Using CRISPR-Cas9 to teach the fundamentals of molecular biology and experimental design
Published online:
Abstract
One of the challenges of inquiry-based research experiences is balancing the instruction on skills and techniques with data generation and analysis. This is especially true in technique heavy disciplines such as molecular biology, where emphasis is often placed on the acquisition of molecular biology skills over more hypotheses driven approaches. Recognizing that molecular technique plays a large role in the process of science though creation and validation of novel reagents, we sought to create a laboratory lesson that reflects how researchers use technique to plan, implement, and execute experiments. CRISPR-based gene edits are one such technique requiring a number of different molecular approaches. Using the CRISPR-Cas9 as a framework, we created a semester long inquiry-based lesson that not only familiarizes students with an emerging technology, but also teaches key molecular techniques and the concepts of deliberate and intentional experimental design. Through collaboration with the instructor, students first identify a gene target based on a specific question. Then students work through the process of creating both a homology directed repair template and a single-guide RNA necessary for this gene edit. Students are also tasked with designing and executing an experiment to test their single-guide RNAs in vitro as proof of principle. Through this process students gain experience with a variety of broadly applicable molecular techniques including PCR, sequence analysis, recombinant DNA technology, and RNA biology. Additionally, students gain an important understanding of the role inquiry plays in the conception and design of experiments.
Citation
Kee, H.L., Kushner, J.K., Deuchler, C.P., Becker, A.K., Clarke, D.G., and Pieczynski, J.N. 2019. Using CRISPR-Cas9 to teach the fundamentals of molecular biology and experimental design. CourseSource. https://doi.org/10.24918/cs.2019.21Society Learning Goals
Biochemistry and Molecular Biology
- Information storage and flow are dynamic and interactive
- What is a genome?
- How does the nucleotide sequence of the gene lead to biological function?
- How are genomes maintained?
- Discovery requires objective measurement, quantitative analysis and clear communication
- What is the scientific process?
- What skills are needed to access, comprehend and communicate science?
- What constitutes a scientific community of practice?
Lesson Learning Goals
Through their study of CRISPR-Cas9, students will learn the fundamental skills of molecular biology techniques involving uses and manipulations of DNA, RNA, and protein. Through learning these basic skills, students will also follow an experiment from conception to design to execution.Lesson Learning Objectives
Module 1- Generate a testable hypothesis that requires a creative design of reagents based on critical reading of and review of prior research.
- Demonstrate proficiency in using molecular cloning software to analyze, manipulate and verify DNA sequences.
- Predict the downstream effect on the mRNA and protein after successfully inserting a DNA repair template into the genome of a cell/organism.
- Compare and contrast the processes of DNA duplication and PCR.
- Demonstrate the ability to design primers to amplify a nucleotide sequence.
- Analyze and evaluate the results of DNA agarose gel electrophoresis.
- Identify the key features in genomic DNA, specifically those required for CRISPR-Cas9 mediated gene edits.
- Explain how compatible ends of DNA are used to produce recombinant DNA in a ligation reaction.
- Explain the chemical principles behind plasmid DNA purification from bacterial cultures.
- Devise a strategy to screen clones based on antibiotic selection and the mechanism of digestion by DNA endonucleases.
- Predict and evaluate the results of a diagnostic digest.
- Explain the chemical principles behind DNA purification using phenol-chloroform extraction and ethanol precipitation.
- Explain the key differences between DNA duplication and transcription.
- Demonstrate the ability to perform lab work with sterile technique.
- Compare and contrast the results of a non-denaturing vs. denaturing agarose gel.
- Evaluate the results of a denaturing agarose gel.
- Design and implement an experiment that tests the CRISPR-Cas9 principle.
- Predict the outcome of a successful in vitro Cas9 digest.
- Summarize important background information on gene of interest from analysis of primary literature.
- Produce figures and figure legends that clearly indicate results.
- Organize and construct a poster that clearly and professionally displays the important aspects of the lesson.
- Demonstrate understanding of the lesson by presenting a poster to an audience in lay terms, mid-level terms, or at an expert level.
- Demonstrate understanding of procedures by writing a formal materials and methods paper.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
One of the unique challenges to teaching a dedicated molecular biology course is the protocol- driven nature of the discipline. A considerable amount of evidence suggests that inquiry-based instruction, instead of verification, "cookbook" type laboratory experiences, are more likely to reach intended learning goals and outcomes (1). Studies overwhelmingly report increases in critical thinking skills, comprehension, and confidence when engaged in authentic, hands-on research in a classroom setting (2,3).
Over the last decade, emphasis has been placed on teaching science by engaging students in the process of doing science. Many studies have been dedicated to understanding how to improve student scientific process skills such as generating hypotheses or evaluating experimental data (2,4). One often overlooked, but demonstratively important, aspect of doing science in the classroom is rational experimental design, or the informed and deliberate preparation that leads to a well-controlled experiment (5). These pre-experimentation planning steps are often the sites where solid fundamentals are critical and where experiments can easily be derailed by confounding variables thus increasing chances of experimental failure (6). Being able to autonomously design and implement an experimental plan leads to increased student engagement and understanding of the laboratory process (7). Although no formal studies have been conducted, we have observed that a large amount of research time in fields utilizing molecular biology is dedicated to careful planning through deliberate reagent preparation in contrast to actually conducting data-generating experiments. Yet we often do not focus on these skills when teaching. As instructors tasked with introducing molecular techniques to students, it is therefore essential to provide a strong foundation in not only practice, but also theory and application of each molecular technique. Questions of why, how, and when to utilize different molecular approaches and what constitutes appropriate controls are invaluable to the design and execution of experiments and allow for the integration of inquiry into the laboratory.
CRISPR-Cas9 gene editing techniques utilize a wide array of molecular biology approaches and can provide a framework for inquiry-driven experiences for students in a molecular biology course. Importantly, CRIPSR systems are broadly applicable across a multitude of systems, all of which utilize the same basic molecular techniques for the generation of reagents. For a typical CRISPR-based gene edit, an RNA guide (sgRNA) will target an endonuclease (Cas enzyme, typically Cas9) to a specific genomic location. The Cas enzyme will produce a break for the cell's endogenous repair machinery to repair. Depending on the need of the researcher, this repair step can be manipulated to insert or change sequences, or even produce null alleles. For an in-depth discussion of CRISPR-Cas systems, see Supporting File 1 (Supporting File 1: Introduction to CRISPR-Cas9 Gene Editing and 8).
By working collaboratively with students to actively design and implement a CRISPR-Cas9 editing strategy, we can begin teaching the important aspects of experimental design and the foundational skills of molecular biology. Students will learn how experimental design skills are essential to generating data that is easily analyzed and limits the potential for bias. Producing the reagents required for a CRISPR-based gene edit requires careful experimental planning and utilizes multiple molecular techniques including basic bioinformatics, primer design/PCR, DNA purification, basic compatible end cloning, and in vitro transcription. To this end, we have developed a semester-long inquiry driven lesson designed to teach and apply core molecular biology skills. In this lesson, students design and produce CRISPR-Cas9 mediated site-specific genome editing reagents that can be tested experimentally both in vitro and in vivo. Our goals are to teach molecular theory and practice so that upon completion of this lesson, students will appreciate the steps involved in elegant experimental design, as well as have a better understanding of how research labs operate. Ideally, the skills and approaches taught here would allow a student to begin work in a research lab with the requisite basic molecular biology skill set.
INTENDED AUDIENCE
This lesson is for intermediate to upper level bioscience majors in a molecular biology or genetics course. We have used this lesson at a small liberal arts college, but it could be scaled for larger institutions or class sizes, albeit with slight modifications such as dedicated support staff or teaching assistants.
REQUIRED LEARNING TIME
This lesson is designed to be taught in 10-14 weeks depending on time available in lab. Laboratory sessions for this lesson will vary from two to five hours depending on the week. Background information on each week's procedures was given to students before each laboratory period in order to prepare. A sample lab schedule is provided in Table 1.
PREREQUISITE STUDENT KNOWLEDGE
Students should have a general understanding of central dogma of molecular biology and general chemistry. Students should also understand how to make solutions using both molarity and percent weight/volumes as well as conversions between metric units.
