The ACTN3 Polymorphism: Applications in Genetics and Physiology Teaching Laboratories
Published online:
Abstract
Inquiry-based undergraduate laboratories provide the opportunity to engage students in a research experience that improves scientific thinking, but such activities can be difficult to develop and implement due to limited time and resources. We have developed a series of inquiry-based laboratory modules that directly address Vision and Change core concepts and competencies, as well as core principles in the fields of genetics and physiology. The laboratory modules focus on a common polymorphism in the alpha-actinin-3 (ACTN3) gene that results in the lack of ACTN3 protein expression in fast twitch muscle fibers in 16% of the human population. This project provides an authentic classroom research experience and addresses the connection between science and society by examining the implications of ACTN3 genetic testing to improve sports training and performance. Modules are broken down into introductory, information literacy and the scientific method, genotyping, and physiology lessons. If all modules are implemented, the study can be completed in 5-8 weeks, but instructors can decide to implement one or more modules independent of each other. We have successfully implemented the modules in First Year Seminar and upper-level physiology classes at Dickinson College and an upper-level genetics class at Georgetown University, indicating that these exercises are adaptable to all levels of the undergraduate biology curriculum, as well as general non-science classes. This article accompanies the Science Behind the Lesson "The Science Behind the ACTN3 Polymorphism."
Citation
Frey TA, Somers DJ, Lehman HL, Hall AE, Hwang EK, Yarden RI. 2019. The ACTN3 polymorphism: Applications in genetics and physiology teaching laboratories. CourseSource. https://doi.org/10.24918/cs.2019.30Lesson Learning Goals
- Understand how a polymorphism in the ACTN3 gene affects protein structure and function (Core Concept in Genetics: How do different types of mutations affect genes and the corresponding mRNAs and proteins? (1); Vision and Change Core Concept: Information Flow, Exchange, and Storage (2)).
- Understand that skeletal muscle function is a complex phenotype that is the result of multiple genes and environmental factors (Core Concept in Genetics: How can one deduce information about genes, alleles, and gene functions from analysis of genetic crosses and patterns of inheritance? (1)).
- Understand how the ACTN3 protein impacts skeletal muscle function (Core Principle of Physiology: Structure-function relationships (3); Vision and Change Core Concept: Structure and Function (2)).
- Apply the scientific method to evaluate the claim that ACTN3 genetic variants affect skeletal muscle function (Vision and Change Core Competency: Ability to apply the process of science (2)).
- Apply quantitative reasoning skills to analyze the relationship between ACTN3 genotype and skeletal muscle phenotypes (Vision and Change Core Competency: Ability to use quantitative reasoning (2)).
- Enhance independent thinking skills and promote feelings of ownership and engagement in the scientific process (Vision and Change Core Competency: Ability to apply the process of science (2)).
Lesson Learning Objectives
- Test hypotheses related to the role of ACTN3 in skeletal muscle function.
- Explain how polymorphic variants of the ACTN3 gene affect protein structure and function.
- List and explain the differences between fast twitch and slow twitch muscle fibers.
- List and explain possible roles of the ACTN3 protein in skeletal muscle function.
- Find and analyze relevant scientific publications about the relationship between ACTN3 genotype and muscle function.
- Formulate hypotheses related to the relationship between ACTN3 genotype and skeletal muscle function.
- Design experiments to test hypotheses about the role of ACTN3 in skeletal muscle function.
- Statistically analyze experimental results using relevant software.
- Present experimental results in writing.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Scientific teaching entails using classroom and laboratory teaching methods that have been scientifically demonstrated to improve learning (4). Moreover, the Vision and Change report (2) calls for the application of scientific teaching in the undergraduate biology curriculum to help all students learn core concepts and develop core competencies. One approach to accomplish these goals is the use of inquiry-based laboratory modules in undergraduate biology classes (5). Inquiry-based laboratory activities can be categorized as guided inquiry, open-ended inquiry, and teacher-collaborative research projects projects (5-7). In guided inquiry, the research question is provided, and students are led to the appropriate experimental design and outcomes. In open-ended inquiry, students come up with the question and experimental design in a more independent fashion. The distinguishing feature of teacher-collaborative research projects is that the students and instructor work together on experimental design and analysis related to a research question where the outcome of the research is not known to the students or instructor, making it a true research experience (6).
We have developed a series of inquiry-based, teacher-collaborative research laboratory modules (Figure 1) that directly address Vision and Change core concepts and competencies as well as core principles in the fields of genetics (1) and physiology (3). These laboratory activities focus on a common polymorphism in the alpha-actinin-3 (ACTN3) gene that results in complete deficiency of the ACTN3 protein in fast twitch muscle fibers. In this genetic variant, the ancestral C nucleotide is replaced by T, resulting in a change from arginine (R) to a stop codon (X) at amino acid position 577 (R577X) (8-10). This genetic change does not cause any obvious muscle disease but is present in nearly 16% of the world’s population (11), making it feasible for study in a college student population. Since fast twitch muscle fibers are primarily responsible for sprint and power-based muscle movement, it has been hypothesized that ACTN3 deficiency would have an effect in athletes competing in sprint or power-based activity. The modules described here provide a unique opportunity for students to study correlations between ACTN3 genotype and skeletal muscle function.
INTENDED AUDIENCE
This laboratory activity was designed for upper-level genetics (300-level) or physiology (300-level) courses (24 students per semester) with a weekly 3-hour lab; however, the modules are adaptable to different classes and levels of the biology curriculum. For example, we have also used the activity in a First-Year Seminar course (3 hours/week, 16 students with a variety of science backgrounds and levels of understanding) and could easily see the modules applied to an introductory biology course or upper level population genetics class. Depending on the course subject and student level, material in the laboratory activities can be adjusted to include more or less complex coverage as needed. Assessments and lesson plans provided here are examples of how the project was implemented by individual instructors.
REQUIRED LEARNING TIME
If all modules are implemented, the study can be completed in 5-8 laboratory sessions (see Table 1), but instructors can decide to implement one or more modules independent of each other (Figure 1).
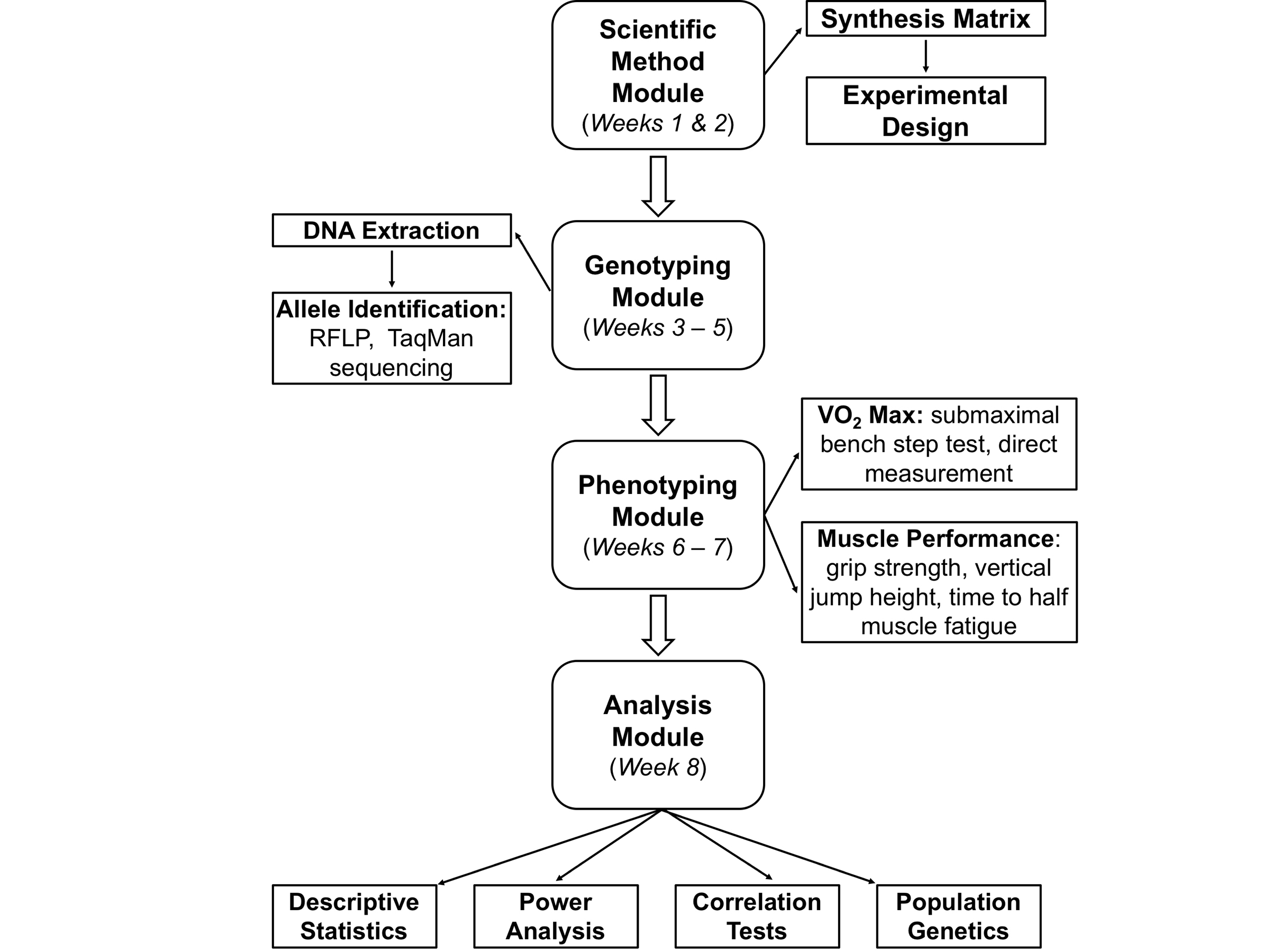
Figure 1. Suggested project workflow for classes. The project involves four modules: a scientific method module, a genotyping module, a physiology module, and an analysis module. Not all activities are necessary for all levels; instructors may choose from several different activities for each module.
