Discovering Prokaryotic Gene Regulation by Building and Investigating a Computational Model of the lac Operon
Published online:
Abstract
The ability to construct and reason through a biological system is an important skill to develop in science education. This level of understanding forces students to recognize the interrelatedness of components within a biological process and appreciate the process as a dynamic system. To develop modeling and computation skills in undergraduate students, we developed a modeling-focused lesson in the context of the lac operon. The lac operon is a common topic in introductory biology, genetics, and molecular biology curricula. The lac operon is a great system for novice modelers as it exemplifies important biological concepts, including the central dogma of molecular biology and gene regulation. The lesson utilizes an online modeling platform called Cell Collective Learn. In our lesson, students explore the important structural and regulatory aspects of the lac operon by constructing a computational model of the system, validating their model through simulations, and then manipulating their model to further explore the system dynamics. The lesson contains all needed resources and question prompts, minimizing the need for instructor guidance.
Citation
Crowther, A., Bergan-Roller, H.E., Galt, N.J., Appleby, L., Dauer, J.T., and Helikar, T. 2019. Discovering Prokaryotic Gene Regulation by Building and Investigating a Computational Model of the lac Operon. CourseSource. https://doi.org/10.24918/cs.2019.4Lesson Learning Goals
Students will:- experience how computational modeling can be used to represent systems.
- evaluate how computational modeling and simulations can be used to observe and semi-quantitatively measure the dynamics of gene expression and regulation.
- value how the regulation of gene expression is influenced by external and internal molecular signals.*
- evaluate how prokaryotic cells maintain homeostasis.**
- evaluate how mutations affect genes and their corresponding mRNA and protein products.***
Lesson Learning Objectives
Students will be able to:- model how the components of the lac operon contribute to gene regulation and expression.
- generate and test predictions using computational modeling and simulations.
- interpret and record graphs displaying simulation results.
- relate simulation results to cellular events.
- describe how changes in environmental glucose and lactose levels impact regulation of the lac operon.
- predict, test, and explain how mutations in specific elements in the lac operon affect their protein product and other elements within the operon.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Biological systems are complex networks of biological entities that work together to carry out specific functions. A hallmark of biological systems is the transfer, storage, and utilization of information (e.g., the flow of genetic information from DNA to protein). As such, it is important that undergraduate biology students understand and trace the flow of information through biological systems (1); students commonly demonstrate weak understanding of ways in which biological pathways exchange and store information, especially at a genetic level (2-5). Students struggle with concepts related to the central dogma of molecular biology, including recognizing the relationship between genes, their protein products, and phenotypic expression (2,4,6-10), as well as the regulation of gene expression (2,10).
The lac operon is a classic example of prokaryotic gene regulation. Showcasing core biological concepts such as the central dogma of molecular biology (10-12), it is commonly taught in undergraduate biology courses ranging from introductory biology and genetics to upper-level molecular biology. Since understanding regulation of the lac operon requires knowing the connection between external stimuli and changes to the intracellular environment, it is a useful system to teach regulatory pathways and dynamic systems behavior. Mastering the mechanisms of the lac operon provides a foundation to tackle more difficult and complex regulatory mechanisms in eukaryotic organisms. A basic challenge for students in understanding gene regulation systems, involves understanding how components in the system are connected (12).
Introducing computational models in the classroom shows promise for improving student learning (13-18). Computational models and simulations allow students to directly interact with and manipulate complex biological systems in order to understand overall biological processes. This approach has been successfully implemented in a variety of science courses, including physics, chemistry, and biology (13,16-18). Classrooms utilizing computational simulations in science education have observed improvements in student comprehension and problem-solving skills (13-18). While there are model-based (19) and interactive, computer-based (20,21) teaching resources on the lac operon available for undergraduate instructors, few resources exist that combine both interactive computer technology and modeling in an accessible classroom activity.
To teach students about core concepts while practicing core competencies in science, we developed a teaching approach that uses a user-friendly education platform, Cell Collective Learn (https://learn.cellcollective.org, [14,22-26]). In this approach, students use freely available lessons to develop their understanding of a biological system by creating a computational model themselves or by investigating a provided computational model of the system through simulations. Students engage in authentic yet accessible computational modeling by relating system components (nodes) with direct relationships (arrows/edges); connecting nodes and edges, students mimic behaviors demonstrated in real biological systems, such as activation and inhibition. Students can then observe, in real-time, the behavior of their computational model through simulations. Students explore complex biological systems by predicting cellular mechanisms, testing their prediction with simulations, reporting their observations, and providing accurate cellular mechanisms based on their simulation results. The lessons developed with this approach are grounded in theories of learning and implement evidence-based best practices specific to modeling and using simulations to support learning science (13,23,27,28). We have previously shown that lessons using the simulation-based portion of this approach helped students learn relevant content and skills (14,22,24,25). Specifically, these lessons were shown to help develop students' systems thinking skills and conceptual understanding of biological systems in the context of cellular respiration and the trp operon (14,22,24,25).
Here we describe a lesson, using a computational modeling and simulation approach, on the regulation of gene expression called Discovering Prokaryotic Gene Regulation by Building and Investigating a Computational Model of the lac Operon. This lesson aims to enhance understanding of the lac operon system, information flow within biological systems, and mechanisms of gene regulation, as well as to develop important skills of modeling, using simulations, and applying the process of science. During the lesson, students first build a computational model of the lac operon system, and then explore the regulatory elements of the lac operon by investigating the impacts of perturbations on the lac operon. We call these exercises Investigations. The lesson is versatile and can be completed individually or within small groups. Students receive all the needed instructions and prompts within the lesson itself and can progress through the lesson without additional help.
Intended Audience
This lesson was designed for undergraduate students at the introductory biology level. The lesson could be implemented in higher-level courses such as genetics by setting higher expectations for responses. The lesson could also be used in higher-level genetics and biochemistry courses as a refresher of basic gene regulation knowledge before students learn about more complicated gene regulatory systems.
