where modifications may be necessary. A pre-activity run-through will also allow the instructor to identify potentially challenging segments within the activity for their specific cohort of students. Additional resources for navigating NCBI and use of NCBI BLAST can be found via the NCBI Help Manual (https://www.ncbi.nlm.nih.gov/books/NBK3831/).
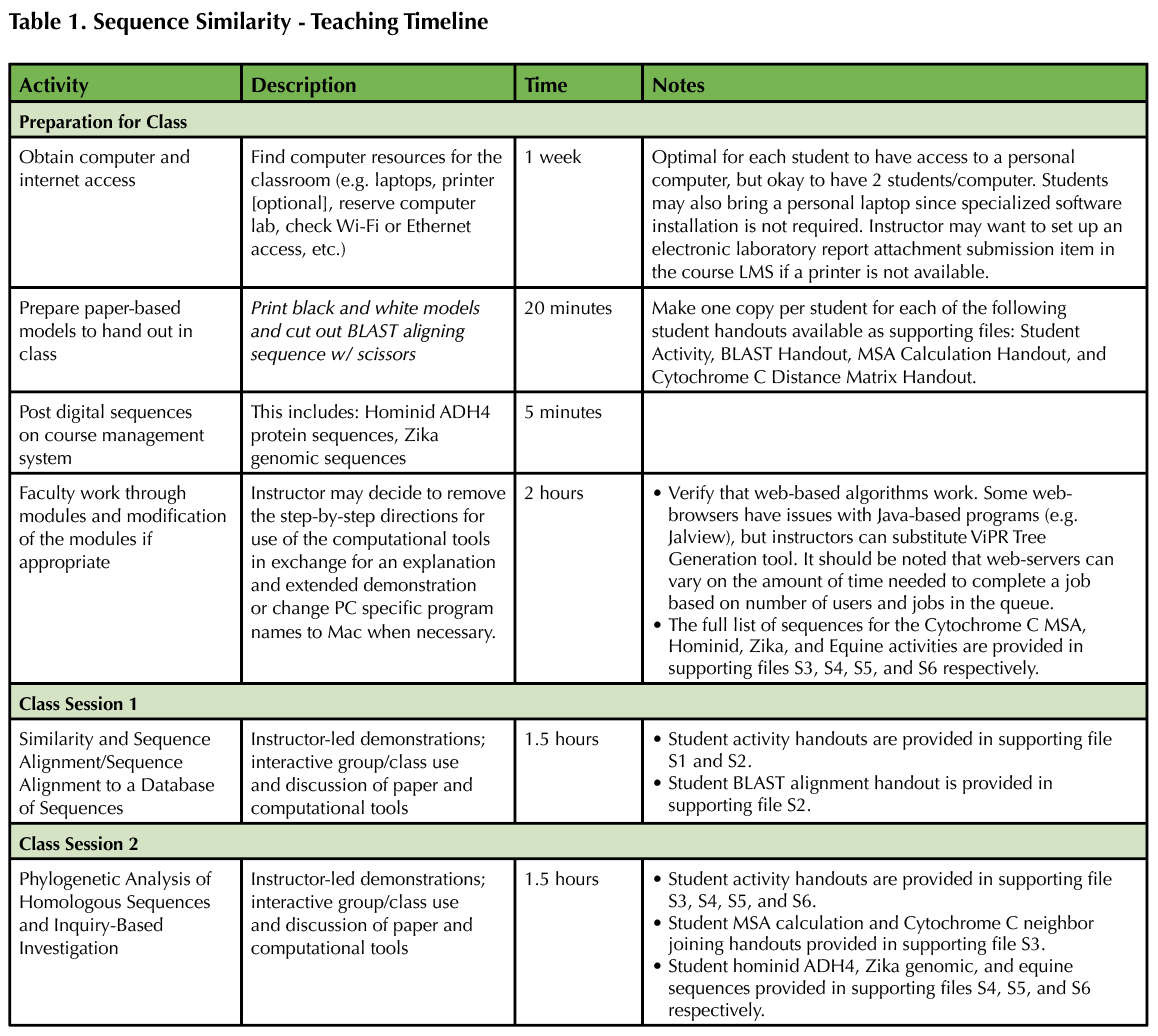
Table 1. Teaching Timeline
After working through Laboratory sessions #1 and 2, instructors should also examine the set of three inquiry-based investigations (Supporting Files S4-S6: Sequence Similarity - Exercises 4.1, 4.2, and 4.3). These scenarios hook student interest through the exploration of the evolution of alcohol metabolism, tracking a Zika Virus outbreak, and a veterinary medicine application. After reviewing the investigations, instructors should consider how to implement them in the classroom. Instructors could select a single case that is most applicable or allot more time and require students to work through all three. Alternatively, instructors could flank the core lab sessions with one of the scenarios, providing a hook for student interest before the BLAST activities followed by an in-depth closing investigation. Alternatively, students could read each scenario and "choose their own adventure."