PREREQUISITE STUDENT KNOWLEDGE
This laboratory activity is designed for students with minimal background in the subject area. Students learn the necessary background information during the introductory lecture and information literacy and scientific method in Module 1.
PREREQUISITE TEACHER KNOWLEDGE AND COMPLIANCE
Instructors should have sufficient competence in basic molecular biology laboratory techniques to instruct students in use of micropipettes and gel electrophoresis. Additional background information summarizing the scientific literature relating to the genetics and physiology behind the ACTN3 polymorphism is provided in the accompanying "Science Behind the Lesson" article.
Institutional Review Board (IRB) approval is required for human subjects research. However, if data generated for this project will only be used for teaching purposes and not for contribution to generalizable knowledge (which is part of the definition of research according to the U.S. Department of Health & Human Services, https://www.hhs.gov/ohrp/regulations-and-policy/decision-charts/index.html#c1), then it qualifies for IRB exemption. Check with your IRB to determine if exemption approval is required by your institution. We have included our Application for Exempt Research (Supporting File S1: IRB Application) and Exemption Notification (Supporting File S2: IRB Exemption) for reference.
In addition to IRB paperwork, Institutional Biosafety Committee (IBC) approval may also be required for this project. DNA is isolated from buccal swabs for ACTN3 genotyping, which involves handling human body fluids. Check with your IBC and obtain approval for this protocol if necessary.
SCIENTIFIC TEACHING THEMES
ACTIVE LEARNING
The laboratory modules outlined here are inquiry-based, teacher-collaborative research modules that can be feasibly used with students of all undergraduate levels. Students work collaboratively in self-selected groups of three to four individuals to first conduct a scientific literature search to identify a primary article that describes ACTN3 and its relation to a phenotype. Students work together in their groups to read articles, identifying specific information to share with the class in a synthesis matrix (described in Module 2 below). As students begin to examine the many approaches to genotyping ACTN3 and research ACTN3-related phenotypes, they can synthesize the information and begin to engage with the topic on a higher-order level. This approach allows students to begin to design their own experiments, thinking about the phenotypes they could measure in class that would be relevant to ACTN3 function and feasible to test in a laboratory setting. The synthesis matrix activity allows students to actively engage with the project and its experimental design, determining the variables to investigate and forming testable hypotheses.
Instructors can determine the amount of autonomy students have in choosing the phenotypes they would like to study; however, brainstorming the feasible options and then deciding which ones to actively study as a class gives the students ownership of the experimental design and allows them to be more invested in the project. Students work in their groups to collect data, using themselves as the study subjects each week. Students then perform each experiment, from isolating their own DNA and genotyping themselves for the ACTN3 gene polymorphism, to testing different muscle-related phenotypes.
ASSESSMENT
In the original implementation, assessment of students during the laboratory modules included multiple elements. Individual lab notebooks were collected and graded for each experiment based on format, inclusion of a hypothesis, reporting of data, analysis and interpretation of results, and overall completion. A brief rubric was given to students to guide them in their lab notebook entries (Supporting File S3: Lab Notebook Rubric), and we found it helpful to provide students with an example of a notebook entry as well. Handouts (provided as supporting files as indicated below) were given to students during each lab that included a protocol for the experiment, but also included questions to encourage students to think about their data collection and analyze their results, particularly how their individual genotypes for ACTN3 related to the outcomes of their physiological experiments.
Students were also required to write lab reports on the ACTN3 project. Depending on the extent of the modules completed for the project, each student was required to write a mid-project lab report and/or an end-of-project lab report. For the mid-project lab report, students were asked to write about one phenotype measured and how it related to what they learned about ACTN3 and its function. For the final lab report, students were asked to integrate background information about ACTN3 from literature, information they learned in class, results they obtained in lab for all the phenotypes they measured, and discuss how the class data related to what has been demonstrated for larger populations in the literature. The rubric given to students to guide them in writing their lab reports is provided in Supporting File S4: Scientific Writing Rubric.
Students were also able to self-assess how they felt about their learning during laboratory sessions. Throughout the entire project, students were invited to give anonymous feedback on the ACTN3 project, where they were asked to openly comment on how they liked or disliked the ACTN3 project and if/how they felt it was a valuable learning experience. At the end of the semester, students were administered a standard class evaluation from the college. One of the questions asked students to assess whether they felt laboratory work was successful as a learning experience. Both of these types of evaluations can be used to assess student feelings about the project and determine if the goals of ownership and engagement in the scientific process was achieved.
INCLUSIVE TEACHING
These laboratory modules have been used successfully in both lower-level and upper-level undergraduate courses for students with varying backgrounds in the biological sciences. For these learning experiences to be successful, it is not required that students have prerequisite knowledge. All required information is given during instruction in lecture and/or laboratory; therefore, it encourages participation of all students regardless of baseline knowledge. Instruction employs handouts, PowerPoint slides and oral instruction, serving the diversity of learning needs in the classroom. As students work in groups in the lab and lecture, they can discuss the experimental protocol and results with their group members to aid in their understanding. We allowed self-selection of groups, but instructors can assign groups to increase diversity in each group and further promote inclusion.
Another way to improve group work and promote inclusion is to provide instruction on effective group work practices at the beginning of the project. This is especially important if you plan to assign a group presentation, poster, or writing assignment related to this project. We have developed a class session on effective group work and planning that can be used with these modules (Supporting File S5: Working in Groups - Lecture Slides). Finally, time is provided during lab classes to complete much of the group work related to this project (e.g., planning, data analysis), in order to be inclusive for students with obligations outside of class, such as work or family commitments.
There is diversity in the prevalence of the different ACTN3 genotypes throughout various populations, genders, and races (10,12-15), and students should observe this diversity when they conduct their literature reviews. Students are able to work collaboratively in their groups to conduct literature searches. Furthermore, classroom collaboration occurs during the development of the synthesis matrix and the overall project, when students have the opportunity to demonstrate their diversity of opinions and engage in productive discussions that ultimately result in the best experimental design.
Lastly, the phenotypic modules, which involve measurement of muscle phenotypes in each student, are applicable to individuals of varying physical capabilities, but are also made voluntary. On average, biological males have higher muscle mass than biological females, and it is therefore important to stratify results by biological sex during analysis of data. In order to account for transgender, non-binary, or other individuals who are uncomfortable disclosing biological sex, an option to not disclose biological sex is included on the demographic information form (Supporting File S10: Demographic Information Form). Collate this information in a manner that keeps it private. While participation in the physical activity and disclosure of biological sex is voluntary, analysis of anonymous results and data is required. We have found that the majority of students opt to participate in the experiments and disclose biological sex.
LESSON PLAN
The recommended timeline for this lesson is in Table 1.
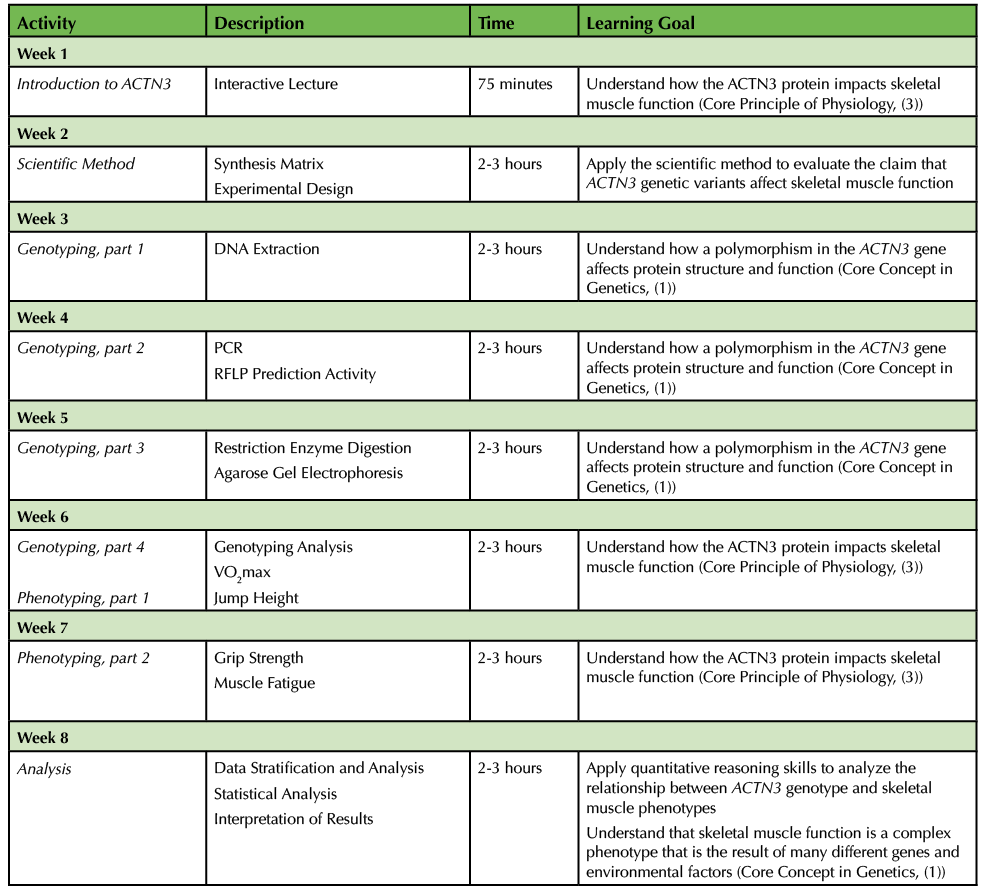
Table 1. ACTN3 Polymorphism - Teaching Timeline
Week 1: Introduction to Project
The first week of the project involves a 75-minute interactive lecture. The goals of this lesson are to introduce ACTN3 as the "gene for speed," to understand how a polymorphism in the ACTN3 gene affects protein structure and function (Learning Goal 1), and to begin to consider how ACTN3 affects skeletal muscle function (Learning Goals 2 and 3).