Required Learning Time
The Discovering Prokaryotic Gene Regulation by Building and Investigating a Computational Model of the lac Operon activity packet (Supporting File S1: Prokaryotic Gene Regulation- Activity packet) could be implemented in-class or as homework. The activity packet (model building and Investigations) should take students about 90 minutes to complete.
Pre-requisite Student Knowledge
Before starting the lesson, students should have basic knowledge about genetics and genetic material (i.e. DNA contains protein-coding regions called genes, definitions of mutation, transcription, translation) and be able to define and distinguish between prokaryotes and eukaryotes. These topics are commonly discussed in introductory biology courses prior to the regulation of gene expression unit. Also, students should have basic computer and internet skills needed to navigate a web browser, type in text responses when prompted, and find and maneuver "play" and "pause" symbols.
While the lesson provides instructions on how to navigate Cell Collective Learn, it may be beneficial for students to first familiarize themselves with the platform in a low-stakes environment. To make students more comfortable with the platform, have them work through the Cell Collective Learn training lesson called Cell Collective Training Module: Factors Influencing Exam Scores (accessible from the home page of Cell Collective Learn at https://learn.cellcollective.org). This training lesson is designed to familiarize new users to the Cell Collective Learn platform by connecting model building and simulation to a relatable context: studying for an exam.
Pre-requisite Teacher Knowledge
Before implementing the lesson in your class, you should have a thorough understanding of the content and skills described above for students. In addition, you should have an understanding of operons and specifically the lac operon, including the biological role of operons (i.e. to coordinate the expression of related genes), the biological role of the lac operon (i.e. to make the enzymes needed for lactose import and breakdown), and the mechanisms regulating lac operon expression (e.g., allolactose binds to and inhibits the lac repressor, allowing RNA polymerase to bind and transcribe the lac operon). This information is provided in the background reading section of the Discovering Prokaryotic Gene Regulation by Building and Investigating a Computational Model of the lac Operon activity packet (Supporting File S1: Prokaryotic Gene Regulation- Activity packet). In addition to the biological background, you should be familiar with Cell Collective Learn. You are encouraged to work through the training module, Cell Collective Training Module: Factors Influencing Exam Scores, in order to better familiarize yourself with the modeling platform. You should also complete the presented lesson activity packet prior to classroom implementation to ensure you are able to provide guidance to your students when needed. Instructors will need to create a Cell Collective Learn account in order to complete the Cell Collective Training Module: Factors Influencing Exam Scores lesson and Discovering Prokaryotic Gene Regulation by Building and Investigating a Computational Model of the lac Operon lesson.
SCIENTIFIC TEACHING THEMES
Active Learning
Students are active participants throughout the entirety of the lesson. As active participants, students will: 1) read about prokaryotic gene regulation and the lac operon in the provided background material, 2) answer short-answer questions aimed to stimulate reading comprehension, 3) describe how the regulatory components of the lac operon are connected, 4) diagram these relationships in order to flesh out their mental model of the lac operon system, and 5) construct, validate, and investigate a computational model of the lac operon. Students are responsible for completing their own work (e.g., reading the background material, building their model, running simulations) and are encouraged to discuss in small groups. Due to the novelty of this type of learning activity (i.e. computational modeling) and type of question probes (i.e. systems thinking), student discussion in this group dynamic is frequently persistent throughout the entire lesson, even though individual student roles are not specified within each group. This type of group structure, apart from ensuring that each student experiences the full value of the lesson, allows technologically-savvy group members to assist students that are struggling with the modeling software, while not domineering the entire lesson.
Assessment
You can assess student understanding throughout the lesson in several ways. For an outline of how you can use our lesson to assess student mastery of the outlined learning objectives, refer to Figure 1. During the building portion of the activity packet (Supporting File S1: Prokaryotic Gene Regulation- Activity packet), students construct and validate a model of the lac operon system. You can evaluate the diagrams students create (Part 6 of Supporting File S1: Prokaryotic Gene Regulation- Activity packet) after model validation in order to evaluate student understanding of the system. Additionally, you can have students create screenshots and submit images of their computational models in order to confirm that they correctly modeled the lac operon. During the investigate portion (pages 8-12 of Supporting File S1: Prokaryotic Gene Regulation- Activity packet), students predict the dynamics of the lac operon model, test their predictions with the simulation, report their observations, and interpret the results. You can use student responses to formatively assess knowledge by evaluating correctness. During the investigate portion, students are prompted to self-assess the correctness of their prediction after running their simulation. During our implementation of the lesson, we formatively assessed the model students built of the lac operon, their observations, and their interpretation of their simulation results.