The instructor should locate the appropriate local resources needed to implement the activity (Table 1). Essential resources include: Internet access, computers/laptops with a web-browser, paper, writing utensils, scissors, and a black and white printer (optional). We request that students bring a personal laptop to the laboratory for this activity, while supplying a few departmental laptops for those students that either do not have a laptop or are not comfortable bringing it to class. Student pairs can share a laptop, but we prefer students gain experience executing the computational steps individually while discussing the tools and analysis in pairs.
The bioinformatics activity requires that students print, annotate screenshots of alignments and phylograms, and attach them to their laboratory report. The ability for students to print may be an issue if the laboratory space is not equipped with printers or if there are logistical issues with students connecting personal laptops with local printers. An alternative to printing these items would be for students to annotate and submit the screenshots electronically through a course learning management system (e.g., Blackboard). To streamline this process, annotated screenshots should be compiled within a single word processing document (e.g., *.doc, *.docx, *.rtf, etc.) and converted to a PDF to allow for rendering in the course learning management system.
The protocols are written from the perspective of using a PC. If the class is working in a computer lab with Macs or students have a personal Mac laptop, instructors can point out comparable programs to use for generating partial screenshots or storing FASTA sequences. For example, Mac users may use TextEdit in place of Windows Notepad. Additionally, Mac users may use the quick keys "command+shift+4" or the Grab application in the utilities folder to take partial screenshots, which automatically save to the desktop, in place of Windows Snipping Tool.
LABORATORY SESSION #1
SIMILARITY AND SEQUENCE ALIGNMENT
The activity starts by having students discuss the idea of similarity using the vases in the first activity figure (Supporting File S1: Sequence Similarity - Exercise 1, questions 1 and 2). The concept of similarity is then extended to the two text passages, and challenges students to conceptualize a way in which similarity for the passages could be quantified (Supporting File S1: Sequence Similarity - Exercise 1, question 3). After a brief two-minute brainstorming session in groups, we bring the students together and introduce the basic method of using rewards (positive integer) and penalties (negative integer) for identities and non-identities, respectively. At this point, we introduce the BLOSUM 62 substitution matrix and demonstrate calculating a numerical score to represent the similarity between two protein sequences. Students are provided time to practice using the BLOSUM 62 matrix (Supporting File S1: Sequence Similarity - Exercise 1, questions 4-8) before a short demonstration on use of a computational algorithm to validate the student's manual calculations (Supporting File S1: Sequence Similarity - Exercise 1, question 9).
SEQUENCE ALIGNMENT TO A DATABASE OF SEQUENCES
The second exercise begins by pointing out that biologists are often interested in comparing a sequence to a large database of sequences that may be of varying lengths (Supporting File S2: Sequence Similarity - Exercise 2). We then briefly explain an abridged overview of the BLAST algorithm and provide students structured time to work through the abridged algorithm via the paper BLAST model exercise (Supporting File S2: Sequence Similarity - Exercise 2, questions 1 and 2). The instructor brings the class back together for a demonstration of the NCBI web portal. We walk students through how to find nucleotide and protein records as well as how to run BLAST on the sequence associated with the database record, along with analysis of the output generated by a BLAST search (Supporting File S2: Sequence Similarity - Exercise 2, questions 3-7).