Materials Required: Access to one or more scientific literature databases.
Instructor Preparation for Class: Be prepared to explain background information about where the ACTN3 protein is expressed (fast twitch vs. slow twitch muscle fibers) and how each polymorphic variant affects ACTN3 protein structure and function. The accompanying "Science Behind the Lesson Article" provides additional background information for the instructor.
Student Preparation for Class: Read a non-scholarly article (examples are available through the links below) and a scholarly review article (e.g., 16 or 17) about the roles of ACTN3 polymorphic variants in athletic performance. Answer questions in Part 1 of the provided guided reading handout (Supporting File S6: Guided Reading Handout) prior to coming to class.
- https://www.scientificamerican.com/article/genes-sports-talent/
- http://anabolicgenes.com/blog/en/actn3-gene-the-strength-and-power-gene/
Class Activity: The class session is broken down into four components with approximate time needed for each component in parentheses:
- Small group activity and class discussion (15 minutes): At the beginning of class, prompt students to consider the validity of the claim that ACTN3 is the "gene for speed" through questions such as, "If you were a parent, would you want to genotype your child for ACTN3 prior to choosing what sports they do?" and "Do you think knowing your ACTN3 genotype would change how you approach athletic training?" Give them 2-5 minutes to discuss within groups and then ask them to share with the class. During the class discussion, make two lists on the board (Yes, it is useful to know ACTN3 genotype/No, it is not useful). At the end of this discussion, point out that speed is a complex phenotype dependent on multiple genes and environment; ACTN3 genotype is a single contribution to this complex phenotype and may only contribute a small amount to skeletal muscle performance.
- Interactive lecture (30 minutes): In order to further consider the impact of ACTN3 genetic variants on skeletal muscle function, discuss the answers to the guided reading questions. Call on students to answer questions and, as necessary, further explain answers to the questions. The PowerPoint slides used to support this discussion are included as Supporting File S7: Guided Reading Interactive Lecture - Lecture Slides.
- End of class brainstorming and class discussion (20 minutes): The 30-minute interactive lecture helps the students understand that the absence of ACTN3 protein in fast twitch muscle fibers makes them behave more like slow twitch muscle fibers. Pose the following question to the class: "If the absence of ACTN3 protein in fast twitch fibers makes them behave more like slow twitch fibers, what phenotypes could be measured in humans to determine the impact of the ACTN3 polymorphisms on skeletal muscle function?" Ask groups of 3-4 students to brainstorm a list of answers (5 minutes) to this question while you walk around and encourage students to recall the major differences between fast twitch and slow twitch fibers. Either ask for responses or call on groups to make a list on the board. Some phenotypes might be obvious to students such as speed, grip strength, and jump height. Some may recall from the review article that slow twitch fibers have a great capacity to use oxygen to make ATP but may not know how to measure that. Point out that VO2max, or the maximal capability of the body to use oxygen with incremental workload, can be measured non-invasively in people.
- Assign homework (10 minutes): Ask students to use the list of possible ACTN3-related human phenotypes to complete an information literacy homework assignment (Part 2 of Supporting File S6: Guided Reading Handout) for the next lab meeting time. In this assignment, student groups are asked to find and analyze primary scholarly publications about how the ACTN3 polymorphism affects muscle performance. We typically had six groups of four students each find one publication on this topic, where each group was asked to submit the paper for instructor approval within 48 hours to avoid overlap, to confirm the paper was appropriate for the assignment, and to allow the instructor to prepare for the next class. Group size should be adjusted based on class size in order to obtain 5-10 publications. At this point, you can guide the literature search more or less depending on the phenotypes the class is able to measure. For example, in initial classes we only wanted to measure VO2max. Therefore, after brainstorming and a class discussion about how ACTN3 might impact muscle function, we told the students to focus their literature search on ACTN3 and VO2max. In a later iteration of the class, we wanted to measure more phenotypes and to allow the students to have more control over choosing which ones to measure. Thus, the literature search assignment asked them to focus on finding specific information in their paper including study population, genotyping method, phenotype(s) measured and methods, and results with statistics (e.g., was there a correlation between ACTN3 genotype and the measured phenotype?). The information students are asked to find in the literature may vary depending on the type of class in which the project is being done.
Anticipated Results: Due to required screening of articles by the instructor, we did not have any problems in our classes at Dickinson College with students finding one primary publication with the requested information without any overlap between groups. A PubMed database search on 5/1/2018 using the keywords ACTN3 and muscle performance yielded 103 articles, 17 of which were review articles. Some groups required guidance to select a primary article with the relevant information. Common mistakes included choosing a review article or a paper that analyzed ACTN3 genotype frequency in different populations instead of one that correlated ACTN3 genotype with muscle function.
Week 2: Scientific Methods Lab
The goals of this 2-3 hour lab session are to further understand how the ACTN3 protein impacts skeletal muscle function (Learning Goal 3) and to apply the scientific method to evaluate the claim that ACTN3 genetic variants affect skeletal muscle function (Learning Goal 4).
Instructor Preparation for Class: You should be familiar with the information in the approved primary research articles that students found as homework for lesson 1. Fill in the synthesis matrix (Supporting File S8: Synthesis Matrix - Blank) with the requested information from the research articles and print a synthesis matrix for each student in the class. If you want to direct the students to additional research articles (for example, if you want them to measure grip strength but they did not find any papers on grip strength), you can find additional papers to add to the matrix.
Student Preparation for Class: Complete Part 2 of the guided reading handout (Supporting File S6: Guided Reading Handout) provided prior to this week's lesson.
Synthesis Matrix Exercise: The goals of this exercise are to help students analyze the results of published studies that tested the hypothesis that ACTN3 genotype impacts skeletal muscle function and to think about how the class can expand upon published results. At the beginning of class, hand out the synthesis matrix template (Supporting File S8: Synthesis Matrix - Blank) and draw the synthesis matrix on the board. You can either start right away or give the student groups 5-10 minutes to review their answers to the guided reading questions related to their paper. Each group reports what they found for each question as you fill in the matrix on the board (see Supporting File S9: Synthesis Matrix - Example to see what a completed matrix looks like). Discuss key elements of the findings, including whether the results are statistically significant. This is a good time to review p values (see the GraphPad Statistics Guide for more information on p-values: https://www.graphpad.com/guides/prism/8/statistics/index.htm). If results are significant, pull up the data from the paper to examine the magnitude of the differences between groups. Since muscle function is a complex phenotype, you can expect differences to be small. This is a good time to remind students of the concept of complex traits. Once all information is on the board, take some time to summarize what the studies found and discuss strengths and weaknesses of the studies. Key points to address are the study populations (e.g., athletes or not, age of participants, biological sex, population size, ethnicity) and how the studies relate to each other. If any studies are conflicting, why might that be? Perhaps they were done on different populations or with different methods. Can any overarching conclusions be made based on these studies?
Study Design: Tell that class that they are going to do their own study to test the hypothesis that ACTN3 genotype impacts skeletal muscle function. Students should work in groups to come up with a plan to test this hypothesis using the class as the study population. Prompt them to use the synthesis matrix information as a guide for experimental design. Once students have thought about the experimental design, discuss it with the whole class. The following steps are needed to complete the study; guide the discussion so that each step is discussed and planned:
- Collect demographic information on the class (What information should we collect?)
- ACTN3 genotyping (How can we genotype for ACTN3?)
- Measure skeletal muscle phenotypes (Which ones can we measure and how?)
- Analyze results (How will we stratify results? Do you think the study population is big enough?)
At this point, you can shape the experimental design to fit the time and resources available. In first-year seminar, genetics, and initial physiology classes, we only wanted to measure VO2max. Therefore, the students were directed to find primary publications on VO2max and the phenotype conversation was directed toward how we can measure VO2max. In a later physiology class, the instructor reserved additional lab time to measure other phenotypes such as grip strength, muscle fatigue, and jump height.