PREREQUISITE TEACHER KNOWLEDGE
Instructors familiar with common molecular biology/biochemistry techniques and applications should have the knowledge to complete all labs for the lesson. Knowledge of molecular biology software to import and analyze genomic DNA sequences, annotate DNA sequences, design primers/oligonucleotides, and align and analyze sequencing data is also needed. Many of these software programs are open-source and readily available online.
SCIENTIFIC TEACHING THEMES
ACTIVE LEARNING
Students are actively involved in every aspect of the lesson and perform every skill. Students work in collaborative groups of two to four to foster discussion, analyze data, answer discussion questions and to provide redundancy in the event that a group member's procedure provides too low yields or fails. To foster autonomy, we use a minimal laboratory set-up providing only standard solutions and equipment, forcing students to collaborate in lab preparation. This lesson should closely mimic a functional research laboratory rather than a teaching laboratory.
ASSESSMENT
Weekly, short pre-lab quizzes of three to four questions were given to assess student preparation (Supporting File S2: Sample Lab Quiz Questions/Answers). Quizzes contain key topics of emphasis for the week, vocabulary, and evaluation of methodology. Quizzes are followed by group discussion, and instantaneous feedback and remediation is given if needed.
To demonstrate an in depth understanding of all procedures, students write a formal materials and methods paper upon completion of the lesson. This assignment is written in the style of a publication quality materials and methods section and is to include all reagents and procedures utilized throughout the lesson (Supporting File S3: Materials, Methods and Figures Rubric). Papers are assessed on the student's ability to write a succinct, yet accurate methodology that includes all important aspects of the lesson.
To assess data presentation, students also create properly formatted figures including figure legends with this written materials and methods assignment. The figures with legends are assessed on clarity, format, and professional appearance.
We use this lesson to teach poster presentation skills. Student groups produce a poster that includes background on their gene of interest with the rationale for their gene edit, the figures produced above, and future directions. Every student is required to present their group's poster at one of three levels; expert, mid-level, or lay person. Students self-select their style within their group, and each group must present in all styles. Students present in their selected style and are assessed on their ability to accurately explain the lesson and answer questions in that style (Supporting File S4: Poster Presentation Rubric).
Students are to keep an accurate laboratory notebook. We then assess their learning using an open lab notebook lab exam/practical (Supporting File S5: Sample Laboratory Exam). This strategy allows us to assess the student's knowledge on the subject and their ability to maintain a proper notebook. The practical portion of lab exam includes questions on general laboratory skills that students should gain from participating fully in the lesson.
This lesson is based on close collaboration between the faculty member/course instructor and the students, where instructors take an active role in helping students evaluate and verify their results. Our assessment methods do not assess student's laboratory success and leave it to instructors on how to evaluate student effort and getting the "correct" answer. Rather our assessment method is based upon whether students learn the theory and application of a skill. The open-endedness of this lesson makes it possible that students might not initially be successful in all phases of the procedure. To increase chances of student success we recommend students work in groups to ensure that individual students have backup samples if needed. In the event that an entire group is unsuccessful in a skill, instructors should adequately prepare back up samples. Note that Module 1 can be run concurrently with Modules 2-4 if needed. Typically, an instructor with previous experience in molecular biology should be able to complete all procedures contained in this lesson within two weeks.
INCLUSIVE TEACHING
We emphasize that the skills and concepts can be applied regardless of future career goals. Given that CRISPR-Cas9 is a recently developed technology, we incorporate the history of the technique's development, especially the significant contributions of women and underrepresented racial minorities in the field.
LESSON PLAN
LESSON BACKGROUND
To incorporate the instructor's research and give students a guided, yet authentic research opportunity, the faculty and students work collaboratively to identify a gene of interest to edit. The first deployment of this lesson created truncation mutants for the C. elegans gene klp-4, but any model or gene target could be used as the instructor sees fit. Although we have included our specific reagents, we have made efforts to make this lesson general so that the lesson may be used in multiple contexts/modifications. A complete list of general materials and solutions required can be found in Supporting Files 6 and 7 (Supporting File S6: Required Lesson Equipment/Reagents and Supporting File S7: Common Solutions).
Each session of this laboratory should begin with a chalk-talk involving key points and processes involved with skill development. We prefer chalk-talks rather than prepared lectures given the varied background knowledge of students and the ability to adjust in "real time" based on student questions and interactions. Chalk-talks enable a focus on the role of each skill in the process of science and how each of these skills have be utilized to achieve research goals (Supporting File S8: Chalk Talk Points and Answers to Discussion Questions). Additionally, the end of each lab handout contains discussion and application questions to reinforce key ideas (Supporting File S8). A sample lesson timeline can be found in Table 1.
MODULE 1: PRINCIPLES OF PCR - HOMOLOGY DIRECTED REPAIR (HDR) TEMPLATE SYNTHESIS
Students perform background research on a gene of interest and consult with the instructor to choose a potential edit that could be made to that gene and design homology directed repair (HDR) knock-in cassettes (Supporting File S9: Guidelines for Consulting on Student Projects). Through this process, students will learn the basics of molecular cloning software, basic bioinformatics skills, and become familiar with the primary literature that they will need for their poster assessment.
Students use molecular cloning software to design primers to amplify the sequence for their HDR knock in cassette, paying attention to the addition of homology arms and maintenance of reading frame upon a successful insertion (Supporting File S10: Lab 1 - PCR and Primer Design and 7). Points of emphasis are the 5'-3' orientation of DNA, the assumption of the opposite strand sequence, and the thermodynamic properties of melting and annealing of complementary DNAs.
After performing their PCR reactions, small aliquots of each reaction are run on agarose gels for analysis (Supporting File S11: Lab 2 - Setting up PCR Reactions, Supporting File S12: Lab 3 - DNA Agarose Gel Electrophoresis and Figure 1A). Students should be able to predict the size of a product and then analyze their data and identify potential positive reactions. Potential positive samples are then purified and sequenced (Supporting File S13: Lab 4 - PCR Purification and Sequence Prep). Using molecular cloning software, students analyze their sequencing data to determine if their PCR reactions were successful (Supporting File S14: Lab 5 - Sequence Analysis, Supporting File S15: KIF17B-1 - .ab1 file, Supporting File S16: KIF17B-2.ape file, Supporting File S17: Unknown.ab1 file, and Figure 1B). Sequence-verified HDR templates are then stored for further downstream use if desired.