LABORATORY SESSION #2
PHYLOGENETIC ANALYSIS OF HOMOLOGOUS SEQUENCES
We begin this session by explaining the concept of a molecular clock. We focus on the cytochrome c protein sequence as an example of a molecular clock in which we can monitor substitution rate in homologous genes over evolutionary time (Supporting File S3: Sequence Similarity - Exercise 3). Students are asked to reflect on what they learned previously in order to brainstorm a way to obtain the human cytochrome c protein sequences and locate similar sequence in other organisms. Students use a general NCBI search (https://www.ncbi.nlm.nih.gov/) followed by use of BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify similar sequences (homologs in most cases). We then explicitly present FASTA file format and demonstrate the use of Windows Notepad to store FASTA formatted sequences (Supporting File S3: Sequence Similarity - Exercise 3, page 2).
For obtaining FASTA-formatted sequences for the multiple sequence alignment, on which a phylogram will be based, Zea mays and Danio rerio are strategically used as example organisms across phylogeny. This comparison gives students practice on manually obtaining FASTA sequences, as they are not included in the default organisms integrated within the HomoloGene (https://www.ncbi.nlm.nih.gov/homologene) database output that students will be introduced to later (Supporting File S3: Sequence Similarity - Exercise 3, page 2). HomoloGene is used to provide students with another example of a useful database within the NCBI portal and reduces the redundancy inherent in gathering FASTA sequences; this keeps engagement levels high during the exercise (Supporting File S3: Sequence Similarity - Exercise 3, pages 2-3). Use of the HomoloGene database can also be used to illustrate that there are multiple ways to access the same information within NCBI.
When using HomoloGene, students may notice that the reference protein sequence for Macaca mulatta (Rhesus macaque) is labeled as an outdated sequence record. Students can still obtain this sequence by clicking on the original accession number for use in the exercise, but the instructor could opt to challenge students to find the updated sequence. Students could utilize BLASTP to search the outdated sequence record against the Refseq database and the organism Macaca mulatta and obtain XP_014992345.1 as the top hit. Students could also use BLASTP to align the Homo sapiens cytochrome c sequence using the Refseq database and the organism Macaca mulatta to obtain XP_014992345.1 as the top hit. For more advanced students, this exercise could also lead to a conversation about the meaning of accession prefixes for reference sequences (XP_ - predicted protein vs. NP_ - experimentally validated protein) as well as a detailed explanation of the utility of the RefSeq database vs. more inclusive databases that are encyclopedic with a level of redundancy (e.g., GenBank).
At this point, we bring the class back together for a brief discussion on multiple sequence alignments (MSAs) where students will conduct all possible pairwise alignments between the list of sequences in order to fill in a distance matrix (Supporting File S3: Sequence Similarity - Exercise 3, question 1). The distance matrix reflects observed substitution rates between sequences that will be used to infer evolutionary distance. The instructor then demonstrates the neighbor-joining distance method for phylogenetic tree generation using the distance matrix (35). Students are then provided with time to walk through the manual neighbor-joining method with cytochrome C (Supporting File S3: Sequence Similarity - Exercise 3, questions 2 and 3). To reduce the manual time needed, yet allow students to get a good idea of what the algorithm does, it is suggested that students look at a subset of the sequences that will be used later with the computational MSA (i.e., the tetrapods and fruit flies as an outgroup) with a portion of the distance matrix pre-filled. This comparison is done to keep student engagement high by reducing tedious work.
After wrapping up the manual neighbor-joining activity, we provide a demonstration of an MSA computational tool (i.e., Clustal Omega - https://www.ebi.ac.uk/Tools/msa/clustalo/) and the use of Jalview and ViPR-based tree generators (Supporting File S3: Sequence Similarity - Exercise 3, question 4). Clustal Omega generates a very basic static cladogram and phylogram within the "Phylogenetic Tree" output tab. Students can generate a more dynamic tree by using the Java-based Jalview program that is integrated in the "Results Summary" output tab in Clustal Omega. Unfortunately, some browsers including Google Chrome and Firefox 52 and above do not support NPAPI-based plugins (e.g., Java), thus alternative web browsers such as Internet Explorer or Safari need to be used. An alternative dynamic tree generator such as the default tool provided by Virus Pathogen Database and Analysis Resource (ViPR - https://www.viprbrc.org/brc/home.spg?decorator=vipr) is an excellent alternative option (Supporting File S3: Sequence Similarity - Exercise 3, questions 5-7).