Anticipated Results: The products of this lab session are the synthesis matrix (Supporting File S9: Synthesis Matrix-Example) and broad experimental design, including a demographic information form (Supporting File S10: Demographic Information Form). At this point, we encouraged the students to use this information to begin the writing assignment related to this project (introduction section of lab report).
Weeks 3 - 6: Genotyping
The goals of this three-week laboratory activity are to understand the polymorphism in ACTN3, learn how to identify the polymorphism using genotyping techniques, and understand how the change in DNA sequence can affect protein structure and function (Learning Goal 1). During these three laboratory sessions, students complete restriction fragment length polymorphism (RFLP) analysis to determine their genotype at the ACTN3 locus. In week 3, students isolate DNA from buccal swabs. In week 4, students PCR amplify a portion of the ACTN3 gene from their DNA and complete an in-class activity to help them predict the results of their RFLP analysis. In week 5, students digest their amplified PCR products using DdeI and run an agarose gel to visualize the results. In week 6, which is carried out concurrently with activities in Module 3: Phenotyping, students analyze the results of their RFLP analysis.
Materials and Equipment Required: Prior to the first lab, obtain the following equipment and materials:
- Cytology brush (Fisher Scientific #22-263358)
- Human gDNA extraction kit (Promega #A2051)
- PCR thermocycler
- ACTN3 primers (Forward: 5' CTGTTGCCTGTGGTAAGTGGG; Reverse: 5' TGGTCACAGTATGCAGGAGGG 3')
- EconoTaq PLUS 2X Master Mix (Lucigen #30035-1)
- DdeI TimeSaver (New England Biolabs #R0175S)
- 37°C incubator
- 100 bp DNA ladder (Thermo Fisher #15628019)
- Agarose
- Agarose gel electrophoresis equipment
- Ethidium bromide (UV light) or SYBR Safe (UV or blue light, Thermo Fisher #SS33102)
- Gel imaging equipment
- Micropipettes and appropriate tips
Week 3: DNA Extraction
During this week's laboratory activity, DNA is isolated from buccal swabs of student's cheek cells using a human genomic DNA miniprep system (Promega). This system uses a four-step method of homogenizing starting material to release the DNA, binding DNA to a ReliaPrepTM binding column, removing impurities from DNA, and eluting purified DNA.
Instructor Preparation for Laboratory: Instructors should be comfortable with the use of micropipettes and prepared to instruct students in the proper use of micropipettes. Prior to the laboratory, divide reagents and materials from the ReliaPrepTM kit among each laboratory table. Please see Supporting File S11: ACTN3 Genotyping Lab Preparation Guide for detailed instructions for laboratory preparation. Preparation will take approximately 1 hour.
Student Preparation for Laboratory: Read the micropipette instruction guide (Supporting File S12: Micropipette Guide) and the laboratory handout (Supporting File S13: ACTN3 Genotyping Lab Handout) prior to lab.
Laboratory Activity: Prior to the start of the laboratory exercise, give a short lecture on RFLP analysis and provide instruction on how to use micropipettes. After completing these two activities, students work in small laboratory groups to complete gDNA extraction following the protocols provided in Supporting File S13: ACTN3 Genotyping Lab Handout. Instruct each student to take two buccal swabs, one from each cheek, to provide a back-up sample.
Anticipated Results: The product from this week's lab is a sample of DNA from each student. Student DNA samples should be stored at -20°C until the next laboratory period. Before DNA samples are used as template for PCR amplification in week 4, assess quality and quantity using a spectrophotometric assay (we use a NanoDrop (Thermo Fisher Scientific), although any UV spectrophotometer can be used). We generally aim for an OD260/280 ratio of 1.8 +/- 0.1 with a concentration of at least 10 ng/µL. With proper instruction, students rarely have trouble isolating DNA using this kit. However, if DNA is not of sufficient quality or quantity to continue with PCR amplification, students can return to redo their DNA extractions. If an instrument for DNA quantification is not available, it is reasonable to continue the activity without quantification.
In general, gDNA extraction from buccal swabs is quite robust, with approximately 94% of Dickinson College students (113/120) successfully extracting DNA of sufficient quality and quantity for subsequent PCR.
Week 4: PCR Amplification of ACTN3
In this week's laboratory activity, students amplify a 291 bp fragment of the ACTN3 DNA using the following primers: 5' CTGTTGCCTGTGGTAAGTGGG 3' (forward) and 5' TGGTCACAGTATGCAGGAGGG 3' (reverse). Once PCR reactions are set up, students complete an activity to predict the results of the RFLP genotyping experiment for each ACTN3 genotype.
Instructor Preparation for Laboratory: Prior to the laboratory, divide reagents and materials for PCR among each laboratory table. In our experience, the most robust results are obtained when some of the components of the PCR reaction are provided pre-mixed. In addition, set up a thermocycler with the following cycling conditions:
- 95°C for 2 min
- 40 cycles of: 95°C for 30 sec, 65°C for 30 sec, and 72°C for 45 sec
- 72°C for 10 min
- 4°C hold
Please see Supporting File S11: ACTN3 Genotyping Lab Preparation Guide for more detailed instructions for laboratory preparation. Preparation will take approximately 30 minutes.
Student Preparation for Laboratory: Read the appropriate section from their textbook on PCR analysis. If such material is not utilized in the course, several appropriate videos exist (e.g., https://www.youtube.com/watch?v=2KoLnIwoZKU). Also read the laboratory handout (Supporting File 13: ACTN3 Genotyping Lab Handout) prior to lab.
Laboratory Activity: Prior to the start of the laboratory exercise, remind students of how this week's laboratory activity connects to the larger project goal. You may also wish to review the principles of PCR and/or micropipette usage. After completing these activities, students work in small laboratory groups to set up PCR reactions following the protocols provided in Supporting File 13: ACTN3 Genotyping Lab Handout. Have students hold their PCR reactions on ice until all groups have completed the set-up, and then load all samples into the thermocycler together.
Once samples are in the thermocycler, students individually complete the RFLP genotyping prediction activity (Supporting File S14: ACTN3 Genotyping Activity). Students are generally able to complete this activity within the time remaining during a 2-3 hour lab period. However, if less time is available, this activity can be assigned for homework.
Anticipated Results: One of the products from this week's lab is an amplified fragment of the ACTN3 gene from each student. PCR amplicons should be stored at -20°C until the next laboratory period. The other product is a completed worksheet predicting the results of genotyping, which will be used to assist students in analysis of their results in week 5. The answers to the worksheet are provided in Supporting File S15: ACTN3 Genotyping Activity Answer Key.
Week 5: DdeI Digestion and Agarose Gel Electrophoresis
In this week's lab, the restriction enzyme DdeI (New England Biolabs) is used to digest PCR-amplified DNA. In individuals with the T allele, two DdeI recognition sites (C|TNAG) are present within the 291 bp ACTN3 PCR amplicon, and DdeI digestion results in three fragments: 108 bp, 97 bp, and 86 bp. In individuals with the C allele, only one DdeI recognition site is present within the ACTN3 amplicon, and DdeI digestion results in two fragments: 205 bp and 86 bp (Figure 2). The full volume (25 µL) of the PCR reaction is used for digestion with 10 U DdeI in a 50 µL reaction at 37°C for 15 minutes, using the TimeSaver version of DdeI.
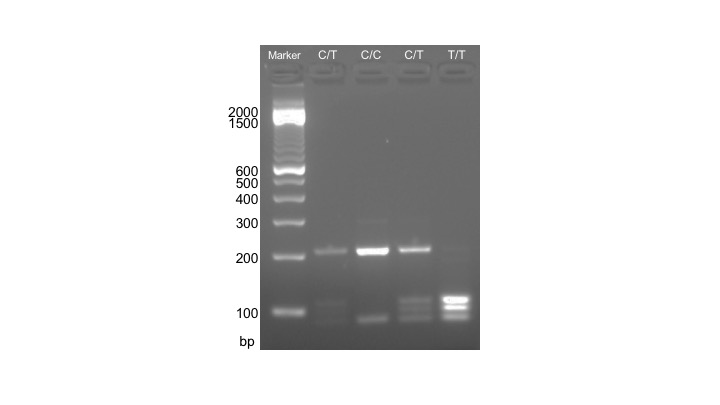
Figure 2. Expected results from RFLP analysis of the ACTN3 polymorphism. Lanes are labeled according to which genotype is displayed: C/T, four bands are visible for individuals carrying both the C and T allele; C/C, two bands are visible for individuals carrying two copies of the C allele; T/T, three bands are visible for individuals carrying two copies of the T allele.
Following digestion, DNA fragments are separated based on size by agarose gel electrophoresis in a 2% agarose gel. DNA fragment sizes are estimated based on comparison to a 100 bp ladder (Invitrogen).
Instructor Preparation for Laboratory: Prior to the laboratory, divide reagents and materials for DdeI digestion and gel electrophoresis among each laboratory table. Please see Supporting File S11: ACTN3 Genotyping Lab Preparation Guide for detailed instructions for laboratory preparation. Preparation will take approximately 1 hour.
Student Preparation for Laboratory: Read the appropriate section from their textbook on restriction enzymes and agarose gel electrophoresis. If such material is not utilized in the course, several appropriate online resources exist (e.g. https://www.dnalc.org/resources/animations/gelelectrophoresis.html). Also read the laboratory handout (Supporting File 13: ACTN3 Genotyping Lab Handout) prior to lab and bring completed RFLP activity from week 4 to lab.