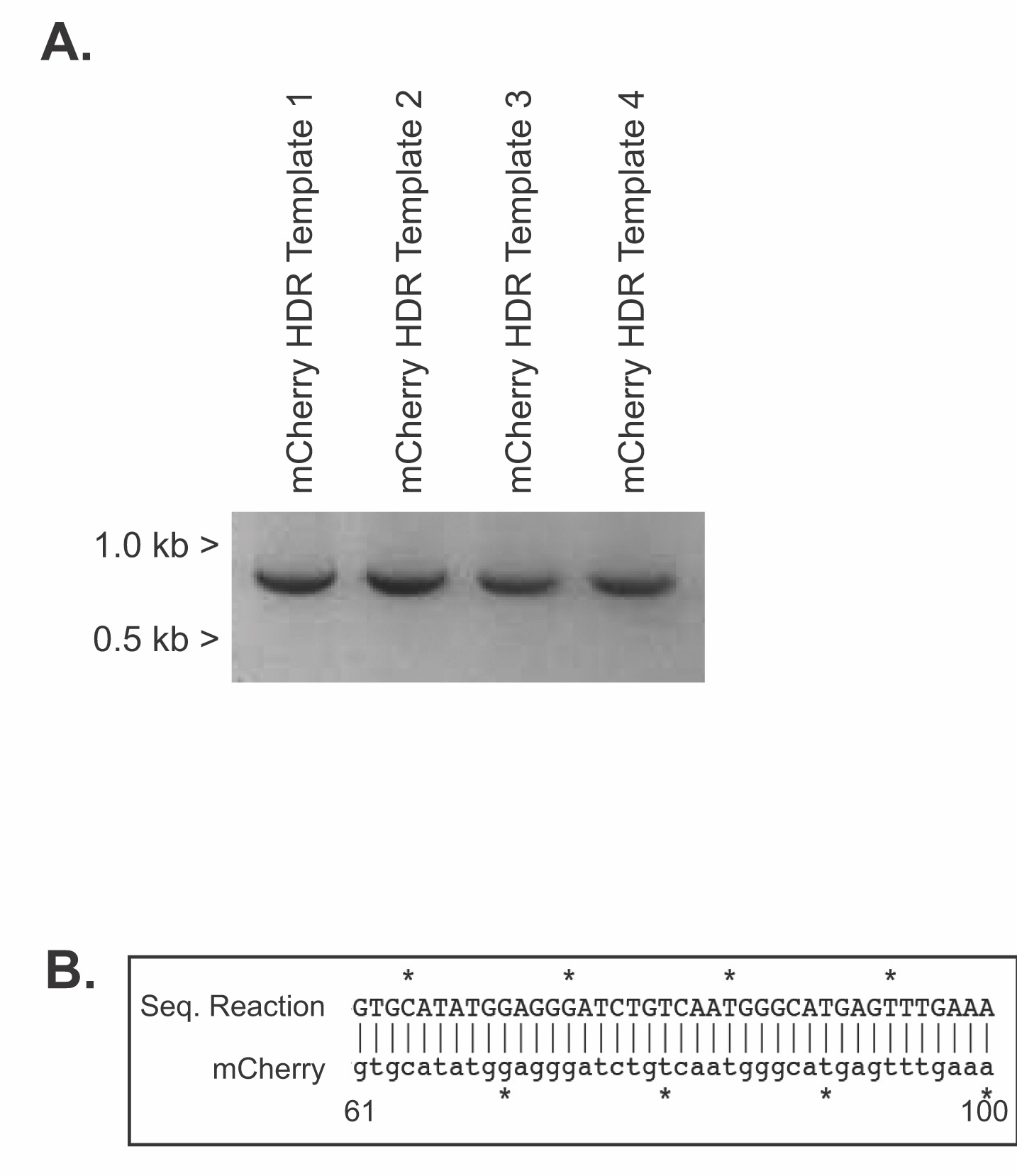
Figure 1. The amplification of mCherry homology directed repair (HDR) templates. (A) Representative agarose gel of PCR reaction products for HDR templates 1-4. Expected size of mCherry PCR products with arms of homology is 935 base pairs. (B) Student generated alignment of sequencing results demonstrating successful amplification of mCherry for HDR Template-3. Nucleotides 61-100 of mCherry are shown for brevity. Sequence alignments were generated using ApE software.
MODULE 2: PRODUCING RECOMBINANT DNA - CONSTRUCTING SINGLE-GUIDE RNA PLASMIDS
Single-guide RNAs (sgRNAs) for site-specific targeting of Cas9 are designed using the annotated genomic sequence of a target gene (Supporting File S18: Lab 6 - Designing crRNA Oligos). Recall that a complete sgRNA contains two linked components; a specific CRISPR RNA (crRNA) that targets a specific DNA sequence and a universal transactivating CRISPR RNA (tracrRNA) that links the crRNA to the Cas9 enzyme. Hwang et al. have designed the plasmid DR274 (Addgene Plasmid #42250, MTA required) that contains an insertion site for crRNAs followed by the tracrRNA sequence. To use this plasmid, users only need to design and anneal complementary oligonucleotides corresponding to a desired crRNA sequence. Importantly, DR274 was designed for directional (one-way) insertion of annealed crRNA oligonucleotides. DR274 contains two BsaI restriction enzyme sites separated by a spacer sequence. The BsaI enzyme recognition sequence is 5' GGTCTC 3', however the enzyme digests one non-specific nucleotide after the 3' end of the GGTCTC recognition sequence (5'GGTCTCN3') and five non-specific nucleotides towards the 5' end on the bottom strand (3'CCAGAGNNNNN5'). The flanking sequences on the crRNA insertion site of DR274 are intentionally different to facilitate directional insertion of annealed crRNA oligonucleotides immediately upstream of the tracrRNA sequence to produce sgRNAs via in vitro transcription (10). Standard ligation reactions use BsaI digested, dephosphorylated DR274 and annealed oligonucleotides (Supporting File S19: Lab 7 - Preparation of sgRNA Plasmids). Ligation reactions are transformed into competent bacteria and plated onto LB-Kanamycin plates for overnight incubation at 37°C.
Students will pick bacterial clones for overnight liquid cultures and receive detailed instruction on the purpose of each solution used in DNA isolation (Supporting File S20: Lab 8 - Minipreps and Diagnostic Digest and 5). Once DNA is successfully isolated, plasmids are sequenced and analyzed for more practice on bioinformatic skills. Plasmids containing crRNA cassettes are diagnostically digested using NheI and HindIII. The digestion produces fragments of 2018 and 143-nucleotides which require a high percentage agarose gel. Success on this diagnostic digest varies, but 50% of the time students are able to produce a band of the correct size. Uncut plasmid can also be run next to digested plasmid to demonstrate supercoiled versus linearized DNA (Figure 2A). Students analyze their gels to determine potential positive clones. Even if diagnostic digests are unsuccessful, students routinely receive positive results from the sequence analysis above (Figure 2B).
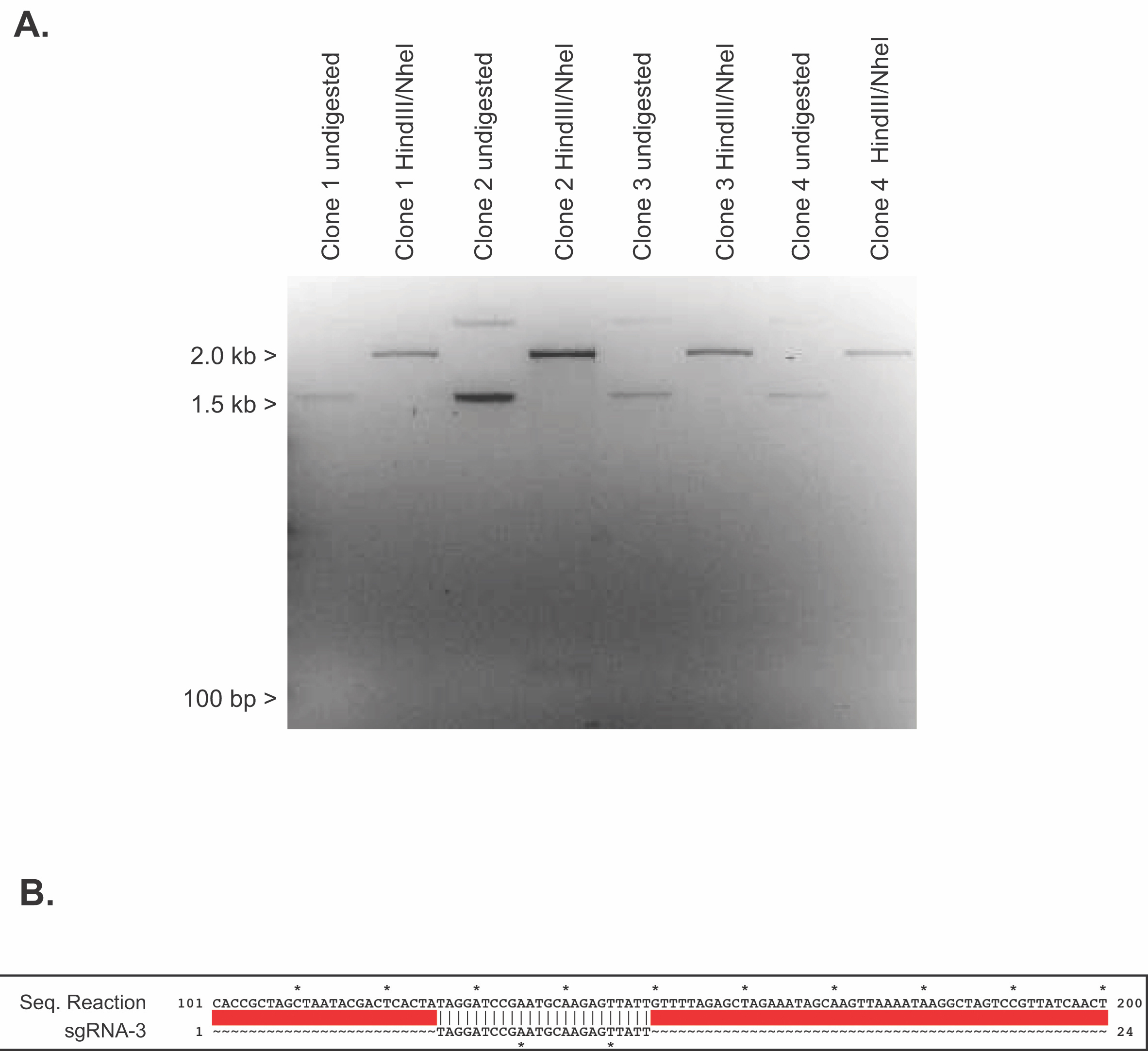
Figure 2. Verification of sgRNA plasmid cloning. (A) Student generated representative agarose gel using HindIII/NheI to perform a diagnostic digest of four individual clones of DR274 + crRNA-3 against undigested, supercoiled sample of the same clone. Expected size of doubly digested positive clones: 2152 base pairs and 134 base pairs (B) Alignment of sequencing results between Clone 2 of DR274 + crRNA-3 and the designed crRNA-3 sequence. Sequence alignments were generated using ApE software.