It is worth noting that the amount of traffic on a web-based server that is hosting a web-based bioinformatics algorithm, can vary considerably depending on the day, time of day, and the computational intensity of the requests submitted at any one time. Thus, the queue for BLAST and MSA searches can range from a few seconds to a few minutes. Where this can become a real issue is for computational-intensive submissions such as an MSA job submission for whole genomic sequences, which may take 15+ minutes to run (e.g., Zika virus genomic MSA as part of one of the inquiry activities). An in-class option to help control for this variable is to consider installing a program such as CLC Sequence Viewer or Molecular Evolutionary Genetics Analysis (MEGA) onto a local machine (36,37). Alternatively, the Virus Pathogen Database and Analysis Resource (ViPR) provides a web-based phylogenetic tree generator with a "Quick Tree" option that uses the FastME algorithm to run a large number or relatively long sequences for quick phylogenetic tree construction (38). Instructors could also consider assigning the computationally expensive MSA alignments as homework, which students could complete asynchronously outside of class or let the algorithm run overnight with analysis being completed the following day.
Inquiry-Based Investigation
To wrap up the series of bioinformatics exercises, the instructor can either guide students to investigate one of the three provided inquiry activities or provide students with the choice of which activity they would like to pursue (Supporting Files S4-S6: Sequence Similarity - Exercise 4.1, 4.2, and 4.3). Exercise 4.1 guides students in an investigation of the evolution of alcohol metabolism in hominids, by comparing the amino acid sequences of an alcohol dehydrogenase enzyme in arboreal and terrestrial hominids. This enzyme catalyzes the oxidation of ethanol to acetaldehyde, the first step in the detoxification of ethanol. Exercise 4.2 leads students in examining the evolution of the Zika virus by comparing the full-length viral genomes of Zika viruses isolated from monkeys, mosquitoes, and humans. Zika is of current interest due to its association with congenital Zika syndrome, a collection of birth defects (notably severe microcephaly) found in babies that have been infected by Zika during pregnancy. Exercise 4.3. takes students on a journey to determine the likely causative agent (bacterial or fungal) of an equine corneal ulcer by constructing phylograms using bacterial 16S DNA and fungal ITS DNA sequences. Based on their results students propose an appropriate treatment.
Suggested solutions to the thought questions are provided at the bottom of each relevant supporting file (Supporting Files S1-S6: Sequence Similarity - Exercise 1, 2, 3, 4.1, 4.2, and 4.3). The instructor may choose to provide students with the "student sequences" for the hominid alcohol dehydrogenase and/or the Zika inquiry exercises to reduce the amount of time students spend retrieving database sequences (Supporting Files S4 and S5: Sequence Similarity - Exercise 4.1 and 4.2). Note that the "student sequences" do not have all the necessary sequence data to answer the inquiry question, thus allowing the students to apply their knowledge on how to retrieve database sequences.
TEACHING DISCUSSION
Overall, students appeared to enjoy the activity and informally expressed interest and excitement, especially while working to solve the inquiry-portion of the learning activity. Grading of student responses to the integrated thought questions suggested that a majority of students satisfied the student learning goals and objectives, and could apply process and conceptual knowledge to the inquiry problems.
The student activity handout integrates a detailed step-by-step tutorial for use of the web-based bioinformatics tools. Since the tools and databases utilized by the activity dynamically change over time it will be essential that instructors check the tutorials and modify them appropriately prior to providing students with the activity handout, however the fundamental alignment concepts introduced by this activity should remain unchanged. With use of the step-by-step directions we have anecdotally observed a subpopulation of students who have trouble thinking beyond the step-by-step instructions and come away from the activity with a rudimentary understanding of the utility of the tools. Since the initial implementation, we have also included three inquiry-based investigations (Supporting Files S4-S6: Sequence Similarity - Exercise 4.1, 4.2, and 4.3) that help address the issue of student depth of understanding and application of the concepts. An alternative approach for instructors to consider would be to remove the step-by-step instructions and walk the class through a demonstration of the web-based tools followed by structured time for the students to experiment with the tools. More students may then become self-sufficient at using the tools.