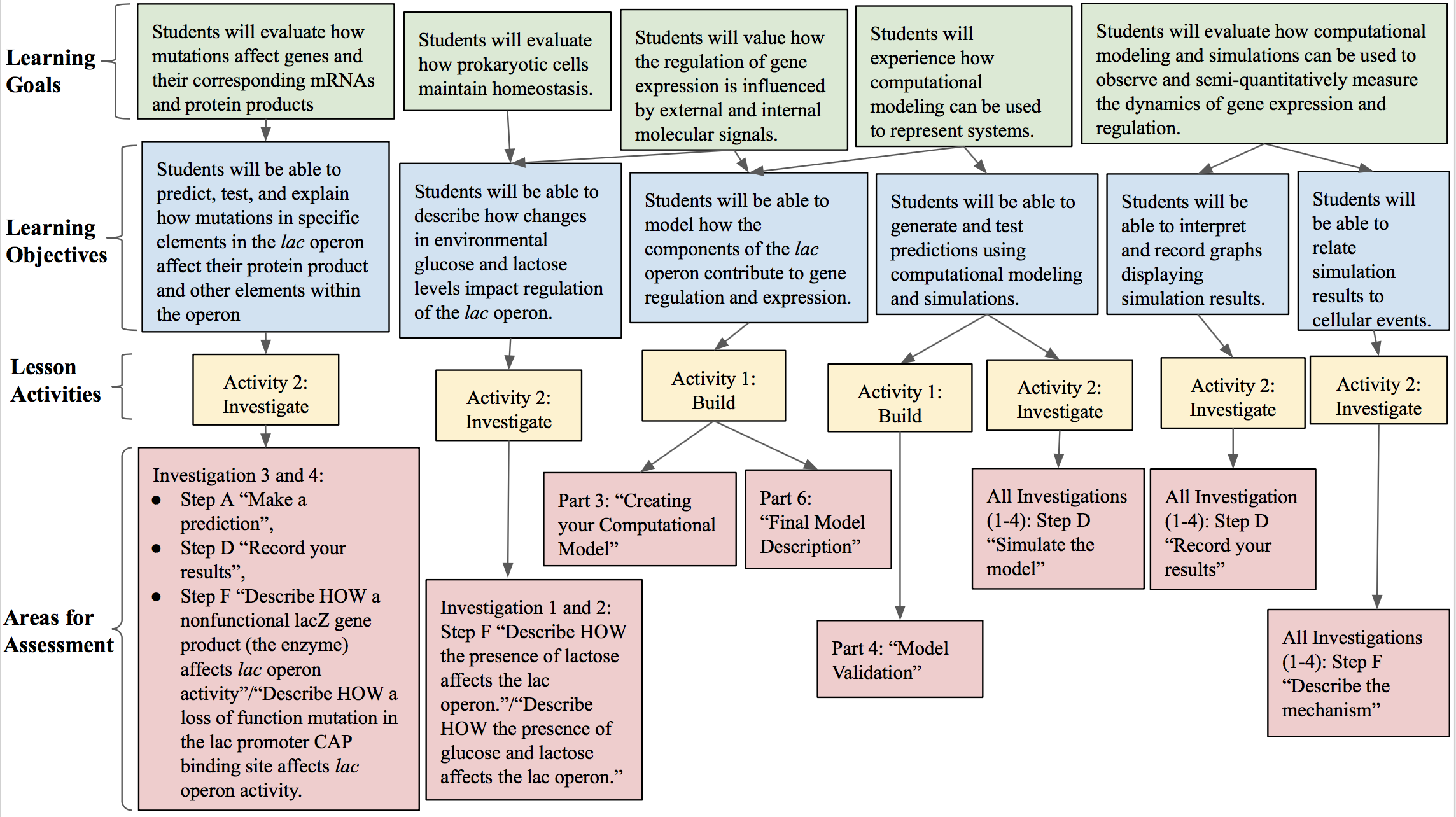
Figure 1: Alignment of Learning Goals. An alignment of learning goals, learning objectives, lesson activities, and assessments.
Inclusive Teaching
During the lesson students can work in small groups of two to four students. Having students work in groups encourages the exchange of ideas among students and offers feedback from peers when individual instructor feedback is not readily available. Working in groups has been shown to be beneficial for fostering student gains in underrepresented student groups, such as first-generation college and black students (29). The lesson also utilizes computational modeling and simulations that are not typically used at the introductory level. This computer-based learning approach may appeal to a variety of students, including those who are otherwise struggling with traditional teaching and/or group work.
LESSON PLAN
An overview and timeline for the lesson plan is provided in Table 1. The learning objectives for this lesson are in part informed by the core concepts for undergraduate biology (1) and specifically introductory biology defined by Khodor, et al. 2004 (30). The lesson is modular, providing flexibility to adapt for different classroom needs. The modules include background information and images about operons and specifically the lac operon, reading comprehension questions, an introduction to Cell Collective Learn, modeling the lac operon, and Investigations of the lac operon. All modules are grouped together in the Discovering Prokaryotic Gene Regulation by Building and Investigating a Computational Model of the lac Operon activity packet (Supporting File S1: Prokaryotic Gene Regulation- Activity packet).
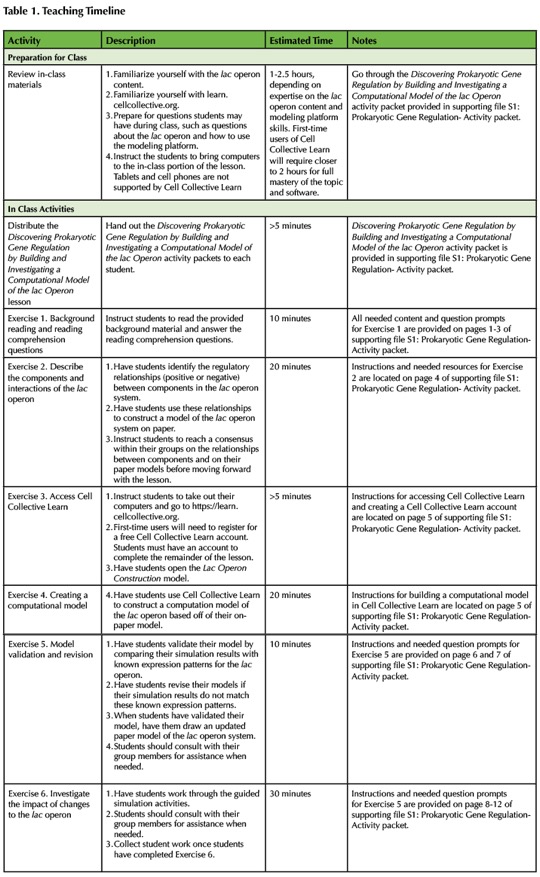
Table 1. Teaching Timeline
Below is a list of materials that are necessary and/or useful for teaching the lesson, all of which are provided as supporting materials, links to websites, or by request from the authors.
- Discovering Prokaryotic Gene Regulation by Building and Investigating a Computational Model of the lac Operon activity packet (Supporting File S1: Prokaryotic Gene Regulation- Activity packet)
- Lac Operon Construction model (accessible at https://learn.cellcollective.org)
- Cell Collective Training Module: Factors Influencing Exam Scores (accessible at https://learn.cellcollective.org)
The Discovering Prokaryotic Gene Regulation by Building and Investigating a Computational Model of the lac Operon activity packet answer key is available by request by contacting the authors.
