Differential Gene Expression during Xenopus laevis Development
Published online:
Abstract
In Developmental Biology classes, students are challenged with understanding how differential gene expression guides embryonic development. It can be difficult for students to realize that genes need to be turned on or off at the right time and place in order for development to proceed normally. In this lab, students working in groups perform experiments with live embryos and visualize differential gene expression allowing them to become invested in their experiment and curious about the results. This lab also addresses the benefits of Xenopus laevis as a model organism and allows students to observe the changes Xenopus embryos undergo during early embryonic stages. After the students have chosen and fixed two stages of Xenopus embryos, they perform an in situ hybridization on the embryos to visualize gene expression at two different developmental stages. They then compare their results with those from other lab groups who analyzed their embryos for different genes. The students self-reported that they better understood the concept of differential gene expression during vertebrate development and enjoyed doing this series of lab experiments working with live materials.
Citation
Shifley ET. 2019. Differential gene expression during Xenopus laevis development. CourseSource. https://doi.org/10.24918/cs.2019.32.Society Learning Goals
Developmental Biology
- Signaling
- How does differential gene expression mediate progressive acquisition of cell fate?
- Experimental Approaches
- How do different organisms help us understand development? And what are their strengths and limitations?
Lesson Learning Goals
From Developmental Biology Learning Framework:- "How do different organisms help us understand development? And what are their strengths and limitations?"
- "How does differential gene expression mediate progressive acquisition of cell fate?"
Lesson Learning Objectives
Students will be able to:- identify different stages of Xenopus development
- contrast the strengths and limitations of the Xenopus model organism
- explain the process and purpose of in situ hybridization
- compare gene expression patterns from different germ layers or organ domains
- compare gene expression patterns from different developmental stages
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
An important concept for undergraduate science majors learning about Developmental Biology is how differential gene expression guides embryonic development. The idea that genes are only turned on at the correct time and location in developing embryos during the differentiation of various cell types, tissues, and organs is often a difficult idea for undergraduates to grasp. A number of active learning techniques help undergraduates become motivated and successful in learning difficult concepts (1,2) and have been identified as a priority for science education (3). This series of lab exercises uses structured inquiry experiments to help students visualize differential gene expression.
Jeff Hardin has discussed how students in Developmental Biology often have trouble thinking in four-dimensions: both the three dimensional embryo as well as developmental timing (4). In these exercises, the students observe Xenopus embryos as they undergo early development, which helps them understand how these embryos have biologically significant three dimensional structures that change over time. Additionally, students from different lab groups are given in situ hybridization probes for different genes expressed in each of the three germ layers so that while they are visualizing differential gene expression, they are also comparing how the germ layers are distinct groups of cells within the three-dimensional embryo.
Other laboratory exercises have used techniques like RT-PCR, qRT-PCR, and flow cytometry in situ hybridization to analyze gene expression differences between cancerous cell lines to help undergraduates learn through inquiry about differences in gene expression (5). Post-graduate students have also used in situ hybridization in Drosophila embryos, manipulating cis-regulatory elements and transcription factors to examine the regulation of gene expression (6). Additionally, there are good examples of other labs using Drosophila to illustrate embryo morphology, mutant phenotypes, loss of function, gain of function, and genetic interaction studies (7). In each of these examples, students are introduced to a number of important techniques while working with different model organisms and participating in on-going research (5-7). In this series of lab exercises after the students have fertilized and observed early Xenopus development, they use their collected embryos to gain experience with whole mount in situ hybridization, a common and useful technique for developmental biologists. The structured inquiry nature of this lab not only allows the students to learn a new technique, it also provides an excellent visualization of differential gene expression in developing Xenopus embryos both spatially and temporally (Figure 1A).