MODULE 3: WORKING WITH RNA - SGRNA PRODUCTION
Run-off in vitro transcription is used to produce sgRNAs (Supporting File S21: Lab 9 - In Vitro Transcription). Sequence verified sgRNA plasmids are first linearized with DraI, followed by phenol-chloroform extraction and ethanol precipitation. Students should take care to prevent RNAse contamination, since DNAs will be used in downstream RNA applications. Typical student yields from this purification procedure range from 10 to 200 ng/μl of linearized template DNA. sgRNAs are then produced by in vitro transcription. As little as 10ng total of linearized template DNA can be used for the in vitro transcription reaction. Samples should be DNase-treated to remove the DNA template after in vitro transcription. In vitro produced sgRNAs are purified and subject to denaturing agarose gel electrophoresis (Supporting File S22: Lab 10 - Purification of sgRNAs). Instead of using traditional denaturing gel techniques, Arnanda and colleagues have developed a simple an effective alternative that use standard agarose gels that include of up to 5% standard household bleach to denature RNAs (7). A sample of 200-250ng of total RNA produces bands of the correct size (~133 nucleotides) and running more than that amount of RNA on the gel leads to RNA secondary structure formation, poor resolution of bands and incorrect size on the gel (Figure 3).
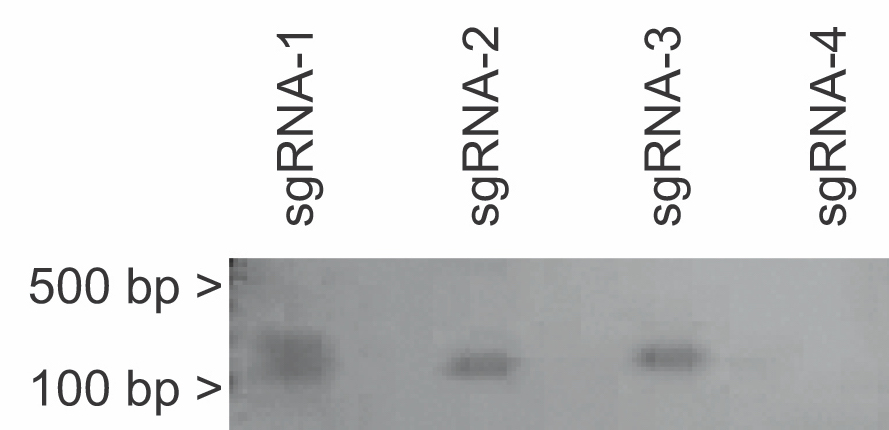
Figure 3. Denaturing bleach gel of in vitro transcribed sgRNAs 1-4. Expected size of product is 133 bases. Note that sgRNA-4 in vitro transcription reaction produced a low yield.
MODULE 4: EXPERIMENTAL DESIGN WITH AN IN VITRO CAS9 CLEAVAGE ASSAY