Before Class
Inform students of the upcoming computer-based activity
You should inform students that they will need a computer for the upcoming class and encourage students to bring their own; tablets and cell phones are currently not supported by Cell Collective Learn. It is ideal for every student to have his or her own computer. However, because the lesson can be done in groups, students without a computer can work with other members of the group who do. Alternatively, you could schedule the activity in a computer lab, if available for your class size and institution.
Review in-class materials
In preparation for the lesson, go through the activity packet (Supporting File S1: Prokaryotic Gene Regulation- Activity packet) as if you were a student. This experience will provide you with a deeper understanding of the lesson content, develop your skills for navigating Cell Collective Learn, and help you prepare for potential issues and questions from your students. All of the necessary materials (Supporting File S1: Prokaryotic Gene Regulation- Activity packet) are provided in this article and are also publicly available on Cell Collective Learn. An answer key is available by request for instructors to prevent easy access by students. For further information, contact Dr. Tomáš Helikar at thelikar2@unl.edu.
Print in-class materials
Each student will also need a copy of the activity packet (Supporting File S1: Prokaryotic Gene Regulation- Activity packet), which includes all background, instructions, and question prompts. You can provide copies for each student or require that students print their own copy and bring it to class.
In-class
Facilitate the Discovering Prokaryotic Gene Regulation by Building and Investigating a Computational Model of the lac Operon in-class activities
First, make sure that each student has a copy of the Discovering Prokaryotic Gene Regulation by Building and Investigating a Computational Model of the lac Operon activity packet (Supporting File S1: Prokaryotic Gene Regulation- Activity packet). This activity packet provides students with all of the background information, directions, and question prompts they need to complete the activity successfully.
Exercise 1. Background reading and reading comprehension questions
Have students read the background information on pages 1-2 and answer the provided reading comprehension questions on page 3 of the activity packet. By reading the background information, students will receive a concise introduction to the structure and function of operons, structure and function of the lac operon, and the basic mechanisms of gene regulation. The accompanying questions on page 3 of the activity packet ensure that students complete the reading and are prepared for the computer-centered portion of the lesson. Reading the background information and answering the questions on page 3 of the activity packet should take students approximately 10 minutes.
Exercise 2. Describe the components and interactions of the lac operon
In the first part of the activity, found on page 4 of the activity packet, students are instructed to identify the positive and negative regulators for each component of the lac operon system and describe the biology behind that relationship (e.g., the lac enzymes are responsible for breaking down lactose). After describing how the different components in the lac operon system impact one another, students translate this information into an on-paper model of the system, using inhibitory arrows (⊣) and activating arrows (→) to denote relationships between components. This on-paper model serves as a draft for the model that students will construct in Cell Collective Learn. In this exercise students are expected to start using their group members as a valuable resource by discussing how each component of the lac operon system plays a role in regulating the lac operon expression and how that translates into a two-dimensional model. Each group should reach a consensus before moving forward. This need to reach a consensus prompts students to conduct thoughtful discussions about the lac operon system. Instructors should allocate 20 minutes for students to complete this portion of the lesson.
Exercise 3. Access Cell Collective Learn
In the second part of the activity, students use their computers to access Cell Collective Learn at https://learn.cellcollective.org, create or log into their account, and open the Lac Operon Construction model from the home page. Instructions on navigating to Cell Collective Lean and selecting the Lac Operon Construction model are provided on page 5 of the activity packet.
Students who need to register for a Cell Collective Learn account will need to select the "Create Account" icon and provide their full name along with their email and their institution of study. Students who already have an account will select the "Login" icon and input their selected email and password. A Cell Collective Learn account is free for all users. Students are able to explore Cell Collective Learn as a guest by selecting the "Explore All Learning Modules" icon, which allows students to simulate pre-built models and download any provided files associated with each lesson. However, students will not be able to utilize all of the features of Cell Collective Learn as a guest, including the ability to create and save new models as well as any other lesson-related work that requires the saving of data on the Cell Collective Learn servers. Thus, students need to have a Cell Collective Learn account in order to complete the Discovering Prokaryotic Gene Regulation by Building and Investigating a Computational Model of the lac Operon lesson.
Exercise 4. Creating a computational model
Students next use Cell Collective Learn to construct a computational model of the lac operon. Students are responsible for adding components (e.g., lac repressor) and the relationship between those components (e.g., allolactose inhibits the lac repressor) in order to correctly model behavior of the lac operon. Instructions on how to construct a computational model in Cell Collective Learn are provided on page 5 of the Discovering Prokaryotic Gene Regulation by Building and Investigating a Computational Model of the lac Operon activity packet (Supporting file S1: Prokaryotic Gene Regulation- Activity packet). Instructors should allocate approximately 20 minutes for students to construct their model of the lac operon. In this exercise, students should assist their fellow group members in navigating the modeling functions and confirming that their constructed computational model accurately reflects the biological relationships between components in the lac operon.
Exercise 5. Model validation and revision
After constructing their model, students must check that the model "behaves" in accordance to known literature on the lac operon. Students are given the literature-supported expression of the lac operon (provided on page 6, Table 2 of the activity packet) based on environmental conditions (e.g., if glucose and lactose are present in the environment, the lac operon will be inactive). Students simulate their model under different environmental conditions (all combinations of presence and absence of glucose and lactose) and compare their simulation results to the expected biological behaviors. If their simulation results do not match the expected behaviors, students are prompted to revisit their model and make revisions as necessary. Students record their revisions on page 7 of the activity packet. This validation and revision step parallels ways in which researchers validate their models before further exploring the system (31,32). Students proceed through the remaining exercises only after achieving the correct model behavior for the lac operon.