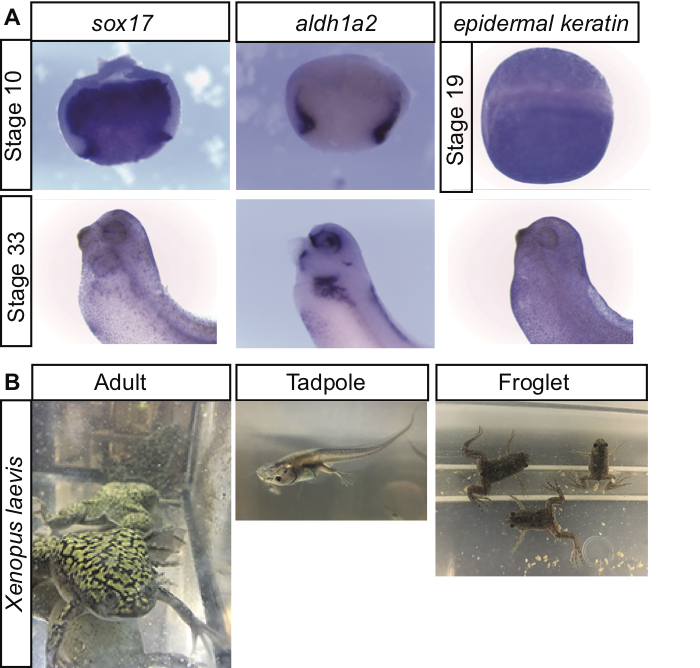
Figure 1. (A) Xenopus laevis differential gene expression. Students fixed embryos at early (Stage 10 or 19) and later (Stage 33) developmental time points. In situ hybridization shows differential gene expression in the embryos with endodermal (sox17), mesodermal (aldh1a2), and ectodermal (epidermal keratin) genes. The Stage 10 embryos were bisected during the in situ hybridization which allows students to see non-overlapping domains of endoderm and mesoderm in early gastrula embryos. Later at Stage 33, sox17 becomes restricted to the foregut endoderm and gall bladder while aldh1a2 is expressed in the eye, kidney, forebrain, and lateral plate mesoderm. The whole Stage 19 embryo is shown in the dorsal view so that students can notice how epidermal keratin is expressed throughout the ectoderm, but is excluded from the neural tube. (B) Xenopus laevis is an excellent model organism for studies in Developmental Biology. Students enjoyed observing not only the early embryonic stages with the in situ hybridization, but also the later stages as tadpoles underwent metamorphosis.
Another important concept for undergraduate science majors to explore are the benefits and limitations of various model organisms used by researchers. Xenopus laevis is an accessible model organism for undergraduates that provides hundreds of embryos in a single day that are large and easily manipulated (8). In the first part of this set of lab exercises, students fertilize Xenopus eggs, de-jelly and sort viable embryos, and observe the first cleavage. Others have successfully used undergraduates to fertilize Xenopus embryos as well as perform animal cap assays and examine oocyte maturation with experiments carried out over two full days of experimentation (9). In contrast, the experiments in this lesson are spread out over three once-a-week three hour lab periods, which might be more typical of an upper level biology class at a university.
INTENDED AUDIENCE
This series of lab exercises was designed for a Developmental Biology class at a primarily undergraduate 4-year university. This was an upper-level elective class for Biology majors and most of the students enrolled were either Juniors or Seniors.
REQUIRED LEARNING TIME
The first week experiment (fertilizing Xenopus laevis eggs) was completed in a 3 hour lab period. The students were required to occasionally check on their embryos outside of the lab period over the following two to three days and fix two different stages of embryos, which took ~15 minutes/day. The second week experiment (first day of in situ hybridization) was completed in a 3 hour lab period. The instructor continued the next two days of in situ washes outside of the lab period. The students were required to place their embryos in staining solution the day before the third lab period, which took ~30 minutes. The third week experiment (photographing and comparing their in situ hybridizations) was completed in ~2 hours of the lab period. The approximate times for each of the lab activities are listed in Table 1, which includes a significant time investment before the labs for the instructor to prepare all reagents and solutions for the experiments.
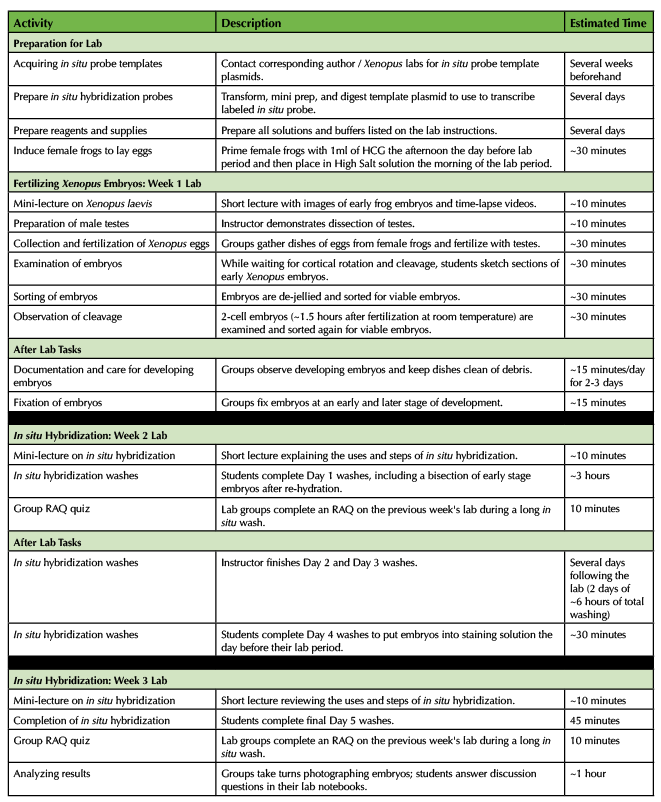
Table 1. Differential Gene Expression during Xenopus laevis Development Labs - Teaching Timeline
PREREQUISITE STUDENT KNOWLEDGE
The students enrolled in this course were required to have completed Introduction to Biology courses and therefore had basic knowledge of genetics and general lab skills such as pipetting and proper use of stereomicroscopes. In lecture, we had covered chapters on identity and patterning, fertilization, differential gene expression, and cell to cell communication before performing the lab, although these exercises could also easily accompany later chapters on amphibian development.