Laboratory Activity: Prior to the start of the laboratory exercise, remind students of how this week's laboratory activity connects to the larger project goal and go over the results of the RFLP activity from week 4. After completing these activities, students work in small laboratory groups to set up restriction digests using the protocols provided in Supporting File S13: ACTN3 Genotyping Lab Handout. Once restriction digests are complete, students load their digested products on a 2% agarose gel and run the gel for approximately 1.5 hours at 100V.
Anticipated Results: The product from this week's lab is an image of a gel, which can be used to infer each student's ACTN3 genotype. The three genotypes of the ACTN3 polymorphism result in distinct banding patterns which are visible by separation on a 2% agarose gel: T/T 108 bp, 97 bp, and 86 bp; C/T 205 bp, 108 bp, 97 bp, and 86 bp; C/C 205 bp and 86 bp (Figure 2). These images should be included as a figure in the final lab report students produce from this project. If gels are not finished running before the end of the laboratory period, instructors may image the gels and provide electronic or hard copies of the gel images to students.
Overall, the genotyping labs are quite robust. The data collected from the Dickinson College student population (n = 113) are consistent with a population of mostly European ancestry C/C, 0.248; C/T, 0.487; T/T, 0.265, which accurately reflects self-reported ancestry in our student population.
Weeks 6, 7, and 8: Phenotyping and Analysis
The goal of these lessons is to analyze the relationship between the ACTN3 genotype of individuals and skeletal muscle phenotypes (Learning Goal 5). As students use themselves as experimental subjects and begin to analyze their physiological results, the goal is for students to be able to tie in skeletal muscle phenotypes with their ACTN3 genotypes and their knowledge of how their genotypes could impact their skeletal muscle function (Learning Goals 3 & 4).
Materials and equipment required: Prior to the first lab, obtain the following equipment and materials:
Submaximal Bench Step Test for VO2max
- 41.3 cm (16.25 in) step
- Metronome
- Stopwatch
- Data sheets
1 mile walk test for VO2max
- Track
- Heart rate monitor (optional)
Vertical jump height
- Chalk
- Tape measures
Grip strength and time to half fatigue
- PC or Mac computer
- LabScribe3 software
- iWorx IXTA data acquisition unit
- USB cable
- IXTA power supply
- ECG cable and electrode lead wires
- Disposable electrodes
- Hand dynamometer
- Alcohol swabs
- Dumbbell or other item of defined weight
Week 6: VO2max and vertical jump height
VO2max
The most accurate way to measure VO2max is by a direct method, which involves measuring inspired and expired gases while the subject is exposed to incremental increases in workload on either a treadmill or cycle ergometer until they reach maximal oxygen consumption as determined by an oxygen consumption plateau despite increased workload (18). However, because this test is not practical in most classroom settings, several indirect methods have been developed based on a linear relationship between VO2max (measured directly) and another easily measurable parameter such as time or heart rate after a submaximal activity. We used two different indirect methods that have both been validated to accurately approximate VO2max direct measurement (19,20). The submaximal bench step test (20) was used at Dickinson College and the 1 mile walk test (19) was used at Georgetown University.
Instructor Preparation for Laboratory: Prior to laboratory, obtain all necessary materials for the lab activity (outlined above). Preparation should take less than 30 minutes.
Student Preparation for Laboratory: Review the lab handout/protocol prior to lab.
Laboratory Activity, Submaximal Bench Step Test: Dickinson College students completed this VO2max test; the protocol can be found in the Laboratory Manual for Exercise Physiology, Exercise 7.1 (21) (https://www.humankinetics.com/AcuCustom/Sitename/DAM/082/Submaximal_Bench_Step_Test.pdf). This test is dependent on heart rate, so tell students to avoid factors that affect heart rate prior to the test: avoid exercise for 24 hours leading up to the test, fast for two hours prior to the test, and avoid food and drugs that alter heart rate prior to the test such as coffee, soda, and energy drinks. At the beginning of the test, subjects should rest for three minutes and determine resting heart rate by palpating the radial pulse for 15 seconds and multiplying by 4 to determine beats/minute. The metronome should be set to 88 beats/minute for women and 96 beats/minute for men, which corresponds to 22 steps/minute and 24 steps/minute, respectively. A foot should contact the step or the floor with each beep. The subject should step up and down for 3 minutes and switch the leading leg at 1.5 minutes. At the end of three minutes, the subject stops and palpates the radial pulse and beats are counted from 3:05-3:20 (15 seconds) and multiplied by 4 to determine the recovery heart rate (HR) in beats/minute. This number is then used to calculate VO2max with the following equations:
- Men: VO2max (ml/kg-min) = 111.33 - (0.42 x HR)
- Women: VO2max (ml/kg-min) = 65.81 - (0.1847 x HR)
Laboratory Activity, 1 Mile Walk Test: Georgetown University students completed this VO2max test; the protocol was obtained from Kline, et al., 1987 (19). Students are instructed to walk as quickly as possible while keeping one foot in contact with the ground for 1 mile around a track. The final heart rate (beats/minute), walk time (minutes), body weight, age, and sex for each participant are recorded, and the following equation is used to calculate VO2max (19):
VO2max = 6.9652 + (0.0091 x WT)-(0.0257 x age) + (0.5955 x sex) - (0.2240 x T) - (0.0115 x HR1-4), where WT = weight (lb), sex: 1 = male, 0 = female, age in years, T = walk time in minutes, and HR1-4 = fourth quarter heart rate
Analysis of VO2max data and the relationship between VO2max and genotype was done in week 8. VO2max is a complex phenotype dependent on both oxygen delivery and utilization and therefore students should be prompted to consider how best to analyze the data based on what we know about VO2max. For example, sex, fitness level, and training type (endurance vs. sprint) all have an impact on VO2max, which can help students determine how to stratify the data for ACTN3 genotype analysis. Information on biological sex, training status, training type, and any health conditions that may impact VO2max was collected at Dickinson College and biological sex information was collected at Georgetown University. Provide students with an excel file containing the anonymous compiled class data, which they can use to stratify the results and calculate average VO2max in different subgroups of students (biological sex, training status, ACTN3 genotype). We used Prism to perform statistical analysis (see the GraphPad Statistics Guide for more detailed explanations of statistical tests: https://www.graphpad.com/guides/prism/8/statistics/index.htm). The choice of statistical test is dependent on how the data are stratified and analyzed. We used two-way ANOVA for VO2max data stratified first by gender then by training status and one-way ANOVA for analysis of VO2max values among genotypes in male and female subgroups.
Anticipated Results: After collecting data in multiple classes, we obtained a total sample size of 120 (78 female, 41 males, 1 undisclosed) at Dickinson College. The sample size was large enough to stratify by biological sex when analyzing VO2max by genotype. However, the sample size was too low (n < 3) in some categories to further stratify by training status within each sex when analyzing VO2max by genotype. Participation in this lab session was optional since the VO2max test required some physical activity and not all students were able or willing to participate. However, we still had a high participation rate of 95% at Dickinson College. Overall, male students (n = 38) had a higher VO2max than female students (n = 75, p < 0.001, Figure 3A) and male trained students (n = 23) had a higher VO2max than male untrained students (n = 15, p = 0.0059, Figure 3B). No significant differences in VO2max between genotypes were observed in either males or females by either the step or walk tests (Table 2). However, the TT genotype did have the highest VO2max by the step test in both males (61.45 ± 3.64 for TT vs. 59.80 ± 2.43 for CT and 59.90 ± 2.78 for CC) and females (43.59 ± 1.08 for TT vs. 42.86 ± 0.71 for CT and 41.61 ± 1.02 for CC). At this point, we prompted the students to compare our class results with the published data we discussed during the synthesis matrix activity and then to consider what the available data mean regarding the impact of ACTN3 genotype on VO2max. If using a publication style lab report as assessment, encourage students to use this information to write about how their data compare to published results and come to an overall conclusion in the discussion section of their papers.