Students set the parameters of the simulation by altering the levels of external components. While the quantitative values of external components (e.g., environmental glucose set to 100%) do not directly correspond to a specific, measurable biological property (such as concentration), students can interpret values (e.g., 100% activity) of external components (e.g., environmental glucose) semi-quantitatively (23). For example, environmental glucose set to 80-100% can be interpreted as "high amounts of environmental glucose" and 0-10% as "low amounts of environmental glucose."
When students simulate the computational model, they observe internal components (e.g., lac operon) as lines on a graph with the x and y axes representing time steps and activity level (ranging from 0 - 100), respectively. The activity level is determined by the regulatory mechanisms of other directly interacting components and also provides a semi-quantitative measure as a probability of a component being active at a given time point. This measure is used to describe the relative activity of a particular model component in response to environmental signals and/or perturbations in the model (23,33,34). Simulation settings have been optimized to provide clear visuals of the system. During this exercise, group discussions should center around simulation interpretation, and well as revisiting discussions from exercise 4 as necessary, such as relating their computational model to real biological interactions.
The time required for students to validate and revise their models varies greatly per group and depends on 1) the amount of revisions required to reach the correct model and 2) student comfort level with Cell Collective Learn. We suggest approximately 10 minutes for students to validate and revise their model.
Exercise 6. Investigate the impact of changes to the lac operon
After confirming the validity of their computational model, students will further explore the lac operon system through simulation activities called Investigations on pages 8-12 of the activity packet. During these Investigations students setup simulations to monitor the outputs and regulatory elements of the lac operon computational model. Because the computational model of the lac operon is based on probabilistic logical modeling (further details on the mathematics can be found in 23, 33, 34 and 35), students should focus on whether and how the activity of a given component is affected positively or negatively (as opposed to specific values) in response to mechanistic changes in the modeled system (this is similar to the interpretation of the output of many biological experiments, including Western Blots, differential gene expression analysis, etc.). Instructors should allocate 30 minutes for students to complete the Investigations.
In Investigations 1 and 2, students explore the mechanism behind lac operon activity in response to the availability of nutrients (i.e., lactose and glucose). These Investigations expand upon the simulations performed during the model validation exercise; now students are responsible for monitoring not only lac operon activity but the activity of the regulatory components within the system. In Investigation 1 students explore how the presence of only lactose in the environment impacts the lac operon system. In Investigation 2, students examine how the presence of lactose and glucose in the environment impacts lac operon system. In Investigations 3 and 4, students will explore the impact of mutations on lac operon regulation. Our goal for these two Investigations is to have students recognize how DNA mutations influence gene regulation by influencing protein function. In Investigation 3 and 4, students explore how a loss-of-function mutation in the CAP binding site and LacZ (gene that codes for an enzyme that breaks down lactose) affects lac operon regulation and expression.
During each Investigation, students engage in the process of science with the following steps: A) making predictions, B) developing a mechanistic explanation to support the prediction, C) testing the prediction using simulations, D) recording the simulation results, E) evaluating the prediction based on simulation results and retesting as necessary, and F) providing an accurate mechanistic explanation supported by simulation results. For example, in Investigation 1 Step A (page 8), students are asked to predict how the presence of environmental lactose in the absence of environmental glucose will affect the activity of the lac operon. In Step B, students support their prediction with a mechanism based on the provided computational model. In Step C, students test their prediction through simulation. In Step D, students report their simulation results. In Step E, students use the simulation results to evaluate the correctness of their prediction. If their simulation results do not align with their prediction, students are prompted to repeat Steps A-E until both their prediction and their results match. Finally, in Step F, students attempt to describe the mechanism responsible for the biological phenomena represented by the simulation results. This organization for the Investigations has previously been shown to be beneficial for student learning of complex biological systems (14,22,24,25).
We designed Steps A - F based on the recommended best practices of using simulations to facilitate science learning (28,36,37) specifically to have students
- explicitly make their conceptions as predictions (Step A) (37),
- provide reasons for their predictions (Step B) (37),
- test their prediction by setting up computer simulations (Step C)
- confront alternative conceptions with simulation results (Step D) (28,36),
- evaluate the consistency of their predictions with their results (Step E) (37), and
- use evidence (from simulation results) to support findings (Step F).
Ways to Facilitate the Discovering Prokaryotic Gene Regulation by Building and Investigating a Computational Model of the lac Operon Activity Packet
While instructor guidance is not required for this lesson, we encourage you to be engaged with your students and be available as they navigate this lesson. When building and simulating their model, encourage students to thoroughly read the provided instructions. All steps needed to build and investigate the model are included, along with additional tips on navigating the Cell Collective Learn platform. You can verify that students are applying mechanistic explanations by reading over their answers to Steps B and F of each Investigation. To do so, look for descriptions that 1) include two or more components, 2) describe the interactions between the components, and 3) provide an explanation of how the phenomena described in the prompt occurs. Be aware that students favor why answers instead of how answers (38). To mediate this kind of thinking, we suggest first checking in with one student within a group and then encouraging group discussion. This strategy provides some feedback to all students in the group, even if indirectly, and promotes peer instruction.
Common areas of difficulty and solutions
Many times, students skip the instructions and become confused or frustrated when the software does not respond as they expected. This behavior is consistent for both the model building and investigation activity. If you suspect frustration from a student, have him or her go back to carefully read and apply the instructions. Retracing steps commonly provides clarity and productive engagement with the software. Additionally, encourage students to ask their peers for help first. If one student skipped an important step in the instruction, their group member might recognize that and guide them to the appropriate action.
During the model building portion of the lesson, the most frequent student issue is setting the appropriate dominance on the lac operon. Dominance refers to the hierarchy of signals regulating a component's behavior (e.g., lac operon). When both the positive regulator (e.g., CAP) and negative regulator (e.g., lac repressor) for the lac operon are active at the same time, you need to decide and tell the model which (activating or inhibiting) signal will determine lac operon activity (i.e., which regulator is dominant). In the instructions, students are shown how to evaluate and set the dominance, but oftentimes students will forget to do this step and become confused when their model does not simulate correctly. This is likely the problem if a student approaches you claiming that they are sure their model is built correctly but is not simulating properly.