PREREQUISITE TEACHER KNOWLEDGE
The instructor should be familiar with in situ hybridization procedures and have some background knowledge in Developmental Biology. For this course we used the 11(th) Edition of Developmental Biology by Gilbert and Barresi as our textbook resource. Other great resources on the Xenopus model organism include Xenbase (8) http://www.xenbase.org/entry/ and Early Development of Xenopus laevis: A Laboratory Manual (10).
SCIENTIFIC TEACHING THEMES
ACTIVE LEARNING
With structured inquiry, students are tasked with fertilizing their own plates of embryos, observing and keeping their embryos alive over several days, and then performing an in situ hybridization on these same embryos (Supporting File S1: Xenopus gene expression - Fertilization instructions and Supporting File S2: Xenopus gene expression - In situ hybridization instructions). This creates a sense of ownership and investment for each lab group in the experiment. The students work in groups of 3 or 4 performing all aspects of the exercises gaining hands-on experience caring for, manipulating, washing, and photographing developing embryos. The groups are assessed together with a Readiness Assessment Quiz (RAQ) every week on the previous week's lab.
ASSESSMENT
The students are required to document their work in an individual lab notebook where they record their procedures, results such as sketches or photographs of the developing embryos, and answers to thought questions at the end of each lab (Supporting File S3: Xenopus gene expression - Lab notebook instructions). The lab notebooks are graded twice during the semester for completion. Additionally, each week the lab groups are given a Readiness Assessment Quiz (RAQ) which tests their basic understanding of the previous week's lab (Supporting File S4: Xenopus gene expression -Readiness Assessment Quiz 1 and Supporting File S5: Xenopus gene expression - Readiness Assessment Quiz 2). Lastly, the students take individual lab exams at three points during the semester testing their understanding of the learning objectives of each of the lab exercises performed (Supporting File S6: Xenopus gene expression - Lab exam questions). At the end of the semester, students responded to a survey questioning their impressions of the lab (approved by the Institutional Review Board of Northern Kentucky University).
INCLUSIVE TEACHING
The syllabus for the course includes a diversity statement as well as a statement listing the instructor’s pronouns and welcoming students to share their own pronouns with the instructor. The syllabus also has a detailed outline of all of the assignments and due dates for the semester to provide a highly structured course including many low stakes assessments throughout the semester, which helps increase student success and decrease the achievement gap (11-13). Students are first introduced to the topic of differential gene expression in an active learning lecture which includes the use of clicker questions and think-pair-share activities, both of which engage a diverse population of students (14,15). During the lab period, the material is accessible in several different formats to facilitate different learning styles including a textbook, a short power-point lecture with diagrams, and videos of early amphibian development. All students are given the opportunity to perform Developmental Biology lab techniques in this series of exercises. They are in permanent lab groups of 3-4 people to foster collaboration and every student is encouraged to take turns at the microscope and with the embryos. Others have found that using guided inquiry work with living organisms to introduce laboratory skills improved their underserved STEM student’s attitudes and success (16). The groups were self-selected on the first day of lab, but could certainly be assigned to encourage a mixture of students.