The inquiry application piece was designed to reinforce use of the tools while engaging students in solving a biological problem. The inquiry activities provide both macro- and microevolutionary and pathology-based investigations for students to address with the appropriate bioinformatics tools. Instructors may present all inquiry questions to students in a "choose your own adventure" style approach to the activity, or only present the class with a single inquiry topic to reflect interests of a majority of the students enrolled in the course or the course content.
Additionally, throughout the activity the authors found it useful to provide students with structured teaching demonstrations combined with group exploration time, capped off with an instructor debrief and follow-up discussion questions for the whole class. A structured environment allowed students to experiment individually, yet kept the class together working on the same content and prevented students from attempting to proceed quickly through the activity without reflection. To facilitate this structured approach, a schedule for the session along with associated time spent on each portion of the activity was written on the whiteboard to keep the whole class on task.
We opted to simplify the BLAST algorithm for introductory purposes as well as considering that the activity focuses more broadly on sequence alignment and similarity and not solely on BLAST. Instructors could add additional information on how a query sequence is broken up into a number of k-letter seed words, where the word length is user defined. The word list is then further expanded to include neighborhood words that are generated by substituting amino acid residues as long as the total substitution score (calculated by using a BLOSUM substitution matrix) is above a user-defined threshold. Walking through the unabridged algorithm would be a good exercise for additional application of substitution matrices for a course with ample time or for students in more advanced bioinformatics courses.
Prior to starting the phylogram, students could be instructed to produce a phylogram based on morphological characteristics (something that is probably more familiar to them). This could entail handing out paper sheets with an imaginary set of organisms with different morphological features (e.g., long/short tail, straight/curly tail, etc.) and could have students quantitate the number of morphological differences to make a basic phylogenetic tree (39, 40, http://www.evolution.wisc.edu/node/38). The morphological difference could be considered analogous to differences in sequence residues, which can be used as a proxy for similarity using molecular data. This may be useful for classes that are transitioning from macroevolutionary principles to understanding life at the molecular level.
The output phylogram for cytochrome c is that of what we would expect for whole-genome phylogeny for the selected organisms in the exercise. Instructors of more advanced courses may find utility in using an alternative protein for this activity that yields a sequence phylogeny that differs from the expected whole-genome organismal phylogeny to showcase the point that not all proteins are good molecular clocks and/or that different proteins evolve at different relative rates over evolutionary time. For example, histone protein sequence has very little flexibility for evolutionary change before the protein becomes nonfunctional. This pattern can be observed by comparing histone H4 protein sequences, which are the same for humans, Rhesus monkey, Xenopus, and zebrafish, while the cattle H4 sequence has a single substitution. Examples of genes that evolve at more rapid rates than cytochrome c include hemoglobin and fibrinopeptide proteins (41). Other factors such as gene duplication events may lead to different selective pressure between compared species on the same homologous genes.
This exercise has been designed as a guided inquiry activity to provide students with real-world applications of bioinformatics, while balancing time investment. This guided-inquiry activity could be adapted to an open-inquiry format if the instructor has more class time, where students could generate their own questions and investigate them by doing some basic bioinformatics data mining. For example, this set of exercises could be coupled with other wet-laboratory investigations such as a DNA barcoding of local organisms to determine if morphologically similar organisms are the same species or if grocery store food products are correctly labeled (42).