Students may haphazardly make changes to the simulation settings which may lead to incorrect simulation results. While students are welcome to explore the model and the modeling platform, correctly working through the lesson requires that students run their simulation with the parameters outlined in the instructions. Running the simulation under different parameters, especially when students are not aware of how those parameters influence the simulation output, may prevent students from confronting their alternative conceptions with correct simulation results. Therefore, it is essential for you to mediate this activity by verifying that students are adjusting the simulation settings correctly. To minimize the need for your intervention, remind students to regularly compare and discuss their simulation results to those of their group mates. Encouraging students to explain to each other why their settings gave observed results builds argumentation skills. This type of discussion will help students detect and resolve any settings errors. Additionally, you can prompt students to focus on simpler predictions about model behavior, such as "if lactose is present, should the lac repressor be on or off?" If these simpler predictions do not match their simulation results, guide them to check the simulation settings.
TEACHING DISCUSSION
Lesson implementation
We have implemented this lesson five times between 2015-2017 in an introductory biology course and are continuing to do so once per semester (Fall, Spring, and Summer). The lesson was provided to introductory biology students during a discussion period (i.e. dry laboratory) that lasted up to three hours. These discussion periods serve to reinforce student knowledge of relevant material in an interactive manner. Students completed the lesson in groups of three to four. All data collected were from consenting students with institutional review board approval.
Student Reaction to the Lesson
During some of the implementations, students who completed the lesson were given an optional survey. Students reported their experience by rating their level of agreement to statements about the lesson using a 1-5 Likert scale.
Introductory biology students from the Spring 2016 implementation (n=124) stated that the lesson helped them understand the biology topic (i.e. the lac operon) (Figure 2A) and helped them learn something new about the lac operon system (Figure 2B).
Figure 2: Student Response To The Lesson. Student responses from the introductory biology Spring 2016 implementation on a survey. Students were asked to A) rate whether the computer model-based activities helped them understand the biology topic. B) rate whether they learned something new about the biology topic through the model-based activities. The y-axis represents the number of student responses. The percentages on the columns represent the percentage of students that selected each response.
Two TAs were interviewed after implementing the lesson in their class. Both TAs stated that they liked the lesson because it fostered student discussion and prompted students to use problem-solving skills. They found the model-building exercise beneficial for student learning, while also noting that this exercise was the most challenging for students. This observation is not surprising considering model building requires the most engagement with the modeling software, which can intimate non-tech savvy students. The TA's also reported that the interactive computer simulations fostered a deeper understanding of the system that is difficult to accomplish from static representations in textbook diagrams and lectures. TA's also observed students occasionally skipping instructions or exercises in the lesson and getting confused later on. Representative comments are presented below.
- "[T]hat one [lesson] taught them how to solve problems in creative ways" (TA 1).
- "[S]tudents had to understand the models at a more fundamental level and keep track of those things in mind when they assembled those elements together and work around the software, which may sometimes be not easily understandable by students as well. [...] I think, despite the second one [model building exercise] being troublesome, students probably learned a bit more" (TA 1).
- "[T]his is a very structurally sound recitation. It starts from very basic all the way to very end. But if anyone were impatient and they jumped straight into the parts where you get to toggle stuff and it looks fun, you will not be able to solve the problems that you're on because you did not have the fundamental basics there" (TA 1).
- "I love these simulations, I think that they allow you to see the pathways in ways that you don't when they're on paper" (TA 2).
- "I think being able sit and play with the levels and manipulate it [the system] really gives you an insight into it that you don't have in a traditional lecture when you're staring at an image of it" (TA 2).
Possible Adaptations
Our lesson is highly flexible and can be adapted to fit a variety of different classroom needs. Students can complete the Discovering Prokaryotic Gene Regulation by Building and Investigating a Computational Model of the lac Operon activity packet individually, and outside of class should timing not allow for an in-class implementation. Alternatively, the background reading and reading comprehension questions could be implemented as homework in order to prime students for the other parts of the lesson. Instructors may wish to split the activity packet into two separate assignments: model building/validation (pages 1-7) and Investigations (pages 8-12). Splitting the lesson this way prevents students from using Figure 8 in the activity packet, which is a pathway diagram of the lac operon system, to replicate this model in Cell Collective Learn during the building exercise. While students with access to this figure did not appear to A) realize this resource existed until after building and validating their model and B) have enough motivation to change their models after-the-fact based on this image, instructors that are grading students on the correctness of their model may wish to separate these two portions of the lesson.
If timing is limited in your class, we have a simulate-only version of this lesson called Regulation of the Lac Operon. This version of the lesson provides students with a pre-constructed model of the lac operon so that students are only responsible for completing the lac operon (simulation) Investigations. The model and corresponding lesson materials are provided publicly on Cell Collective Learn (https://learn.cellcollective.org).
Because providing mechanistic explanations is novel for many students, we suggest having students practice creating a simple mechanistic explanation with feedback from you and their peers before attempting the lac operon Investigations. This type of feedback is essential for supporting student learning (39) and can provide instructors with valuable information on student understanding of mechanisms and on writing mechanistic explanations.
Some instructors and students may prefer teaching this material along with other concepts. For example this lesson could be linked to readings and/or lectures on gene regulation. Additionally, you could couple this lesson with discussions about cellular respiration in order to connect how bacterial cells use the glucose generated from lactose breakdown.
For advanced courses, instructors can challenge students to consider the limitations of the lac operon model and to provide possible ways to improve this model to better represent the biology. For example, students could suggest expanding the model by adding beta-galactosidase, the enzyme responsible for lactose breakdown, or RNA polymerase to the lac operon model. This type of thinking requires strong modeling skills and mastery of the biology. While a potentially valuable experience, we caution instructors teaching introductory courses that students at this level may not have the skills needed to benefit from this type of task. Furthermore, we are currently developing a new lesson designed specifically for senior-level biochemistry students, which will be available in the near future on Cell Collective Learn. Finally, you could add a summative post-assessment after the lesson to gauge what information students retained from the lesson. This post-assessment could focus on content related to the lac operon, gene regulation, and/or mechanistic descriptions. For example, the post-assessment can ask about lac operon expression if the lac repressor gene has a gain- or loss-of-function mutation.