LESSON PLAN
XENOPUS LAEVIS AND OTHER SUPPLIES
If your institution has a colony of Xenopus laevis frogs available, you would need approximately 1 male and 4 female frogs for each lab section. Using four female frogs means that even if some of the females do not lay viable eggs, you will have plenty as long as one of the other females lays viable eggs. If your institution does not have Xenopus, you can purchase breeding pairs of frogs from Carolina Biological, Ward's Scientific, Nasco, or Xenopus Express for approximately $60. However, the husbandry and upkeep for the adult frogs may not be worth the time and expense if they are only going to be used for this series of lab exercises once a year. Another convenient option is to purchase fertilized embryos, which are available from Nasco, Xenopus Express, and Carolina Biological for approximately $40. Although the students would not be able to observe the cortical rotation or early cleavage, they would still be able to practice staging the embryos, to observe the embryos develop and change three-dimensionally over time, and to fix them for in situ hybridization. This would also not require the submission of an IACUC protocol if you are not using adult Xenopus or keeping older tadpoles.
There are a several solutions required for the fertilization and in situ hybridization lab exercises. These will need to be prepared over the course of several days before the lab, and are listed at the end of the student lab instructions (Supporting File S1: Xenopus gene expression - Fertilization instructions and Supporting File S2: Xenopus gene expression - In situ hybridization instructions).
Additionally, before running this lab, the instructor will need to acquire in situ hybridization probes. The Xenopus research community is incredibly generous in sharing the plasmids for in situ probes if you contact them. The ones used in this lab (like sox17, aldh1a2, and epidermal keratin) are available by contacting the author (shifleye1@nku.edu). There are also many other excellent options of probes that give interesting expression patterns, especially if your students want to look at a specific region or organ domain of the developing embryo. For instance, some additional good options of genes with distinct expression patterns include mesodermal markers nkx2.5, foxf1, or tbxt and ectodermal markers sox9, foxi1e, or snail2 and endodermal markers hhex, endodermin, or mixer. If you search on http://www.xenbase.org/entry/ for any gene of interest, it will provide a page with lots of information on your gene and under the expression tab, you can view in situ hybridization images that have been generated from different labs and publications including the corresponding author (8). After acquiring the plasmids with your genes of interest, you will need to digest them to use as a template to make the in situ probes using the instructions included in Supporting File 7 (Supporting File S7: Xenopus gene expression -In situ hybridization probe instructions).
XENOPUS LAEVIS FERTILIZATION AND EARLY DEVELOPMENT - LAB WEEK 1
The afternoon before the lab period, the instructor primes the female frogs by injecting near their nuptial pads 1ml of Human Chorionic Gonadotropin (HCG) to stimulate the frog to begin laying eggs. Females will start to lay eggs ~9 hours after injection if they are kept at RT or ~14 hours after injection if they are kept at 15°C, and will continue to lay eggs for ~8 hours (10). The morning of the lab period, the female frogs are placed in a high salt solution to keep them happy and healthy (10).
At the beginning of lab, the instructor gives a short ~10 minute power-point presentation introducing the benefits of Xenopus laevis as a model organism as well as the three-dimensional structure of the early cleavage and gastrula stage embryos using images and time-lapse videos (Supporting File S8: Xenopus gene expression -Introduction to Xenopus PowerPoint). The instructor also demonstrates how to remove the testes from an anestisized male frog. The groups of students then each collect a petri dish full of embryos from the high salt tanks holding the egg-laying female frogs and fertilize them with a piece of the testes following the lab procedure (Supporting File S1: Xenopus gene expression - Fertilization instructions). Once the embryos have undergone cortical rotation, the students de-jelly the embryos and examine them under a dissecting microscope to begin to sort viable from inviable embryos. Students are instructed to remove white, marbled, or unfertilized eggs from their plates. It can be challenging initially for students to find the dead or dying embryos with their transfer pipet while looking through the dissecting microscope, but encourage them to take turns within their group. While the students are waiting for their remaining viable embryos to develop, they can sketch and label prepared sections of the various stages of frog embryos (available from Carolina Biological or Anatomy Warehouse).
Towards the end of the lab period (~90 minutes after fertilization at room temperature), the embryos should undergo their first cleavage and the students can again sort their plates, removing embryos with irregular cleavage planes. Students then place their plates of embryos at 16-18°C at the end of the lab period for overnight incubation. Each group is tasked with sketching or photographing their developing embryos, clearing their plates of dead embryos, and replacing the culture medium for the next 2-3 days. They can move their plates of embryos out to room temperature the next day or keep them at cooler temperatures to slow their development (a chart outlining the speed of development at different temperatures is included in their instructions). Each group is instructed to fix a set of embryos at two different developmental stages (ideally at least 6 embryos per stage). They pipet the embryos into a collection jar or vial, remove most of the culture media, and then cover the embryos with MEMFA fixative and place at 4°C.