SUPPORTING MATERIALS
- S1. Sequence Similarity - Exercise 1
- S2. Sequence Similarity - Exercise 2
- S3. Sequence Similarity - Exercise 3
- S4. Sequence Similarity - Exercise 4.1
- S5. Sequence Similarity - Exercise 4.2
- S6. Sequence Similarity - Exercise 4.3
ACKNOWLEDGMENTS
We would like to thank the American Society for Microbiology for sponsoring a genomics education workshop in which the initial concept and writing of this activity began. Participating workshop faculty, especially Komal Vig, helped guide the conceptual basis of the activity. We would also like to thank the Network for Integrating Bioinformatics into Life Sciences Education (NIBLSE) Incubator facilitator (Michael Sierk) and participants (Sebastian Galindo and Hayley Orndorf) for critical feedback and help refining the activity.
References
- Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ, Iyer R, Schatz MC, Sinha S, Robinson GE. 2015. Big data: astronomical or genomical? PLoS Biol 13:e1002195. doi: 10.1371/journal.pbio.1002195
- Marx V. 2013. Biology: The big challenges of big data. Nature 498:255-260. doi: 10.1038/498255a.
- Barone L, Williams J, Micklos D. 2017. Unmet needs for analyzing biological big data: A survey of 704 NSF principal investigators. PLoS Comput Biol 13:e1005755. doi: 10.1371/journal.pcbi.1005755
- Jagodnik KM, Koplev S, Jenkins SL, Ohno-Machado L, Paten B, Schurer SC, Dumontier M, Verborgh R, Bui A, Ping P. 2017. Developing a framework for digital objects in the Big Data to Knowledge (BD2K) commons: Report from the Commons Framework Pilots workshop. J Biomed Inform 71:49-57. doi: 10.1016/j.jbi.2017.05.006.
- Miller M, Zhu C, Bromberg Y. 2017. clubber: removing the bioinformatics bottleneck in big data analyses. J Integr Bioinforma 14. doi: 10.1515/jib-2017-0020
- Canela-Xandri O, Law A, Gray A, Woolliams JA, Tenesa A. 2015. A new tool called DISSECT for analyzing large genomic data sets using a Big Data approach. Nat Commun 6:10162. doi: 10.1038/ncomms10162.
- Levine AG. 2014. An explosion of bioinformatics careers. Science 344:1303-1306. doi: 10.1126/science.344.6189.1303
- Dinsdale E, Elgin SC, Grandgenett N, Morgan W, Rosenwald A, Tapprich W, Triplett EW, Pauley MA. 2015. NIBLSE: A network for integrating bioinformatics into life sciences education. CBE-Life Sci Educ 14:le3. doi: 10.1187/cbe.15-06-0123.
- Lopatto D, Hauser C, Jones CJ, Paetkau D, Chandrasekaran V, Dunbar D, MacKinnon C, Stamm J, Alvarez C, Barnard D. 2014. A central support system can facilitate implementation and sustainability of a classroom-based undergraduate research experience (CURE) in genomics. CBE-Life Sci Educ 13:711-723. doi: 10.1187/cbe.13-10-0200.
- Williams J, Drew J, Galindo-Gonzalez S, Robic S, Dinsdale E, Morgan W, Triplett E, Burnette J, Donovan S, Elgin S. 2017. Barriers to Integration of Bioinformatics into Undergraduate Life Sciences Education. bioRxiv 204420. doi: https://doi.org/10.1101/204420
- Pevzner P, Shamir R. 2009. Computing has changed biology--biology education must catch up. Science 325:541-542. doi: 10.1126/science.1173876.
- Magana AJ, Taleyarkhan M, Alvarado DR, Kane M, Springer J, Clase K. 2014. A survey of scholarly literature describing the field of bioinformatics education and bioinformatics educational research. CBE-Life Sci Educ 13:607-623. doi: 10.1187/cbe.13-10-0193
- Elgin SC, Hauser C, Holzen TM, Jones C, Kleinschmit A, Leatherman J. 2017. The GEP: crowd-sourcing big data analysis with undergraduates. Trends Genet 33:81-85. doi: 10.1016/j.tig.2016.11.004.
- Madlung A. 2018. Assessing an effective undergraduate module teaching applied bioinformatics to biology students. PLoS Comput Biol 14:e1005872. doi: 10.1371/journal.pcbi.1005872.