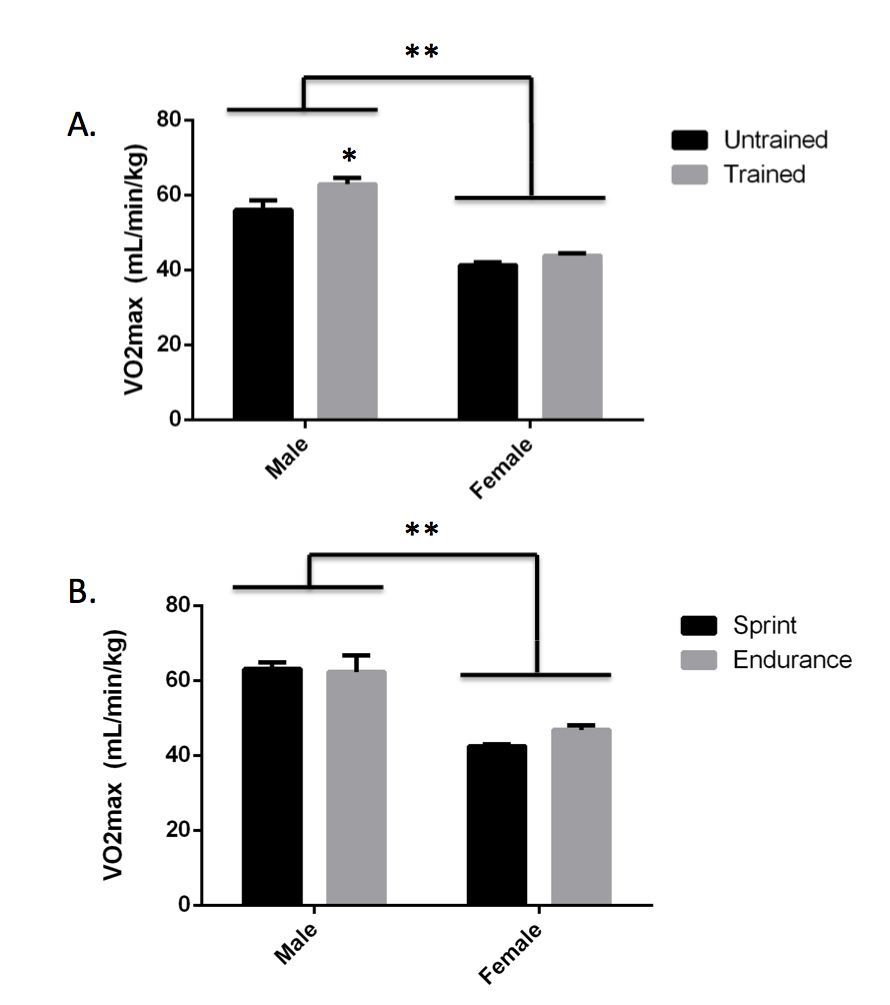
Figure 3. Effect of Sex and Training Status on VO2max. VO2max was approximated by the submaximal bench step test. Male students (n = 38) have higher VO2max than female students (n = 75, **p < 0.001). (A) Trained male students (n = 23) have higher VO2max than untrained male students (n = 15, *p = 0.0059). No difference in VO2max was observed between trained female students (n = 43) and untrained female students (n = 32, p = 0.3017). (B) Among the trained students, a trend toward higher VO2max was observed in female endurance athletes (n = 13) as compared to female sprint athletes (n = 30, p = 0.0992). No significant difference in VO2max was observed in male endurance athletes (n = 6) as compared to male sprint athletes (n = 17, p = 0.9916). Statistical analysis was done by two-way ANOVA. All bars represent average +/- standard error.
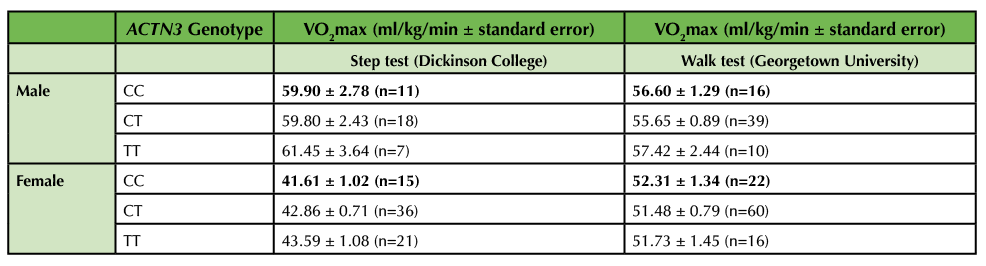
Table 2. VO2max as a function of biological sex and ACTN3 genotype. Genotype was determined by PCR-RFLP analysis. VO2max was determined indirectly by either the submaximal step test (Dickinson College) or the walk test (Georgetown University). Results are displayed as average ± standard error. No significant differences in VO2max were observed between genotypes in either males or females by one-way ANOVA.
Vertical Jump Height
During this lab activity students complete multiple trials of a countermovement jump (CMJ) in an effort to jump vertically as high as possible.
Instructor Preparation for Laboratory: Obtain necessary materials prior to the laboratory activity. Crush the chalk in a plastic bag that students can reach into to coat their fingers with chalk dust prior to jumping. Find a location that has an unobstructed wall with a high ceiling, where students can easily stand adjacent to the wall, jump, and tap. Preparation should take less than 30 minutes.
Student Preparation for Laboratory: Prior to performing any countermovement jumps, students watch a brief video that demonstrated the countermovement jump method (https://www.youtube.com/watch?v=MshBlueDsDk). They should practice the method a few times to become comfortable with it prior to beginning the experiment and making measurements.
Laboratory Activity: The lab handout for this test is included as Supporting File S16: Jump Height Lab Handout. Students work in pairs for this activity. To begin, individuals stand with their sides to the wall and feet flat on the ground. Subjects coat the hand on the arm closest to the wall in chalk dust, reach as high as possible and touch the wall. Using a tape measure, the highest point of the mark on the wall is measured as the standing reach height. From the same standing position, the subject then performs a countermovement jump, as demonstrated by the instructor, and hits the wall at the highest point of the jump. This mark is measured in the same manner as the initial standing reach. Subjects repeat their jumps and measurements five times. To calculate vertical jump height, subtract the standing reach measurement from each jumping reach measurement. Add each subject's highest vertical jump to the class data sheet for analysis.
Anticipated Results: After performing literature searches and the synthesis matrix exercise, our students identified jump height as a muscle performance exercise sometimes used by study groups examining physiological correlations with ACTN3 genotypes. This vertical jump height activity was performed during one semester in an upper level physiology class with a total class size of 22 students (16 females, 6 males). Participation in this lab session was optional; however, 19/22 (86%) students participated and those who did not participate had injuries preventing them from doing so.
Analyses of these and other data were completed during week 8 of the project. On average, males (n = 5) were able to jump higher than females (n = 14; p = 0.001; Figure 4A). The sample size was too low (n < 3) to stratify by each genotype; however, analysis was completed of ACTN3-expressing vs. ACTN3-deficient subjects and the results showed no statistically significant differences (Table 3).
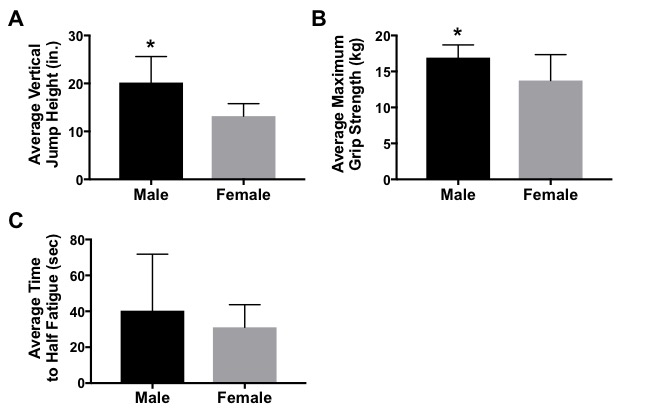
Figure 4. Effect of Sex on Muscle Performance. (A) Students performed a countermovement jump and measured the highest of five vertical jumps. Average vertical jump height is greater in males (n = 5) compared to females (n = 14) (*p<0.05). (B) Students squeezed a hand dynanometer as tightly as possible to measure grip strength. Males (n = 6) have a greater maximum grip strength than females (n = 6)(*p < 0.05). (C) Students squeezed a hand dynanometer as tightly and as long as possible for this test. The time it took students to fall half way below their maximum grip strength was measured. Time to half fatigue was not significantly different between males (n = 6) and females (n = 15)(p = 0.33). All bars represent average +/- standard error.
Week 7: Grip strength and time to half fatigue
The goal of this lesson is to analyze the relationship between the ACTN3 genotype of individuals and skeletal muscle phenotypes; specifically muscle strength and muscle fatigue time (Learning Goal 5). As students use themselves as experimental subjects and begin to analyze their physiological results, the goal is for students to be able to tie in skeletal muscle phenotypes with their ACTN3 genotypes and their knowledge of how their genotypes could impact their skeletal muscle function (Learning Goals 3 & 4).
Instructor Preparation for Laboratory: This laboratory activity requires the use of computers and LabScribe3 software, as well as iWorx Systems, Inc. equipment. Students work in groups of 3-4 with a computer and full equipment setup per group. We recommend setting up and calibrating the equipment and preparing the LabScribe3 program prior to lab (see Supporting File S17: iWorx Grip Strength Calibration Protocol). Setting up computers and equipment for six groups took one instructor approximately 60-90 minutes.
Note that cell phones and laptops interfere with the electrodes and acquisition unit and may cause background noise in the recordings. We suggest that students keep cell phones and laptops away from the workspace to avoid this issue.
Student Preparation for Laboratory: Review the lab handout/protocol prior to the laboratory.
Laboratory Activity: The lab handout for this test is included as Supporting File S18: iWorx Grip Strength Lab Handout. Grip strength is measured using the IXTA data acquisition unit and hand dynamometer from iWorx Systems, Inc. LabScribe3 software is used to record and analyze data using a slightly modified version of the Human Muscle Grip Strength and Electromyogram (EMG) Activity experimental protocol (Experiment HM-1; iWorx Systems, Inc.). Subjects use a hand dynamometer to measure grip strength as the EMG activity of the forearm muscles used to generate the subject's grip are recorded. Per the protocol, the raw EMG data is mathematically transformed so that the area under the graph of the absolute integral of the EMG is linearly proportional to the strength of the muscle contraction. To measure maximum grip strength as well as the amount of time it takes the forearm muscles to reach half maximum strength (half-maximum fatigue time), subjects connect electrocardiogram electrodes to their dominant forearms. Subjects sit with their forearms resting on the table while a baseline EMG recording is conducted for ten seconds. Subjects then squeeze the bulb of the hand dynamometer as tightly and as long as possible. The EMG recording is stopped once the subject's muscle force drops to half the initial maximum force. Record each subject's maximum grip strength and half-maximum fatigue time, and add them to the class data sheet for analysis.