To demonstrate the principle of CRISPR-Cas9 and the process of experimental design, we use an in vitro method (Supporting File S23: Lab 11 - In vitro Cas9 Assay and 8, also see https://www.neb-online.de/wp-content/uploads/2015/06/LocusModificationDetectionCas9_Protocol_0515.pdf for basis of assay ). We have built our own synthetic target construct by designing a custom dsDNA containing each group's 20-nucleotide target followed by a PAM sequence. The custom dsDNA block is ligated into the multiple cloning site of pET28a(+). The pET vector based synthetic target is first linearized with EcoRV to provide linear substrate for our assay. Using this synthetic target substrate, a successful cleavage by the sgRNA-Cas9 duplex will result in products of ~4 kb and 1.5 kb (Supporting File S24: Lesson Verified Reagents and Methods and Figure 4). Presumably, any double stranded DNA (plasmid, cDNA clone, PCR product, etc.) containing the specific 20-nucletide target sequence followed by the PAM sequence could be used as a substrate for this assay. Using the synthetic target has the advantage of only requiring one common substrate for all student groups instead of producing or obtaining a number of different targets. We recommend using linearized template for this assay to make the interpretation of the results easier for students. Students need to maintain a nuclease free environment during the assay. If time permits, once students have demonstrated successful design of their reagents, the option exists to use these reagents in vivo or ex vivo.

Figure 4. Cleavage of site-specific target in an in vitro Cas9 assay. Representative student data of the in vitro Cas9 assay for sgRNAs 1-4 against a synthetic target substrate. No sgRNA and no sgRNA/no Cas9 were used as controls. Successful cleavage produces products of ~4000 base pairs and 1500 base pairs.
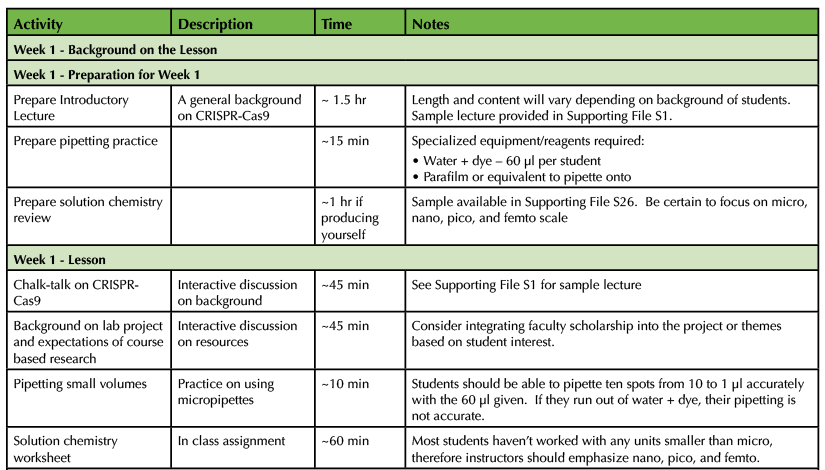
Table 1. CRISPR-Cas9 Teaching Timeline - Week 1