Working with live organisms always carries a risk that things may not go as planned. By having 4 females lay eggs, there are likely to be hundreds of viable eggs, but it is possible that none of the females lay good eggs on the day of the lab. If they do not, students can at least sketch the prepared slides of Xenopus embryos and watch the videos showing the early development. If possible, it would be worthwhile for the instructor to have some reserve embryos (~50 young stages, ~50 tailbud stages) fixed and stored in ethanol at -20°C so that if the frogs do not lay eggs, the groups can still use these extra embryos for the in situ hybridization part of the lab.
IN SITU HYBRIDIZATION - LAB WEEK 2
The fixed embryos are dehydrated with ethanol and can be stored at -20°C for any amount of time until the instructor is ready to start the in situ hybridization lab. For this week of lab, the instructor again gives a short ~10 minute presentation explaining how in situ hybridization works and why the results of in situ hybridization are useful (Supporting File S9: Xenopus gene expression -Introduction to in situ hybridization PowerPoint). The students follow the procedure for Day 1 of in situ hybridization on their Xenopus embryos (Supporting File S2: Xenopus gene expression - In situ hybridization instructions). As part of day 1, the instructor demonstrates how early stage Xenopus embryos can be bisected for better staining during the in situ hybridization. This is the most technically challenging part of the lab because the student must position a round embryo on an agarose coated plate and cut it in half with a razor blade. The instructor can rotate through to help each group. Letting each student try to bisect an embryo is a good experience. Usually there are so many embryos it's not a problem if most of them get crushed rather than cut. Each group really only needs one or two successfully bisected embryos to get a good picture. I would also suggest each group leaves several young embryos whole in case none of the bisected embryos make it through the in situ. The whole embryos will still stain with the protocol (for instance epidermal keratin in Figure 1A), but if the students are successful in bisecting an embryo, they are neat to photograph because cavities like the archenteron are apparent and it is easier to compare mesodermal and endodermal genes (for instance sox17 and aldh1a2 in Figure 1A).
Each group is assigned a different gene and given a prepared Digoxigenin (Dig) labeled probe. During some of the long washing steps of this lab, students can also work through an additional excellent exercise by Dr. Cebra-Thomas making Play-doh models of the different stages of gastrulation (17) https://www.swarthmore.edu/NatSci/sgilber1/DB_lab/Frog/frog_gast_model.html. The students should at least get their embryos into the pre-hybridization wash before the end of the lab period depending on how long their bisection takes. After this lab period, the instructor or a teaching assistant can take over the in situ washes, performing Day 2 and Day 3 washes on subsequent days. If you have labs that meet more than once a week, your students could be involved with some of these washes as well. The embryos are left in the last 1X MAB wash at 4°C over the weekend, and each group is tasked with completing the Alkaline Phosphatase washes on the day before their next lab period. This takes ~30 minutes for a student to complete. The embryos remain in the staining solution overnight at room temperature.
IN SITU HYBRIDIZATION - LAB WEEK 3
After a brief review of the steps of in situ hybridization that have been completed, students view their embryos under a microscope to determine if a specific staining pattern has appeared. The probes provided to each group typically give strong staining. They complete several washes to stop the staining and then each group takes turns photographing their embryos with a microscope imaging system. We used a Leica dissecting scope with a camera. If this is not available the students could hold their phone cameras carefully above a microscope eyepiece to take a picture of their embryos. All the photographs from each of the lab groups are shared online so that the students can compare their gene's expression pattern with others and students also post their photographic results in their individual lab notebooks. This last part of the lab does not take the entire 3 hour period and could accompany another lab exercise or activity within the same lab period. The students can also use the extra time to research their gene expression pattern, make comparisons between groups, and write conclusions in their lab notebooks.
ASSESSMENT
Lab groups are assessed with a Readiness Assessment Quiz (RAQ) each week in lab on the previous week’s material (Supporting File S4: Xenopus gene expression -Readiness Assessment Quiz 1 and Supporting File S5: Xenopus gene expression - Readiness Assessment Quiz 2). Every student is responsible for keeping an individual laboratory notebook documenting the procedures, results, and answers to discussion questions for the lab (Supporting File S3: Xenopus gene expression - Lab notebook instructions). Additionally, students have individual lab exams assessing material from the labs (Supporting File S6: Xenopus gene expression - Lab exam questions). In the final grade calculations, the group RAQs were worth 8%, the individual lab notebook was worth 5%, and the lab exams were worth 15%. For the RAQ and lab exams, the questions are mostly at remember or understand levels of Bloom’s taxonomy, although some questions ask them to apply what they had learned to a new situation. The questions at the end of each lab exercise that the students answer in their lab notebooks also require analyzing.