Anticipated Results: The grip strength exercises were performed at Dickinson College during one semester in an upper level physiology class with a total class size of 22 students (16 females, 6 males). Participation in the grip strength and muscle fatigue experimental sessions was optional; however, 21/22 (95%) students participated and the student who did not participate had injuries preventing them from doing so.
Analyses of these data were completed during week 8 of the project. Class data demonstrated that, on average, males (n = 6) had greater maximum grip strength than females (n = 15; p = 0.027; Figure 4B). The sample set was stratified by genotype; however, due to our small n, no statistically significant differences were found between CT, CC, or TT genotypes for maximum grip strength (Table 3). Similarly, analysis of data demonstrating the time it took for students to reach half muscle fatigue showed that males (n = 6) took a longer time to reach half fatigue than females (n = 15; p = 0.33; Figure 4C); however, these results were not statistically significant. Likewise, analysis of the sample set by genotype in relation to half fatigue time showed no significant differences between genotypes (Table 3).
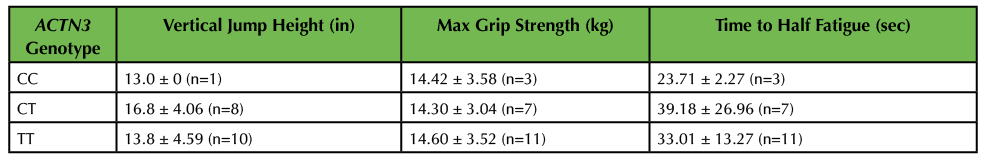
Table 3. Muscle performance as a function of ACTN3 genotype. Genotype was determined by PCR-RFLP analysis. Three measurements of muscle performance were completed, including vertical jump height, maximum grip strength, and time to half muscle fatigue. Results are displayed as average ± standard error. No significant differences were observed between genotypes by one-way ANOVA analysis.
Week 8: Data Analysis
During week 8, have students analyze all results, and encourage them to return to the literature to compare the published data on ACTN3-related muscle phenotypes to the recorded class results. The majority of VO2max publications use a direct measurement of VO2max; encourage students to consider any limitations of using either the step test or walk test to measure VO2max. Similarly, for vertical jump height, students focused on studies that used a countermovement jump; encourage them to look at studies that analyzed jump height using different techniques so they are able to discuss the strengths and weaknesses of the methods. Prompt students to think about how ACTN3 may contribute to the various muscle phenotypes. For VO2max, review the role of ACTN3 in muscle metabolism, and for the countermovement jump, consider the molecular and gross components of the muscles. In addition, encourage students to think critically about how an individual's ACTN3 genotype may influence their grip strength ability and the time it takes for an individual to reach muscle fatigue. Ask if their hypothesis regarding ACTN3 genotype and grip strength would be the same or different than their hypothesis for ACTN3 genotype and time to half fatigue, and why or why not. Task them with including a discussion of these points and how their data fit into published data sets in their lab reports.
TEACHING DISCUSSION
Vision and Change calls for the use of scientific teaching in the undergraduate biology curriculum to help students learn an established set of core concepts and competencies (2). Inquiry-based laboratory experiences are one way to achieve the goals set forth by Vision and Change (4). The series of inquiry-based teacher-collaborative laboratory modules presented here can be used at different levels of the biology curriculum to directly address Vision and Change core competencies, including the ability to apply the scientific process, use quantitative reasoning skills, tap into the interdisciplinary nature of science, communicate and collaborate across disciplines (orally and in writing), and make connections between science and society. In addition to these Vision and Change core competencies that apply to all classes using these modules, the modules also touch on core concepts set forth by Vision and Change including evolution, structure and function, information flow, exchange, and storage, and pathways and transformation of energy and matter. These core concepts can be emphasized to varying degrees, depending on the course.
The design of the laboratory modules laid out here lends itself to many different options and variations of the project. The outlined project is presented as an example of the workflow, though instructors could adapt and construct it to fit individual class needs. Certainly, we could envision the use of the scientific method module in any course, as we believe the synthesis matrix exercise is invaluable for students to make connections in the literature and design experiments. We can also envision the use of just the genotyping module, where students could take a deeper dive into the genetics of ACTN3 in themselves and the population. For example, in a genetics or population genetics class, students could be instructed to calculate allele and genotype frequencies from compiled class data and use a Chi-square test to determine whether the class population is in Hardy-Weinberg equilibrium at this allele, or whether the class allele or genotype distribution fits within different population distributions. If there is no interest in physiology, a class could work through weeks 1-5 and week 8, with the possibility of combining weeks 3 and 4 and/or weeks 5 and 8, for example. In addition, the order of the physiology modules presented here is a suggestion for a timeline of completion, but could certainly be done in any order or combination depending upon the class focus. If an instructor wants to complete the entire project in a shorter timeframe he or she can manipulate our suggested module outline as desired. For example, both weeks 1 and 2 could be done during lecture to introduce ACTN3 and plan experiments. Weeks 3 and 4 could be completed in the same lab session. Weeks 5 and 6 could be completed in the same lab session if the instructor sets up the restriction enzyme digests for the class to save time. Instructors may be interested in performing only one physiology experiment and could choose one described here or another that may come out of the synthesis matrix exercise. Additionally, analysis could be done in any given week(s) as the project progresses instead of in a separate lab session as suggested. We see the flexibility and adaptability of these modules as a major strength and benefit of this project and its effective use in the laboratory.
As we have continued to use the synthesis matrix activity as part of this project, the modules have evolved through student input into the types of muscle performance experiments they would like to conduct. You may find that as students go through the synthesis matrix exercise, they may find other physiological tests that could be completed as part of this project as well, such as sprinting. The synthesis matrix allows students to take ownership of the project. Allowing them to do literature searches to discover the different ways to perform genotyping and/or the different phenotypes that other groups have measured, and then make decisions about how to design their own experiments and what physiological tests to perform, worked to engage students and have them look forward to the laboratory activity each week. Students gave this feedback during anonymous surveys throughout the project, which supports that this learning goal was achieved.
One of the learning goals of these modules is for students to understand that skeletal muscle function is a complex phenotype that is the result of many different genes and environmental factors. Through the analysis of published results and class data it is obvious that ACTN3 contributes a small amount to skeletal muscle phenotypes, if at all, in a college student population. Based on published information, you can have the students do a power analysis (more information about power analysis can be found in the GraphPad Statistics Guide: https://www.graphpad.com/guides/prism/8/statistics/index.htm) at the beginning of the experiment to determine how large of a sample size is needed for various phenotypes. Unless you are teaching a large class, it is unlikely that a large enough sample size will be collected in one semester, especially if the data are stratified by sex and training status. Given that the phenotypes are measured by the same method, data can be compiled over the years for analysis. Supporting File S19: ACTN3 Data Spreadsheet contains the data we collected from 5 classes using the submaximal bench step test for VO2max (n = 120) and the grip strength/fatigue (n = 21) and jump height data from 1 class (n = 20).
We are considering improvements in a few areas for future iterations of the modules. As mentioned previously, we collected student information on biological sex, training status, training type, and any health conditions that may impact VO2max, jumping, or grip strength at the beginning of project. We categorized students as trained vs. untrained, where they were considered trained if they participated in a collegiate sport and untrained if not. Similarly, we stratified those trained students as having either sprint or endurance training. Moving forward we will consider being more specific and clearer in our definitions of trained vs. untrained and sprint vs. endurance. Many students argued that they exercised daily and so should be considered trained, which we need to take into consideration. Also, most student athletes had trouble categorizing themselves as sprint vs. endurance and felt they had a mixture of both types of training. If we want to use this information to stratify the study set and analyze data, we will need to refine our definitions. Additionally, we would be more mindful in future iterations to connect the phenotypic laboratory exercises with corresponding topics in the lecture setting. Since this project is run over 8 weeks, at times the lecture topics do not correspond with the topics of the lab activities. In the future, we would do more pre-laboratory instruction/lecture to help students connect the larger concepts and the structure and function relationship of ACTN3 within the given lab exercise.