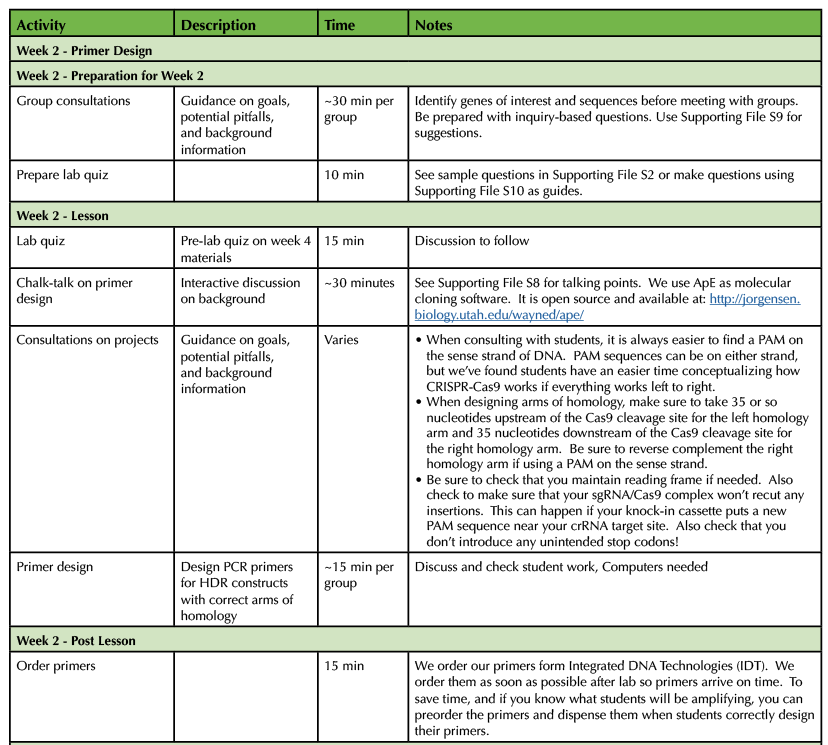
Table 1. CRISPR-Cas9 Teaching Timeline - Week 2

Table 1. CRISPR-Cas9 Teaching Timeline - Week 3
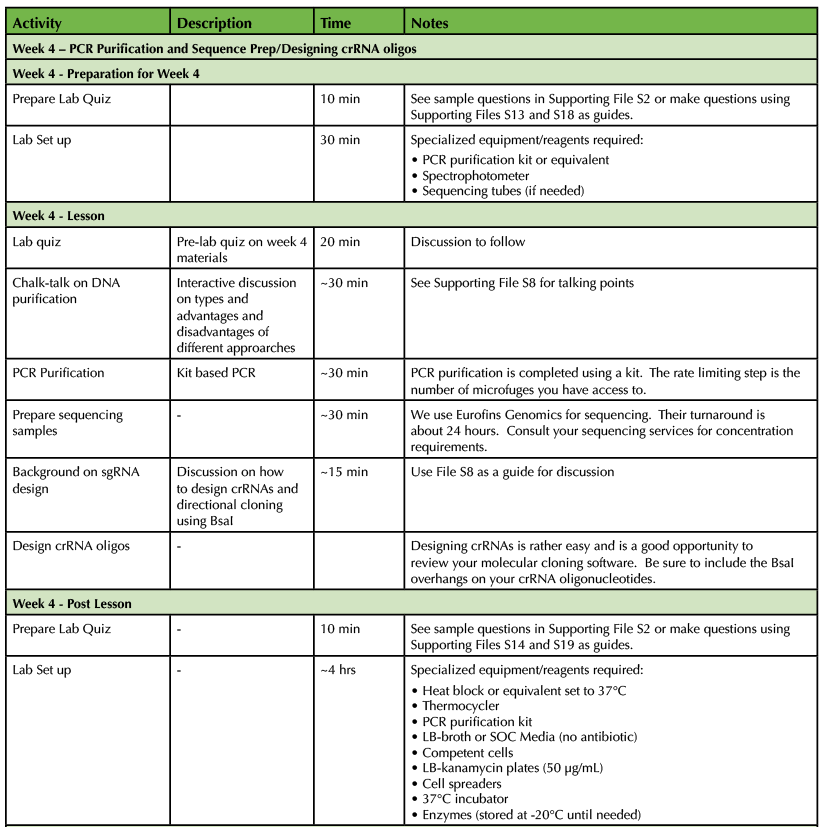
Table 1. CRISPR-Cas9 Teaching Timeline - Week 4
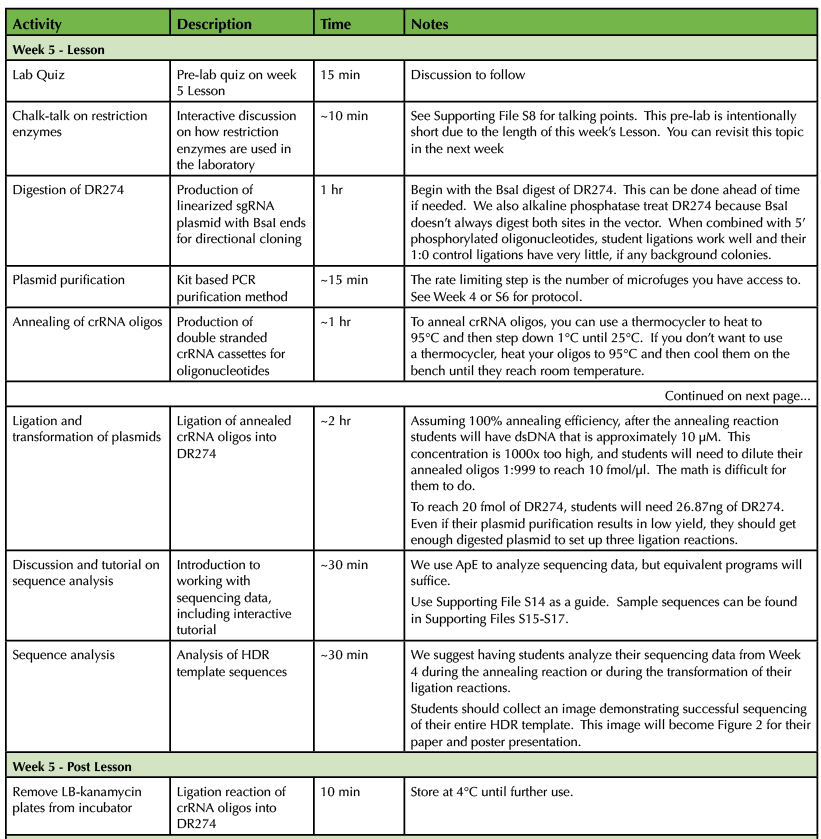
Table 1. CRISPR-Cas9 Teaching Timeline - Week 5
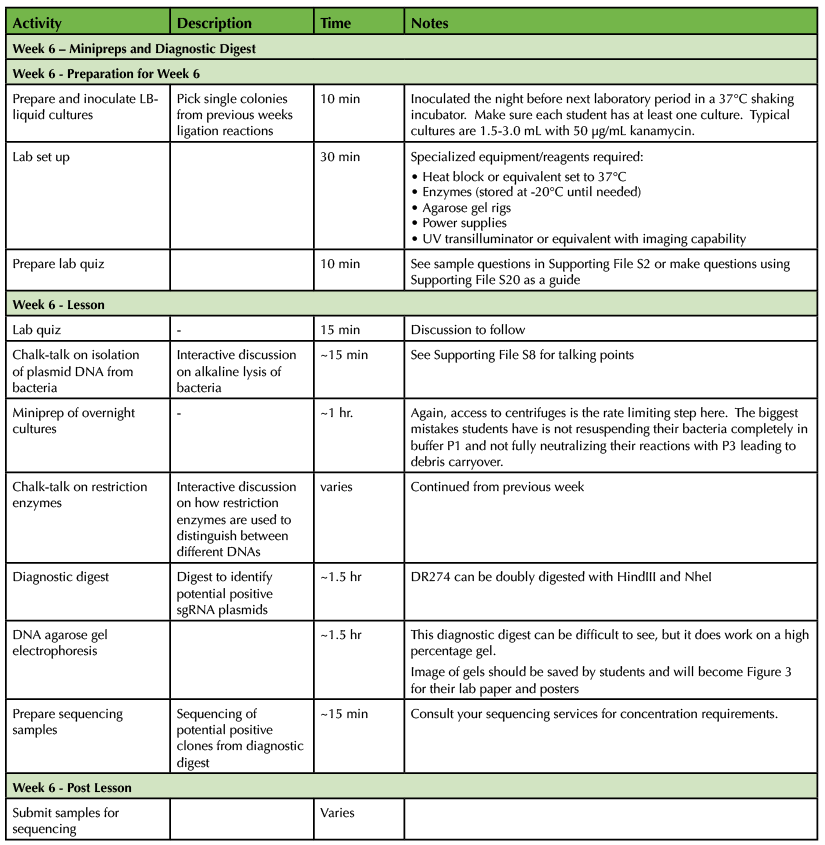
Table 1. CRISPR-Cas9 Teaching Timeline - Week 6
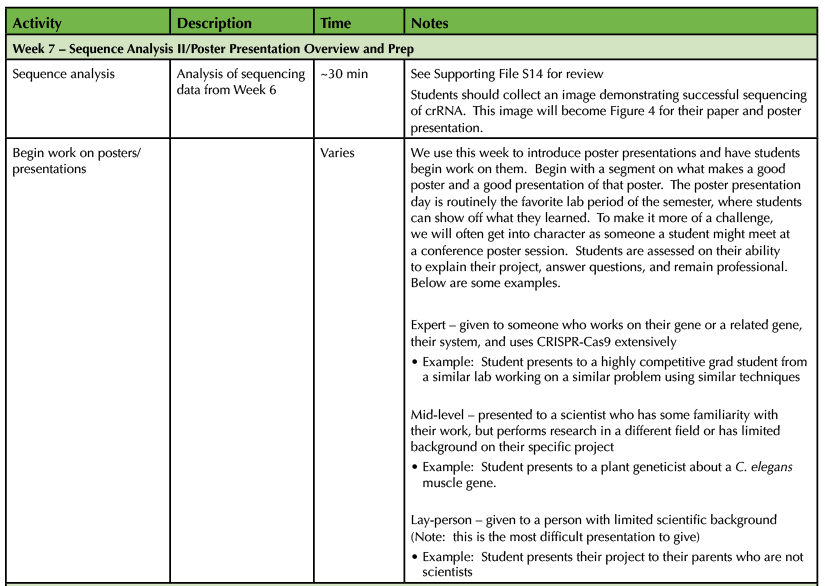
Table 1. CRISPR-Cas9 Teaching Timeline - Week 7
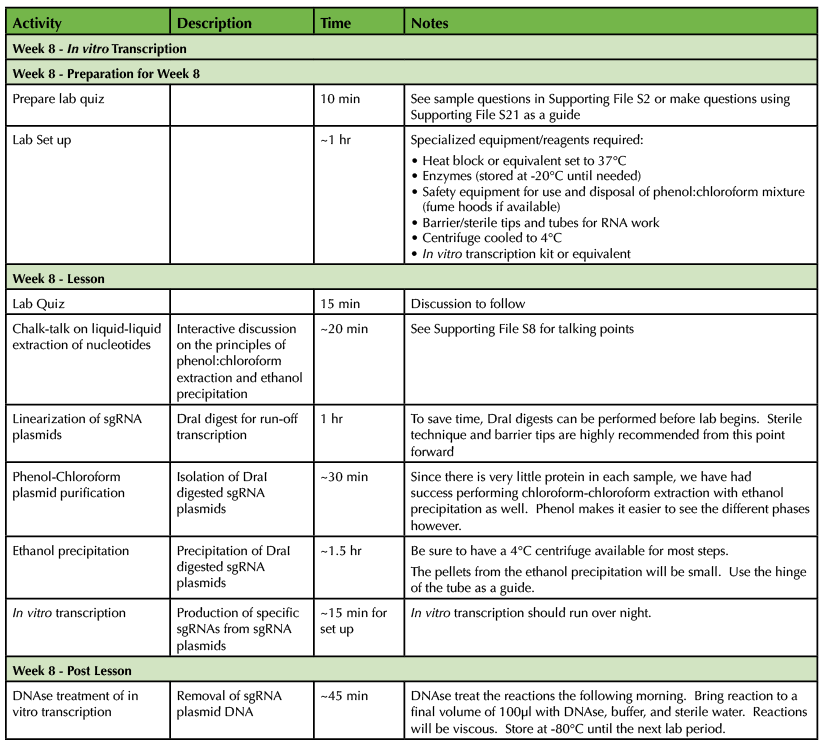
Table 1. CRISPR-Cas9 Teaching Timeline - Week 8
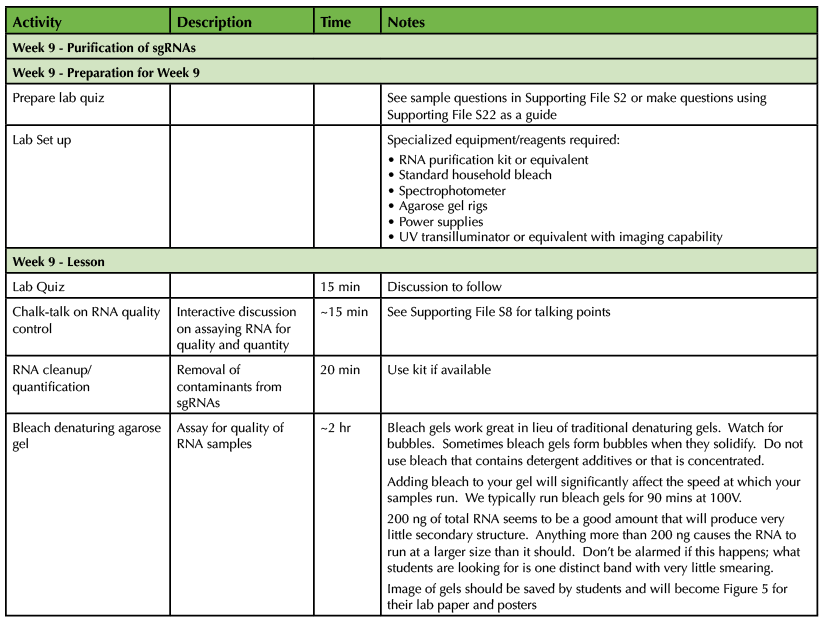
Table 1. CRISPR-Cas9 Teaching Timeline - Week 9
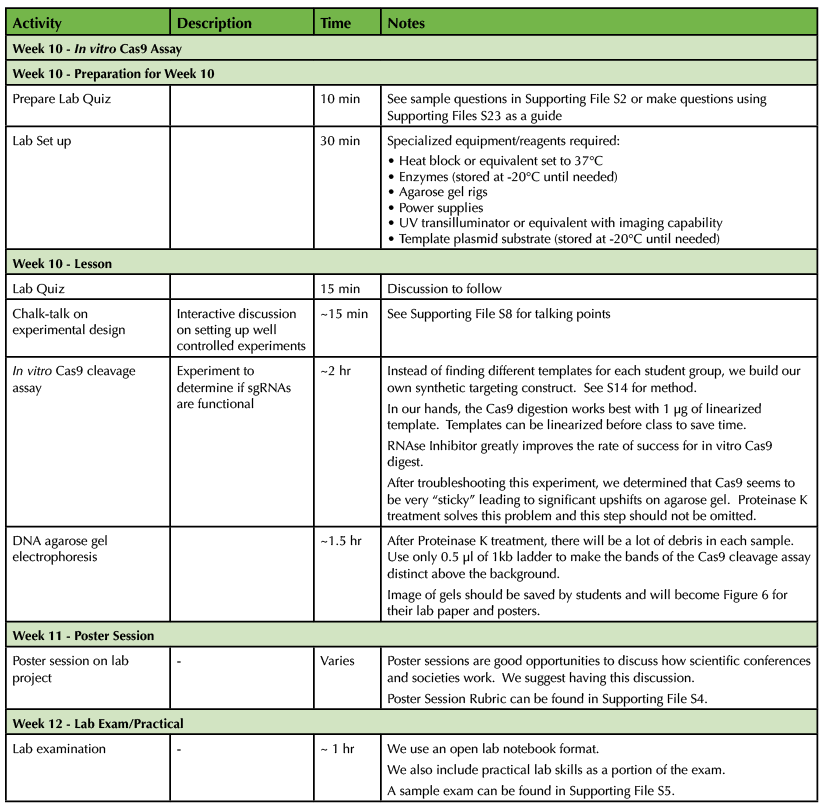
Table 1. CRISPR-Cas9 Teaching Timeline - Weeks 10-12
TEACHING DISCUSSION
During primary literature analysis, we found that students understood the experiments and the data presented but struggled to conceptualize how an experiment was "built" from an idea or question. For example, students were unable to articulate how a researcher produced a DNA for a stable cell line expressing a GFP-tagged protein but could describe the observed differences in protein localization before and after drug treatment. This knowledge gap seemed to be ubiquitous across multiple biology subdisciplines. Consistent with the practice of having students learn by performing science as outlined by Vision and Change (http://visionandchange.org/finalreport/) we also focus on the early parts of the scientific process: how researchers conceptualize ideas, generate hypotheses and then build the reagents to execute an experiment that answers their specific question. This reinforces the importance of the construction phase of novel research that is often undervalued in classroom-based reading of primary literature or laboratory experiments.