TEACHING DISCUSSION
This series of lab exercises worked well in giving the students experiences with living organisms, specifically Xenopus laevis. The ownership and responsibility the students felt towards their embryos during the lab exercises was encouraging to see. The students were also able to learn about and participate in a common experiment used by Developmental Biologists, in situ hybridization, to examine differential gene expression. Visualizing the different layers or groups of cells staining dark purple with endodermal, mesodermal, or ectodermal genes was a great way to help students see that as different germ layers form and cells differentiate in the embryo, they express different genes (Figure 1A). Additionally, depending on the gene assigned and the stages they fixed, students were able to see temporal changes in gene expression over developmental time as well (Figure 1A). For instance, SRY-box 17 alpha (Sox17a) is expressed throughout the endoderm in early stage embryos where it helps specify the endoderm, but at later tailbud stages its expression becomes restricted to the foregut endoderm and gall bladder (18-20). Aldehyde dehydrogenase 1 family member A2 (alhd1a2) is a retinoic acid generating enzyme that is expressed in the gastrulating Xenopus mesoderm (21) and plays diverse roles during vertebrate development (for instance (22)) while epidermal keratin is expressed in the non-neural ectoderm (23-25). When the students compared their gene expression pattern with other groups, they were able to contrast these different gene expression patterns and see how they changed over developmental time.
STUDENT IMPRESSIONS
The students who were enrolled in this class came in with a mixture of experience or no experience in undergraduate research as well as a mixture in their level of interest in Developmental Biology (Figure 2A). Nearly all students reported being more interested and more informed about the types of experiments Developmental Biologists perform at the end of the semester (Figure 2A). The students clearly enjoyed the Xenopus series of lab exercises. Many of them wrote that their favorite part was “working with live organisms” and “getting to see all of the stages of development starting from fertilization.” They also reported learning more from this lab than a typical lab and agreed or strongly agreed that the lab helped them visualize embryonic development and differential gene expression (Figure 2B). One aspect of the lab that was a concern was the requirement that the students come to the lab outside of the regularly scheduled lab period to document, fix, or wash embryos. The student responses show that this was a problem for 20% of the class (Figure 2B). Indeed, at our institution ~40% of the undergraduates are first-generation college students and many are working outside jobs at the same time as attending school or commuting. We talked about trying to coordinate within their group of 3 or 4 students to share this responsibility and the instructor also offered to cover a certain day if they needed help. However, the goal was still to encourage them to observe the developing embryos themselves, if possible, on the 2-3 weekdays following the fertilization. This aspect could also be addressed at the beginning of the semester when the groups are first formed to make sure each group has someone who can come into lab outside of class time. I also included in the IACUC protocol for this lab exercise to house extra tadpoles from our fertilization over the course of the semester and the students loved seeing the tadpoles start metamorphosis by the end of the semester, which was something they could observe from week to week in lab and did not require outside class time.

Figure 2. Student evaluation responses. (A) Student evaluation responses on their experience and interests. The students reported an increased interest and understanding of Developmental Biology research after having completed the course. (B) Student evaluation responses to the Xenopus series of lab exercise. Overall, the students enjoyed the lab, especially working with live materials. The students felt that the learning objectives were achieved. Some students had a problem coming into lab outside of the regularly scheduled period.
EXTENSIONS
In this lab, the students only observed the normal development of Xenopus laevis and normal gene expression patterns. One great way to expand on this lab would be to allow groups of students to perform different manipulations on their developing embryos. Others have outlined inquiry-based labs where students were tasked with researching the impacts of environmental toxins on development, writing and screening their research proposals, and then carrying out their manipulations on sea urchins or chick embryos and finally writing a report on their results (26). This series of labs was found to increase the students motivation and performance (26). Students have also done genetic and environmental manipulations with developing zebrafish and Caenorhabditis elegans models (27). For Xenopus, if the female frogs are laying a good number of eggs, each group of students could perform any number of environmental manipulations on half of their embryos, keeping the other half as controls. One example is to treat some of the embryos with Lithium Chloride as outlined by Justin Spenillo (28) https://www.swarthmore.edu/NatSci/sgilber1/DB_lab/Frog/JA_Spenillo/justin2.html). There are also a large number of small molecule inhibitors that Xenopus embryos can be cultured in if there is a specific genetic signaling pathway the students would like to manipulate in their embryos (29) http://www.xenbase.org/common/jsp/showWiki.jsp?Small_Molecules_for_Xenopus_Research. Having students commit time before performing these manipulations to do background research into primary literature would be important and would give the instructor time to order any reagents necessary. Gehring and Eastman describe how they scaffolded information literacy assignments in a Developmental Biology class, which helped their students identify and apply valid sources to their own inquiry-based labs (30). Another great extension of this lab would be to spend more time instructing the students on the imaging of the Xenopus embryos. Watson and Lom describe a series of labs where their students created image portfolios and posters, rather than written lab reports, which helped their students hone their scientific communication skills throughout the semester (31).