- Feig AL, Jabri E. 2002. Incorporation of bioinformatics exercises into the undergraduate biochemistry curriculum. Biochem Mol Biol Educ 30:224-231. doi: 10.1002/bmb.2002.494030040093
- Inlow JK, Miller P, Pittman B. 2007. Introductory bioinformatics exercises utilizing hemoglobin and chymotrypsin to reinforce the protein sequence-structure-function relationship. Biochem Mol Biol Educ 35:119-124. doi: 10.1002/bmb.30.
- Zhang X. 2011. Exploring cystic fibrosis using bioinformatics tools: a module designed for the freshman biology course. Biochem Mol Biol Educ 39:17-20. doi: 10.1002/bmb.20460.
- Rahman SJ, Charles TC, Kaur P. 2016. Metagenomic approaches to identify novel organisms from the soil environment in a classroom setting. J Microbiol Biol Educ 17:423. doi: 10.1128/jmbe.v17i3.1115
- Terrell CR, Listenberger LL. 2017. Using molecular visualization to explore protein structure and function and enhance student facility with computational tools. Biochem Mol Biol Educ 45:318-328. doi: 10.1002/bmb.21040.
- Sigismondi L. 1989. A paper model of DNA structure and replication. Amer Biol Teach 51:422-423. doi: 10.2307/4448969
- Priano C. 2013. Shaping tRNA. Am Biol Teach 75:708-709. doi: 10.1525/abt.2013.75.9.14
- Vrentas CE, Adler JJ, Kleinschmit AJ, Massimelli J. 2018. Riboflavin Riboswitch Regulation: Hands-On Learning about the Role of RNA Structures in the Control of Gene Expression in Bacteria. J Microbiol Biol Educ, 19(2). doi: 10.1128/jmbe.v19i2.1501
- Weisstein AE, Gracheva E, Goodwin Z, Qi Z, Leung W, Shaffer CD, Elgin SCR. 2016. A Hands-on Introduction to Hidden Markov Models. CourseSource. doi: 10.24918/cs.2016.8
- Handelsman J, Ebert-May D, Beichner R, Bruns P, Chang A, DeHaan R, Gentile J, Lauffer S, Stewart J, Tilghman SM, others. 2004. Scientific teaching. Science 304:521-522. doi: 10.1126/science.1096022
- Brewer CA, Smith D. 2011. Vision and change in undergraduate biology education: a call to action. American Association for the Advancement of Science, Washington, DC. ISBN 978-0-87168-741-8
- Weaver G, Russell CB, Wink DJ. 2008. Inquiry-based and research-based laboratory pedagogies in undergraduate science. Nat Chem Biol 4:577-580. doi: 10.1038/nchembio1008-577.
- Boyle JA. 2004. Bioinformatics in undergraduate education: practical examples. Biochem Mol Biol Educ 32:236-238. doi: 10.1002/bmb.2004.494032040376.
- Bednarski AE, Elgin SC, Pakrasi HB. 2005. An inquiry into protein structure and genetic disease: introducing undergraduates to bioinformatics in a large introductory course. Cell Biol Educ 4:207-220. doi: 10.1187/cbe.04-07-0044
- Cummings MP, Temple GG. 2010. Broader incorporation of bioinformatics in education: opportunities and challenges. Brief Bioinform 11:537-543. doi: 10.1093/bib/bbq058.
- Schneider MV, Watson J, Attwood T, Rother K, Budd A, McDowall J, Via A, Fernandes P, Nyronen T, Blicher T. 2010. Bioinformatics training: a review of challenges, actions and support requirements. Brief Bioinform 11:544-551. doi: 10.1093/bib/bbq021.
- Sayres MAW, Hauser C, Sierk M, Robic S, Rosenwald AG, Smith TM, Triplett EW, Williams JJ, Dinsdale E, Morgan W, Burnette I, Donovan S, Drew J, Elgin SCR, Fowlks E, Galindo-Gonzalez S, Goodman A, Grandgenett N, Goller C., Jungck J, Newman J, Pearson W, Ryder E, Tosado-Acevedo R, Tapprich W, Tobin T, Toro-Martinez A, Welch L, Wright R, Barone L, Ebenback D, McWilliams M, Olney K, Pauley M. 2018. Bioinformatics Core Competencies for Undergraduate Life Sciences Education. PloS one, 13(6) e0196878. doi: 10.1371/journal.pone.0196878.