Major questions in STEM education revolve around how to accurately define, let alone assess, an inquiry-based research experience. Auchincloss and colleagues proposed five core components of course-based research: scientific practice, discovery, relevance, collaboration, and iteration (14). Further studies have classified labs into different subcategories based upon level of student inquiry and structure provided by the curriculum (15,16). These proposed classification schemes provide a structural framework for implementing inquiry-based practices in a science curriculum. Factors influencing the level of inquiry can include class size, institutional space, resources, time, student level and most importantly, course content. Since molecular biology draws heavily on protocol, we were forced to think critically about how to implement the inquiry component. Although not inherently generating novel data, we are instead focusing on how scientists logically and systematically develop specific tools that will ultimately allow the researcher to answer novel questions. To further increase the amount of inquiry involved in this lesson, we also include a number of discussion and/or application questions associated with each week's lab activities and have students work collaboratively to address key principles associated with these techniques. Additionally, since students are responsible for purposefully designing each novel reagent, they become familiar with the iterative nature of laboratory work.
CRISPR-Cas9 is quickly becoming a ubiquitous technique across many different fields of biology (17-19). Deploying CRISPR-Cas9 utilizes many different foundational molecular techniques that provide ample opportunity for students to learn and practice the skills that reinforce core tenants of the central dogma. CRISPR-Cas9 has been successfully deployed in other teaching contexts using varied models, demonstrating its value as teaching tool (20-22). During the first iteration of this laboratory, the instructor selected the targets focused on moving their own research forward. The practice of incorporating personal scholarship, especially in technique-to-product driven course like molecular biology, is beneficial to both faculty and students (23). However, as we have expanded the scope of this lesson, we chose to take a more general approach by allowing significant student input into choice of targets and reagent design. By allowing more student input, we have vastly expanded our students' understanding of both the advantages and limitations of not only CRISPR-Cas9 technology, but experimental design as a whole. Even in direct comparison between two CUREs, the experience that provided the most student input to experimental design resulted in larger gains in learning (24).
Considering the ubiquitous nature of CRISPR-Cas9, we envision this lesson to be accessible to even large enrolled courses, where students might have diverse backgrounds. For example, students might be able to form groups around a central process or disease and then develop reagents to systematically test hypotheses related to a larger defined problem. Likewise, students could also organize around different model organisms, allowing them to form collaborative groups that share common research interests. Obviously, deploying this lesson in larger courses requires more logistics, therefore one alternative might be to narrow student choice to a subset of known genes or use pre-engineered reagents. Where space, resources, time and/or level of success are issues, instructors can choose to eliminate certain aspects of the lesson, or use our lesson verified reagents found in Supporting File S24. We have successfully used this lesson in four different semesters at our institute and used this methodology to incorporate the research of other faculty members using varied systems such as C. elegans, zebrafish, and cultured cell lines. In all iterations of this course, students were successful in producing all the reagents for their respective gene edits, and the success rate of the in vitro cleavage assay is approximately 50%, with at least one group member being successful in most iterations of the lab. Although this is a guided inquiry-based exercise, there is the possibility that students might not ultimately be successful in their creation of reagents. Despite this chance of "failure" it has been demonstrated that learning gains can still be achieved even when defined laboratory research goals are not (25).
EFFECTIVENESS OF LESSON
Although we have not formally assessed this lesson, informal conversations with students and anecdotal evidence suggests that this lesson is effective in fulfilling both our teaching goals and objectives. Students have informally reported that they enjoyed learning and actively participating in a lab that uses an emerging technology. Students have also commented that after engaging in this lesson, their confidence and understanding in other laboratories, both in courses or in other formal undergraduate research experiences, is improved. Students also commented that this lesson improved their appreciation for novel research. They also expressed excitement for learning about new advances in biotechnology and related that this experience piqued their interest in new scientific topics. Importantly, this lesson also creates space for important student-led ethical discussions about the use of CRISPR-Cas9 in research and healthcare (Supporting File S25: Bioethics of Gene Editing - lecture slides).
MODIFICATIONS AND ADAPTATIONS
This lesson is appropriate for upper division Molecular Biology or Genetics courses. Although originally deployed in a Molecular Biology context, if this lesson were applied to a Genetics course, modules could be added to include genotyping and inheritance patterns once edits are successfully made. For larger courses, considerations might need to be made on group sizes, number of replicate samples, or limitations put on what can be proposed by student groups. We have found that groups of three students work the best, and student projects involving the generation of combination knockout/reporter alleles created by knocking in the cDNA sequence for a fluorescent protein in the first exon of a gene are the most straight forward when time or class sizes are limiting.
SUPPORTING MATERIALS
- S1. Using CRISPR-Cas9 - Introduction to CRISPR-Cas9 Gene Editing lecture slides. Lecture slides providing a brief background on the history and uses of gene editing.
- S2. Using CRISPR-Cas9 - Sample Lab Quiz Questions. Low level, prelab quizzes with answers to gauge student preparation.
- S3. Using CRISPR-Cas9 - Materials and Methods Paper Rubric. Rubric for assessment of formal paper associated with lesson.
- S4. Using CRISPR-Cas9 - Poster Presentation Rubric. Rubric for assessment of student generated posters.
- S5. Using CRISPR-Cas9 - Sample Laboratory Examination. Lab notebook based practical exam taken at the end of lesson.
- S6. Using CRISPR-Cas9 - Required Lesson Equipment and Reagents. Suggested vendors and notes included.
- S7. Using CRISPR-Cas9 - Common Solutions . Recipes for solutions used in this lesson.
- S8. Using CRISPR-Cas9 - Chalk Talk Points and Answers to Discussion Questions. Key points for each prelab discussion and a guide to discussion and application question found in each lab.
- S9. Using CRISPR-Cas9 - Guidelines of Consultation on Projects. Key points for helping students develop a good research plan.
- S10. Using CRISPR-Cas9 - Lab 1 PCR and Primer Design
- S11. Using CRISPR-Cas9 - Lab 2 Setting up PCR Reactions
- S12. Using CRISPR-Cas9 - Lab 3 DNA Agarose Gel Electrophoresis
- S13. Using CRISPR-Cas9 - Lab 4 PCR Purification and Sequence Prep
- S14. Using CRISPR-Cas9 - Lab 5 Sequence Analysis
- S15. Using CRISPR-Cas9 - Lab 5 sequencing tutorial file KIF17B-1.ab1
- S16. Using CRISPR-Cas9 - Lab 5 sequencing tutorial file KIF17B-2.ape
- S17. Using CRISPR-Cas9 - Lab 5 sequencing tutorial file Unknown.ape
- S18. Using CRISPR-Cas9 - Lab 6 Designing crRNA Oligos
- S19. Using CRISPR-Cas9 - Lab 7 Preparation of sgRNA Plasmids
- S20. Using CRISPR-Cas9 - Lab 8 Minipreps and Diagnostic Digest
- S21. Using CRISPR-Cas9 - Lab 9 In vitro Transcription
- S22. Using CRISPR-Cas9 - Lab 10 Purification of sgRNAs
- S23. Using CRISPR-Cas9 - Lab 11 In vitro Cas9 Assay
- S24. Using CRISPR-Cas9 - Lesson verified reagents and methods. Reagents available upon request
- S25. Using CRISPR-Cas9 - Bioethics of Gene Editing Lecture Slides
- S26. Using CRISPR-Cas9 - Solution Chemistry Practice Problems and Answers
ACKNOWLEDGMENTS
The authors would like to acknowledge Rollins College BIO341 students for their work in streamlining this lesson. The authors would also like to acknowledge Dr. Susan Walsh for critical feedback of both the lesson and manuscript.
References
- 1. Murthy PPN, Thompson M, Hungwe K. 2014. Development of a semester-long, inquiry-based laboratory course in upper-level biochemistry and molecular biology. J Chem Educ 91:1909-1917.
- Boyd-Kimball D, Miller KR. 2018. From Cookbook to Research: Redesigning an Advanced Biochemistry Laboratory. J Chem Educ 95:62-67.