Conclusions
The Discovering Prokaryotic Gene Regulation by Building and Investigating a Computational Model of the lac Operon lesson introduces students to basic concepts relating to gene regulation and the lac operon using a free, interactive, web-based modeling platform, Cell Collective Learn (https://learn.cellcollective.org). The model building and model simulation activities encourages student learning in an interactive manner while also exposing students to computational practices in biological system investigations. This approach is well received in the classroom, with both students and teaching assistants claiming that the lesson was beneficial to student learning. The lesson can be modified to fit a range of classroom needs including varying time constraints, student familiarity with computational modeling and specifically with Cell Collective Learn, and student familiarity with the lac operon.
SUPPORTING MATERIALS
-
S1. Discovering Prokaryotic Gene Regulation by Building and Investigating a Computational Model of the lac Operon - Activity packet
ACKNOWLEDGMENTS
We thank Dr. Steven Harris, James Buescher, and the teaching assistants of the LIFE 120 course for their assistance in implementation and their feedback on the lesson. We thank Taylor Uhlir for processing the student response data. We thank Erica Pribil for assisting with the student response analysis. This work was supported by the National Science Foundation under Grant No. IUSE #1432001 to T. Helikar and J. Dauer.
References
- American Association for the Advancement of Science. 2011. Vision and change in undergraduate biology education: a call to action. Washington, D.C.
- Kate Wright L, Nick Fisk J, Newman DL. 2014. DNA->RNA: What do students think the arrow means? CBE Life Sci Educ 13:338-348.
- Smith MK, Knight JK. 2012. Using the Genetics Concept Assessment to document persistent conceptual difficulties in undergraduate genetics courses. Genetics 191:21-32.
- Smith MK, Wood WB, Knight JK. 2008. The genetics concept assessment: A new concept inventory for gauging student understanding of genetics. CBE Life Sci Educ 7:422-430.
- Klymkowsky MW, Underwood SM, Garvin-Doxas K. 2010. Biological Concepts Instrument (BCI): A diagnostic tool for revealing student thinking. arXiv:1012.4501
- Banet E, Ayuso E. 2000. Teaching genetics at secondary school: A strategy for teaching about the location of inheritance information. Sci Educ 84:313.
- Dauer JT, Momsen JL, Speth EB, Makohon-Moore SC, Long TM. 2013. Analyzing change in students' gene-to-evolution models in college-level introductory biology. J Res Sci Teach 50:639-659.
- Dauer JT, Long TM. 2015. Long-term conceptual retrieval by college biology majors following model-based instruction. J Res Sci Teach 52:1188-1206.
- Reinagel A, Speth EB. 2016. Beyond the central dogma: Model-based learning of how genes determine phenotypes. CBE Life Sci Educ 15.
- Cooper RA. 2015. Teaching the Big Ideas of Biology with Operon Models. Am Biol Teach 77:30-39.
- Santillan M, Mackey MC. 2001. Dynamic regulation of the tryptophan operon: a modeling study and comparison with experimental data. Proc Natl Acad Sci U S A 98:1364-1369.
- Stefanski KM, Gardner GE, Seipelt-Thiemann RL. 2016. Development of a Lac operon concept inventory (LOCI). CBE Life Sci Educ 15: ar24.
- Smetana LK, Bell RL. 2012. Computer Simulations to Support Science Instruction and Learning: A critical review of the literature. Int J Sci Educ. 34:1337-1370.
- Appleby L, Bergan-Roller HE, Galt NJ, Pribil E, Chizinski C, Dauer JT, Helikar T. In preparation. A computational model-based lesson improves students' systems thinking in the context of cellular respiration. JMBE
- Bergan-Roller HE, Galt NJ, Pribil E, Chizinski C, Helikar T, Dauer JT. 2018. Using concept maps to characterise cellular respiration knowledge in undergraduate students. J Biol Educ. DOI:10.1080/00219266.2018.1541001
- Bell RL, Smetana LK. 2008. Chapter 3. Using Computer Simulations to Enhance Science Teaching and Learning Technology in the secondary science classroom.
- Rutten N, Van Joolingen WR, Van Der Veen JT. 2012. The learning effects of computer simulations in science education. Comput Educ 58:136-153.
- Kulik CLC, Kulik JA. 1986. Effectiveness of computer-based education in colleges. AEDS J 19:81-108.
- Stefanski KM. 2015. The Use Of a Hands-On Model In Learning The Regulation Of An Inducible Operon And The Development Of a Gene Regulation Concept Inventory. Master of Science. Middle Tennessee State University. Tennessee, USA.
- Stevens R, Johnson DF, Sollerz A. 2005. Probabilities and Predictions: Modeling the Development of Scientific Problem-Solving Skills. Cell Biol Educ 4: 42-57.
- Raineri D. 2001. Virtual laboratories enhance traditional undergraduate biology laboratories. Biochem Mol Biol Educ 29: 160-162
- Bergan-Roller HE, Galt NJ, Chizinski CJ, Helikar T, Dauer JT. 2018. Simulated Computational Model Lesson Improves Foundational Systems Thinking Skills and Conceptual Knowledge in Biology Students, BioScience (68) 8:612–621, https://doi.org/10.1093/biosci/biy054
- Helikar T, Cutucache CE, Dahlquist LM, Herek TA, Larson JJ, Rogers JA. 2015. Integrating Interactive Computational Modeling in Biology Curricula. PLoS Comput Biol 11.