- Welch L, Lewitter F, Schwartz R, Brooksbank C, Radivojac P, Gaeta B, Schneider MV. 2014. Bioinformatics curriculum guidelines: toward a definition of core competencies. PLoS Comput Biol 10. doi: 10.1371/journal.pcbi.1003496.
- Burgstahler S. 2011. Universal design: Implications for computing education. ACM Trans Comput Educ TOCE 11:19. doi: 10.1145/2037276.2037283
- Darby A. 2010. Understanding universal design in the classroom. Natl Educ Assoc URL http://www.nea.org/home/34693.htm.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406-425. doi: 10.1093/oxfordjournals.molbev.a040454
- Qiagen. 2008. CLC Sequence Viewer 7.8.1.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870-1874. doi: 10.1093/molbev/msw054.
- Desper R, Gascuel O. 2002. Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. In International Workshop on Algorithms in Bioinformatics (pp. 357-374). Springer, Berlin, Heidelberg. doi: 10.1089/106652702761034136
- Schneider B, Strait M, Muller L, Elfenbein S, Shaer O, Shen C. 2012. Phylo-Genie: engaging students in collaborative 'tree-thinking' through tabletop techniques, p. 3071-3080. In Proceedings of the SIGCHI Conference on Human Factors in Computing Systems. ACM. doi: 10.1145/2207676.2208720
- University of Pittsburgh. 2017. Tree-Thinking Group.
- Nei M, Suzuki Y, Nozawa M. 2010. The neutral theory of molecular evolution in the genomic era. Annu Rev Genomics Hum Genet 11:265-289. doi: 10.1146/annurev-genom-082908-150129.
- Micklos DA, Hilgert U, Nash B. 2013. Genome science: a practical and conceptual introduction to molecular genetic analysis in Eukaryotes. Cold Spring Harbor Laboratory Press. ISBN 978-1-621821-09-0
Article Files
Login to access supporting documents
Kleinschmit-Sequence Similarity- An inquiry based and under the hood approach for incorporating molecular sequence alignment in introductory undergraduate biology courses.pdf(PDF | 159 KB)
S1. Sequence Similarity - Exercise 1.docx(DOCX | 397 KB)
S2. Sequence Similarity - Exercise 2.docx(DOCX | 317 KB)
S3. Sequence Similarity - Exercise 3.docx(DOCX | 122 KB)
S4. Sequence Similarity - Exercise 4.1.docx(DOCX | 57 KB)
S5. Sequence Similarity - Exercise 4.2.docx(DOCX | 140 KB)
S6. Sequence Similarity - Exercise 4.3.docx(DOCX | 1 MB)
- License terms

Comments
Comments
1 Like
Adam Kleinschmit @ on (Edited: @ on )
Validated Assessment Instrument and Learning Resource Effectiveness
A collaborative group of faculty applied an iterative co-design process to develop a validated objective learning gains assessment instrument for this resource. The fifteen-item pre-/post-assessment instrument consists of a combination of multiple-choice and multiple-select questions to allow for scaling data collection within large enrollment courses.
The instrument has been utilized in diverse course implementation contexts (e.g., lower-division courses, upper-division courses, various course-content emphases) and institution types (e.g., research-intensive, primarily undergraduate institutions). Data collected as part of an educational research study indicated learning gains were observed across diverse life science courses, thus supporting the effectiveness of this learning resource.
This dataset can be found via the publication cited below. The validated assessment instrument can be found within the articles “supporting information” section associated with the research article for classroom use:
Kleinschmit AJ, Ryder EF, Kerby JL, Murdoch B, Donovan S, Grandgenett NF, et al. (2021). Community development, implementation, and assessment of a NIBLSE bioinformatics sequence similarity learning resource. PLoS ONE, 16(9): e0257404. https://doi.org/10.1371/journal.pone.0257404
Copy link Report abuse