- Pedwell RK, Fraser JA, Wang JTH, Clegg JK, Chartres JD, Rowland SL. 2018. The beer and biofuels laboratory: A report on implementing and supporting a large, interdisciplinary, yeast-focused course-based undergraduate research experience. Biochem Mol Biol Educ 46:213-222.
- DiBartolomeis SM. 2011. A semester-long project for teaching basic techniques in molecular biology such as restriction fragment length polymorphism analysis to undergraduate and graduate students. CBE Life Sci Educ 10:95-110.
- Brownell SE, Wenderoth MP, Theobald R, Okoroafor N, Koval M, Freeman S, Walcher-Chevillet CL, Crowe AJ. 2014. How students think about experimental design: Novel conceptions revealed by in-class activities. Bioscience 64:125-137.
- Hiebert SM. 2007. Teaching simple experimental design to undergraduates: do your students understand the basics? AJP Adv Physiol Educ 31:82-92.
- Silva T, Galembeck E. 2017. Developing and supporting students' autonomy to plan, perform, and interpret inquiry-based biochemistry experiments. J Chem Educ 94:52-60.
- Wang H, La Russa M, Qi LS. 2016. CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem 85:227-264.
- Paix A, Folkmann A, Rasoloson D, Seydoux G. 2015. High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9ribonucleoprotein complexes. Genetics 201:47-54.
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh J-RJ, Joung JK. 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31:227-229.
- Ahn SC, Baek BS, Oh T, Song CS, Chatterjee B. 2000. Rapid mini-scale plasmid isolation for DNA sequencing and restriction mapping. Biotechniques 29:466-8.
- Aranda PS, Lajoie DM, Jorcyk CL. 2012. Bleach gel: A simple agarose gel for analyzing RNA quality. Electrophoresis 33:366-369.
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A Programmable Dual-RNA - Guided. Science (80- ) 337:816-822.
- Auchincloss LC, Laursen SL, Branchaw JL, Eagan K, Graham M, Hanauer DI, Lawrie G, McLinn CM, Pelaez N, Rowland S, Towns M, Trautmann NM, Varma-Nelson P, Weston TJ, Dolan EL. 2014. Assessment of course-based undergraduate research experiences: a meeting report. CBE Life Sci Educ 13:29-40.
- Brownell SE, Kloser MJ. 2015. Studies in Higher Education Toward a conceptual framework for measuring the effectiveness of course-based undergraduate research experiences in undergraduate biology. Stud High Educ 40:525-544.
- Buck LB, Bretz SL, Towns MH. 2008. Characterizing the Level of Inquiry in the Undergraduate Laboratory. J Coll Sci Teach. 38:52-58.
- Dong Y, Simões ML, Marois E, Dimopoulos G. 2018. CRISPR/Cas9 -mediated gene knockout of Anopheles gambiae FREP1 suppresses malaria parasite infection. PLoS Pathog 14.
- Generoso WC, Gottardi M, Oreb M, Boles E. 2016. Simplified CRISPR-Cas genome editing for Saccharomyces cerevisiae. J Microbiol Methods 127:203-205.
- Wang F, Shi Z, Cui Y, Guo X, Shi YB, Chen Y. 2015. Targeted gene disruption in Xenopus laevis using CRISPR/Cas9. Cell Biosci. BioMed Central.
- Sehgal N, Sylves ME, Sahoo A, Chow J, Walker SE, Cullen PJ, Berry JO. 2018. CRISPR Gene Editing in Yeast: An Experimental Protocol for an Upper-Division Undergraduate Laboratory Course. Biochem Mol Biol Educ 46:592-601.
- Adame V, Chapapas H, Cisneros M, Deaton C, Deichmann S, Gadek C, Lovato TL, Chechenova MB, Guerin P, Cripps RM. 2016. An undergraduate laboratory class using CRISPR/Cas9 technology to mutate drosophila genes. Biochem Mol Biol Educ 44:263-275.
- Jay M. Bhatt AKC. 2018. First Year Course-Based Undergraduate Research Experience (CURE) Using the CRISPR/Cas9 Genome Engineering Technology in Zebrafish. J Microbiol Biol Educ 2129:1-9.
- Nam S-C. 2018. Integration of a faculty's ongoing research into an undergraduate laboratory teaching class in developmental biology. Biochem Mol Biol Educ 46:141-150.
- Laungani R, Tanner C, Brooks TD, Clement B, Clouse M, Doyle E, Dworak S, Elder B, Marley K, Schofield B. 2018. Finding Some Good in an Invasive Species: Introduction and Assessment of a Novel CURE to Improve Experimental Design in Undergraduate Biology Classrooms. J Microbiol Biol Educ 19.
- Gin LE, Rowland AA, Steinwand B, Bruno J, Corwin LA. 2018. Students Who Fail to Achieve Predefined Research Goals May Still Experience Many Positive Outcomes as a Result of CURE Participation. CBE Life Sci Educ 17:ar57.
Article Files
Login to access supporting documents
Using CRISPR-Cas9 to teach the fundamentals of molecular biology and experimental design(PDF | 509 KB)
S1. Using CRISPR-Cas9 - Introduction to CRISPR-Cas9 Gene Editing lecture slides.pptx(PPTX | 1024 KB)
S2. Using CRISPR-Cas9 - Sample Lab Quiz Questions.docx(DOCX | 25 KB)
S3. Using CRISPR-Cas9 - Materials and Methods Paper Rubric.xlsx(XLSX | 13 KB)
S4. Using CRISPR-Cas9 - Poster Presentation Rubric.xlsx(XLSX | 11 KB)
S5. Using CRISPR-Cas9 - Sample Laboratory Examination.docx(DOCX | 21 KB)
S6. Using CRISPR-Cas9 - Required Lesson Equipment and Reagents.docx(DOCX | 24 KB)
S7. Using CRISPR-Cas9 - Common Solutions .docx(DOCX | 17 KB)
S8. Using CRISPR-Cas9 - Chalk Talk Points and Answers to Discussion Questions.docx(DOCX | 75 KB)
S9. Using CRISPR-Cas9 - Guidelines of Consultation on Projects.docx(DOCX | 20 KB)
S10. Using CRISPR-Cas9 - Lab 1 PCR and Primer Design.docx(DOCX | 68 KB)
S11. Using CRISPR-Cas9 - Lab 2 Setting up PCR Reactions.docx(DOCX | 22 KB)
S12. Using CRISPR-Cas9 - Lab 3 DNA Agarose Gel Electrophoresis.docx(DOCX | 40 KB)
S13. Using CRISPR-Cas9 - Lab 4 PCR Purification and Sequence Prep.docx(DOCX | 26 KB)
S14. Using CRISPR-Cas9 - Lab 5 Sequence Analysis.docx(DOCX | 29 KB)
S15 - KIF17B-1.ab1_.zip(ZIP | 156 KB)
S16 - KIF17B-2.txt(TXT | 1 KB)
S17 - Unknown.ab1_.zip(ZIP | 190 KB)
S18. Using CRISPR-Cas9 - Lab 6 Designing crRNA Oligos.docx(DOCX | 24 KB)
S19. Using CRISPR-Cas9 - Lab 7 Preparation of sgRNA Plasmids.docx(DOCX | 27 KB)
S20. Using CRISPR-Cas9 - Lab 8 Minipreps and Diagnostic Digest.docx(DOCX | 27 KB)
S21. Using CRISPR-Cas9 - Lab 9 In vitro Transcription.docx(DOCX | 39 KB)
S22. Using CRISPR-Cas9 - Lab 10 Purification of sgRNAs.docx(DOCX | 21 KB)
S23. Using CRISPR-Cas9 - Lab 11 In vitro Cas9 Assay .docx(DOCX | 22 KB)
S24. Using CRISPR-Cas9 - Lesson verified reagents and methods.docx(DOCX | 21 KB)
S25. Using CRISPR-Cas9 - Bioethics of Gene Editing Lecture Slides.pptx(PPTX | 216 KB)
S26. Using CRISPR-Cas9 - Solution Chemistry Practice Problems and Answers.docx(DOCX | 24 KB)
- License terms

Comments
Comments
There are no comments on this resource.