Most students seemed to enjoy the laboratory sessions related to this project. Many students self-reported on anonymous class surveys and course evaluations that they liked the aspect of using themselves as test subjects, exploring their own genotypes and phenotypes and thinking about the connections between the two by using the knowledge learned in class and laboratory. At the end of the lab and while writing the discussion sections of their final papers, students were pushed to consider what the data meant. In our experience, many students were used to labs with a pre-defined outcome where they are supposed to find a difference or clear answer to the questions being investigated. However, analysis of class data alongside published results on how ACTN3 contributes to muscle physiology forces the students to conclude that a single gene contributes very little, if at all for some phenotypes, to the complex trait of muscle function in a college student population. Some students wanted to conclude that the experiment didn't work, which is a simplistic and inaccurate interpretation of the results. Therefore, students should be prompted to think more deeply about the results and what they mean for genetic testing of ACTN3 in athletes, which brings the whole project back to the non-scholarly articles on ACTN3 being the "gene for speed." Students should conclude for themselves whether this is an accurate statement based on the data. In our experience, a class discussion about the conclusions that can be drawn from the results is required for most students to realize that the experiment worked on a technical level, but the data don't support a strong contribution of ACTN3 to muscle performance in a college student population. In addition, depending on the course topic, the instructor can encourage students to think about how the results of this project relate to the concept of precision medicine. Most human diseases are complex in nature, and therefore this project can be used as a jumping-off point to discuss where the field of precision medicine currently stands and the challenges associated with personalized diagnoses and therapies.
ACKNOWLEDGEMENTS
This project was reviewed and approved by the Institutional Review Board (IRB) at Dickinson College (protocol #340) and Georgetown University (protocol # 212/00000336).
SUPPORTING MATERIALS
- S1. ACTN3 Polymorphism: IRB Application
- S2. ACTN3 Polymorphism: IRB Exemption
- S3. ACTN3 Polymorphism: Lab Notebook Rubric
- S4. ACTN3 Polymorphism: Scientific Writing Rubric
- S5. ACTN3 Polymorphism: Working in Groups - Lecture Slides
- S6. ACTN3 Polymorphism: Guided Reading Handout
- S7. ACTN3 Polymorphism: Guided Reading Interactive Lecture - Lecture Slides
- S8. ACTN3 Polymorphism: Synthesis Matrix-Blank
- S9. ACTN3 Polymorphism: Synthesis Matrix-Example
- S10. ACTN3 Polymorphism: Demographic Information Form
- S11. ACTN3 Polymorphism: ACTN3 Genotyping Lab Preparation Guide
- S12. ACTN3 Polymorphism: Micropipette Guide
- S13. ACTN3 Polymorphism: ACTN3 Genotyping Lab Handout
- S14. ACTN3 Polymorphism: ACTN3 Genotyping Activity
- S15. ACTN3 Polymorphism: ACTN3 Genotyping Activity Answer Key
- S16. ACTN3 Polymorphism: Jump Height Lab Handout
- S17. ACTN3 Polymorphism: iWorx Grip Strength Calibration Protocol
- S18. ACTN3 Polymorphism: iWorx Grip Strength Lab Handout
- S19. ACTN3 Polymorphism: ACTN3 Data Spreadsheet
References
- Genetics Society of America Education Committee. 2015. Core Concepts and Core Competencies in Genetics. cited 2018; Available from: http://www.genetics-gsa.org/education/GSAPREP_CoreConcepts_CoreCompetencies.shtml.
- American Association for the Advancement of Science. 2011. Vision and Change in Undergraduate Biology Education: A Call to Action. cited 2018; Available from: http://visionandchange.org.
- Michael J, McFarland J. 2011. The core principles ("big ideas") of physiology: Results of faculty surveys. Adv Physiol Educ 35:336-341.
- Handelsman J, Ebert-May D, Beichner R, Bruns P, Chang A, DeHaan R, Gentile J, Lauffer S, Stewart J, Tilghman SM, Wood WB. 2004. Scientific teaching. Science 304:521-522.
- Beck C, Butler A, Burke da Silva K. 2014. Promoting inquiry-based teaching in laboratory courses: Are we meeting the grade? CBE Life Sci Educ 13:444-452.
- D'Avanzo C. 1996. Three ways to teach ecology labs by inquiry: Guided, open-ended, and teacher collaborative. Bull Ecol Soc Am 77:92-93.
- Weaver GC, Russell CB, Wink DJ. 2008. Inquiry-based and research-based laboratory pedagogies in undergraduate science. Nat Chem Biol 4:577-580.
- North KN, Yang N, Wattanasirichaigoon D, Mills M, Easteal S, Beggs AH. 1999. A common nonsense mutation results in alpha-actinin-3 deficiency in the general population. Nat Genet 21:353-354.
- MacArthur DG, North KN. 2007. ACTN3: A genetic influence on muscle function and athletic performance. Exerc Sport Sci Rev 35:30-34.
- Yang N, MacArthur DG, Gulbin JP, Hahn AG, Beggs AH, Easteal S, North K. 2003. ACTN3 genotype is associated with human elite athletic performance. Am J Hum Genet 73:627-631.
- Moran CN, Yang N, Bailey ME, Tsiokanos A, Jamurtas A, MacArthur DG, North K, Pitsiladis YP, Wilson RH. 2007. Association analysis of the ACTN3 R577X polymorphism and complex quantitative body composition and performance phenotypes in adolescent Greeks. Eur J Hum Genet 15:88-93.
- Friedlander SM, Herrmann AL, Lowry DP, Mepham ER, Lek M, North KN, Organ CL. 2013. ACTN3 allele frequency in humans covaries with global latitudinal gradient. PLoS One 8:e52282.
- Pickering C, Kiely J. 2017. ACTN3: More than just a gene for speed. Front Physiol 8:1080.
- Scott RA, Irving R, Irwin L, Morrison E, Charlton V, Austin K, Tladi D, Deason M, Headley SA, Kolkhorst FW, Yang N, North K, Pitsiladis YP. 2010. ACTN3 and ACE genotypes in elite Jamaican and US sprinters. Med Sci Sports Exerc 42:107-112.
- Yang N, MacArthur DG, Wolde B, Onywera VO, Boit MK, Lau SY, Wilson RH, Scott RA, Pitsiladis YP, North K. 2007. The ACTN3 R577X polymorphism in East and West African athletes. Med Sci Sports Exerc 39:1985-1988.
- Berman Y, North KN. 2010. A gene for speed: The emerging role of alpha-actinin-3 in muscle metabolism. Physiology 25:250-259.
- Del Coso J, Hiam D, Houweling P, Perez LM, Eynon N, Lucia A. 2019. More than a 'speed gene': ACTN3 R577X genotype, trainability, muscle damage, and the risk for injuries. Eur J Appl Physiol 119:49-60.
- Weiglein L, Herrick J, Kirk S, Kirk EP. 2011. The 1-mile walk test is a valid predictor of VO2max and is a reliable alternative fitness test to the 1.5-mile run in U.S. Air Force males. Mil Med 176:669-673.
- Kline GM, Porcari JP, Hintermeister R, Freedson PS, Ward A, McCarron RF, Ross J, Rippe JM. 1987. Estimation of VO2max from a one-mile track walk, gender, age, and body weight. Med Sci Sports Exerc 19:253-259.
- McArdle WD, Katch, F.I., Pechar, G.S., Jacobson, L., Ruck, S. 1972. Reliability and interrelationships between maximal oxygen uptake, physical work capacity and step-test scores in college women. Med Sci Sports 4:182-186.
- Haff GG, Dumke C. 2012. Laboratory Manual for Exercise Physiology. 1st Edition. Champaign, IL: Human Kinetics.
Article Files
Login to access supporting documents
The ACTN3 Polymorphism: Applications in Genetics and Physiology Teaching Laboratories(PDF | 454 KB)
S1.ACTN3 Polymorphism-IRB Application.pdf(PDF | 144 KB)
S2.ACTN3 Polymorphism-IRB Exemption.pdf(PDF | 55 KB)
S3.ACTN3 Polymorphism-Lab Notebook Rubric.docx(DOCX | 18 KB)
S4.ACTN3 Polymorphism-Scientific Writing Rubric.docx(DOCX | 24 KB)
S5.ACTN3 Polymorphism-Working in Groups - Lecture Slides.pptx(PPTX | 56 KB)
S6.ACTN3 Polymorphism-Guided Reading Handout.docx(DOCX | 17 KB)
S7.ACTN3 Polymorphism - Guided Reading Interactive Lecture - Lecture Slides.pptx(PPTX | 532 KB)
S8.ACTN3 Polymorphism-Synthesis Matrix-Blank.docx(DOCX | 43 KB)
S9.ACTN3Polymorphism-Synthesis Matrix-Example.docx(DOCX | 96 KB)
S10.ACTN3 Polymorphism-Demographic Information Form.docx(DOCX | 54 KB)
S11.ACTN3 Polymorphism-Genotyping Lab Preparation Guide.docx(DOCX | 19 KB)
S12.ACTN3 Polymorphism-Micropipette Guide.docx(DOCX | 87 KB)
S13.ACTN3 Polymorphism-ACTN3 Genotyping Lab Handout.doc(DOC | 47 KB)
S14.ACTN3 Polymorphism-Genotyping Activity.docx(DOCX | 39 KB)
S15.ACTN3 Polymorphism-ACTN3 Genotyping Activity Answer Key.docx(DOCX | 63 KB)
S16.ACTN3 Polymorphism-Jump Height Lab Handout.docx(DOCX | 103 KB)
S17.ACTN3 Polymorphism- iWorx Grip Strength Calibration Protocol.docx(DOCX | 80 KB)
S18.ACTN3 Polymorphism-iWorx Grip Strength Lab Handout.docx(DOCX | 146 KB)
S19.ACTN3 Polymorphism-ACTN3 Data Spreadsheet.xlsx(XLSX | 43 KB)
- License terms

Comments
Comments
There are no comments on this resource.