- Bergan-Roller HE, Galt NJ, Dauer JT, Helikar T. 2017. Discovering Cellular Respiration with Computational Modeling and Simulations. Course Source. https://doi.org/10.24918/cs.2017.10
- Crowther A, Bergan-Roller HE, Galt NJ, Dauer JT, Helikar T. 2018. Discovering Prokaryotic Gene Regulation with Simulations of the trp Operon. Course Source.https://doi.org/10.24918/cs.2018.9
- Cutucache CE, Helikar T. 2014. Cancer biology: an inquiry-based approach. Kendall Hunt. Place of publication not identified.
- Gilbert JK. 2004. Models and modelling: Routes to more authentic science education, p. 115-130. In International Journal of Science and Mathematics Education.
- Louca LT, Zacharia ZC. 2012. Modeling-based learning in science education: Cognitive, metacognitive, social, material and epistemological contributions. Educ Rev 64:471-492.
- Eddy S, Hogan K. 2014. Getting Under the Hood: How and for Whom Does Increasing Course Structure Work? CBE Life Sci Educ. 13: 453-468.
- Khodor J, Halme DG, Walker GC. 2004. A Hierarchical Biology Concept Framework: A Tool for Course Design. Cell Biol Educ 3:111-121.
- Brodland GW. 2015. How computational models can help unlock biological systems. Semin Cell Dev Biol. 47-48:62-73
- Loman N, Watson M. 2013. So you want to be a computational biologist? Nat Biotechnol 31:996-998.
- Helikar T, Konvalina J, Heidel J, Rogers JA. 2008. Emergent decision-making in biological signal transduction networks. Proc Natl Acad Sci 105:705088105.
- Helikar T, Kowal B, McClenathan S, Bruckner M, Rowley T, Madrahimov A, Wicks B, Shrestha M, Limbu K, Rogers JA. 2012. The Cell Collective: toward an open and collaborative approach to systems biology. BMC Syst Biol. 6:96.
- Helikar T, Rogers JA. 2009. ChemChains: a platform for simulation and analysis of biochemical networks aimed to laboratory scientists. BMC Syst Biol 3:58.
- Duit R, Treagust DF. 2003. Conceptual change: A powerful framework for improving science teaching and learning. Int J Sci Educ 25:671-688.
- Liew C-W, Treagust DF. 1998. The Effectiveness of Predict-Observe-Explain Tasks in Diagnosing Students' Understanding of Science and in Identifying Their Levels of Achievement., p. 224-234. In Annual Meeting of the American Educational Research Association.
- Abrams E, Southerland S. 2001. The how's and why's of biological change: How learners neglect physical mechanisms in their search for meaning. Int J Sci Educ 23:1271-1281.
- Bell B, Cowie B. 2001. The characteristics of formative assessment in science education. Sci Educ 85:536-553.
Article Files
Login to access supporting documents
Discovering Prokaryotic Gene Regulation by Building and Investigating a Computational Model of the lac Operon(PDF | 766 KB)
S1. Discovering Prokaryotic Gene Regulation - Activity packet.pdf(PDF | 690 KB)
- License terms

Comments
Comments
0 Like
Mike Klymkowsky @ on
I read with interest your CourseSource paper and have a few comments.
First, the lac operon is not homeostatic (at least as I understand the term) and it is inducible rather than repressible (as the expression lacI, which encodes the lac repressor is constitutive), under particular conditions, specifically the absence of a more efficient energy source. This might be addressed by introducing some historical background on diauxie – a recent review by Blaiseau & Holmes (2021) might be useful.
Second, induction of the lac operon relies upon stochastic (noisy) processes that arise from the small number of repressor molecules per cell. This has been nicely modeled in the paper by Vilar et al (2003) that you might find useful; I used their model in presenting the lac operon in biofundamentals (chapter 9, pp. 192-195).
Difficulties in understanding such "random" (i.e. stochastic) processes are documented and discussed in Garvin-Doxas et al (2008) In this context, I have found the experiments by Elowitz et al (2002) particularly useful in introducing students into the concept of stochasticity (as well as the molecular mechanisms involved). More generally, I wonder whether referring to stochastic events as due to "leakiness" helps students understand the molecular level processes involved? A leaky faucet is different, mechanically from one that turns on and off sporadically (which is what the lac operon appears to do) - see Vilar et al (2003)
An appreciation of the ubiquity of stochastic processes at the cellular level is central for developing an understanding of differential gene expression revealed most dramatically by single cell RNA SEQ and related studies (Kærn et al., 2005; Nishimura et al., 2015). A recent paper by Braun (2021) makes this point in the context of determinism and free will (see also Rolls and Deco, 2010).
Literature cited:
Blaiseau, P. L. and Holmes, A. M. (2021). Diauxic inhibition: Jacques Monod's Ignored Work. Journal of the History of Biology 54, 175-196.
Braun, H. A. (2021). Stochasticity Versus Determinacy in Neurobiology: From Ion Channels to the Question of the “Free Will”. Frontiers in Systems Neuroscience 15, 39.
Elowitz, M. B., Levine, A. J., Siggia, E. D. and Swain, P. S. (2002). Stochastic gene expression in a single cell. Science 297, 1183-1186.
Garvin-Doxas, K. and Klymkowsky, M. W. (2008). Understanding Randomness and its impact on Student Learning: Lessons from the Biology Concept Inventory (BCI). Life Science Education 7, 227-233.
Kærn, M., Elston, T. C., Blake, W. J. and Collins, J. J. (2005). Stochasticity in gene expression: from theories to phenotypes. Nature Reviews Genetics 6, 451-464.
Nishimura, K., Tsuru, S., Suzuki, H. and Yomo, T. (2015). Stochasticity in gene expression in a cell-sized compartment. ACS Synthetic Biology 4, 566-576.
Rolls, E. T. and Deco, G. (2010). The noisy brain: stochastic dynamics as a principle of brain function: Oxford university press Oxford.
Vilar, J. M., Guet, C. C. and Leibler, S. (2003). Modeling network dynamics: the lac operon, a case study. J Cell Biol 161, 471-476
Copy link Report abuse