CONCLUSION
Overall, this series of lab exercises is an excellent way to get undergraduate students working with live organisms and observing early stages of development. Additionally, using the in situ hybridization protocol, students can visualize and document how differential gene expression occurs during different stages of vertebrate development.
SUPPORTING MATERIALS
- S1: Xenopus gene expression - Fertilization instructions
- S2: Xenopus gene expression - In situ hybridization instructions
- S3: Xenopus gene expression - Lab notebook instructions
- S4: Xenopus gene expression - Readiness Assessment Quiz 1
- S5: Xenopus gene expression - Readiness Assessment Quiz 2
- S6: Xenopus gene expression - Lab exam questions
- S7: Xenopus gene expression - In situ hybridization probe instructions
- S8: Xenopus gene expression - Introduction to Xenopus PowerPoint
- S9: Xenopus gene expression - Introduction to in situ hybridization PowerPoint
ACKNOWLEDGMENTS
I would like to thank the undergraduate students of BIO305/L: Developmental Biology at Northern Kentucky University spring semester 2018 for their participation in the course and their thoughts on the lab exercise. I would like to acknowledge and thank Dr. Michelle McWhorter (mmcwhorter@wittenberg.edu) for providing her lab notebook instructions for Developmental Biology labs. Additionally, I appreciate the support of Xenbase.org and the images they provided as well as all of the Xenopus protocols from the Zorn lab (https://www.cincinnatichildrens.org/research/divisions/d/dev-biology/labs/zorn). I appreciate the help of EDS and BVB in reviewing the manuscript. This work was supported by the Kentucky Biomedical Research Infrastructure Network NIGMS grant #8P20GMI03436.
References
- Handelsman JM, Sarah; Pfund, Christine. 2007. Scientific Teaching. Roberts and Company; W.H. Freeman and Company, New York, NY.
- Ambrose SAB, Michael W.; DiPietro, Michele; Lovett, Marsha C.; Norman, Marie K. 2010. How Learning Works. Jossey-Bass, San Francisco.
- Anonymous. 2011. Vision and Change in Undergraduate Biology Education: A Call to Action. AAAS.
- Hardin J. 2008. The missing dimension in developmental biology education. CBE Life Sci Educ 7:13-16.
- Hargadon KM. 2016. A model system for the study of gene expression in the undergraduate laboratory. Biochem Mol Biol Educ 44:397-404.
- Llamusi B, Munoz-Soriano V, Paricio N, Artero R. 2014. The use of whole-mount in situ hybridization to illustrate gene expression regulation. Biochem Mol Biol Educ 42:339-347.
- Nam SC. 2018. Integration of a faculty's ongoing research into an undergraduate laboratory teaching class in developmental biology. Biochem Mol Biol Educ 46:141-150.
- Xenbase. Xenbase; RRID:SCR_003280. http://www.xenbase.org/entry/. Accessed 1/4/2019.
- Olive M, Thiebaud P, Landry M, Duvert M, Verna A, Barillot W, Theze N. 2003. Using Xenopus as a model system for an undergraduate laboratory course in vertebrate development at the University of Bordeaux, France. Int J Dev Biol 47:153-160.
- Sive HL, Grainger RM, Harland RM. 2000. Early development of Xenopus laevis : a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- Haak DC, HilleRisLambers J, Pitre E, Freeman S. 2011. Increased structure and active learning reduce the achievement gap in introductory biology. Science 332:1213-1216.
- Penner MR. 2018. Building an Inclusive Classroom. J Undergrad Neurosci Educ 16:A268-A272.
- Tanner KD. 2013. Structure matters: twenty-one teaching strategies to promote student engagement and cultivate classroom equity. CBE Life Sci Educ 12:322-331.
- Freeman S, Eddy SL, McDonough M, Smith MK, Okoroafor N, Jordt H, Wenderoth MP. 2014. Active learning increases student performance in science, engineering, and mathematics. Proc Natl Acad Sci U S A 111:8410-8415.
- Eddy SL, Hogan KA. 2014. Getting under the hood: how and for whom does increasing course structure work? CBE Life Sci Educ 13:453-468.
- Fromherz S, Whitaker-Fornek JR, Sharp AA. 2018. Classroom-Based Research Experiences to Support Underserved STEM Student Success: From Introductory Inquiry to Optogenetics in the Embryonic Chicken. J Undergrad Neurosci Educ 17:A97-A110.
- Cebra-Thomas J. 2003. Gastrulation Models, on Swarthmore College. https://www.swarthmore.edu/NatSci/sgilber1/DB_lab/Frog/frog_gast_model.html. Accessed 1/7/2019.
- Costa RM, Mason J, Lee M, Amaya E, Zorn AM. 2003. Novel gene expression domains reveal early patterning of the Xenopus endoderm. Gene Expr Patterns 3:509-519.
- Sinner D, Rankin S, Lee M, Zorn AM. 2004. Sox17 and beta-catenin cooperate to regulate the transcription of endodermal genes. Development 131:3069-3080.
- Hudson C, Clements D, Friday RV, Stott D, Woodland HR. 1997. Xsox17alpha and -beta mediate endoderm formation in Xenopus. Cell 91:397-405.
- Chen Y, Pollet N, Niehrs C, Pieler T. 2001. Increased XRALDH2 activity has a posteriorizing effect on the central nervous system of Xenopus embryos. Mech Dev 101:91-103.
- Dubey A, Rose RE, Jones DR, Saint-Jeannet JP. 2018. Generating retinoic acid gradients by local degradation during craniofacial development: One cell's cue is another cell's poison. Genesis 56.
- Jonas E, Sargent TD, Dawid IB. 1985. Epidermal keratin gene expressed in embryos of Xenopus laevis. Proc Natl Acad Sci U S A 82:5413-5417.
- Jonas EA, Snape AM, Sargent TD. 1989. Transcriptional regulation of a Xenopus embryonic epidermal keratin gene. Development 106:399-405.
- Plouhinec JL, Medina-Ruiz S, Borday C, Bernard E, Vert JP, Eisen MB, Harland RM, Monsoro-Burq AH. 2017. A molecular atlas of the developing ectoderm defines neural, neural crest, placode, and nonneural progenitor identity in vertebrates. PLoS Biol 15:e2004045.
- Madhuri M, Broussard C. 2008. "Do I need to know this for the exam?" Using popular media, inquiry-based laboratories, and a community of scientific practice to motivate students to learn developmental biology. CBE Life Sci Educ 7:36-44.
- Nicole L. Jacobs-McDaniels EMM, R. Craig Albertson, and Jason R. Wiles. 2013. Using model organisms in an undergraduate laboratory to link genotype, phenotype, and the environment. Journal of Biological Education 47:52-59.
- Spenillo JA. 2201. Developmental Effects of Lithium Chloride on Xenopus Embryos, on Swarthmore College. https://www.swarthmore.edu/NatSci/sgilber1/DB_lab/Frog/JA_Spenillo/justin2.html. Accessed 1/7/2019.
- Molecules XS. 2018. Small molecules for Xenopus research, on Xenbase. http://www.xenbase.org/common/jsp/showWiki.jsp?Small_Molecules_for_Xenopus_Research. Accessed 1/7/2019.
- Gehring KM, Eastman DA. 2008. Information fluency for undergraduate biology majors: applications of inquiry-based learning in a developmental biology course. CBE Life Sci Educ 7:54-63.
- Watson FL, Lom B. 2008. More than a picture: helping undergraduates learn to communicate through scientific images. CBE Life Sci Educ 7:27-35.
Article Files
Login to access supporting documents
Differential Gene Expression during Xenopus laevis Development(PDF | 477 KB)
S1. Xenopus gene expression - Fertilization instructions.docx(DOCX | 438 KB)
S2. Xenopus gene expression - In situ hybridization instructions.docx(DOCX | 657 KB)
S3. Xenopus gene expression - Lab notebook instructions.doc(DOC | 29 KB)
S4. Xenopus gene expression -Readiness Assessment Quiz 1.docx(DOCX | 2 MB)
S5. Xenopus gene expression - Readiness Assessment Quiz 2.docx(DOCX | 270 KB)
S6. Xenopus gene expression - Lab exam questions .docx(DOCX | 4 MB)
S7. Xenopus gene expression In situ hybridization probe instructions.docx(DOCX | 17 KB)
S8. Xenopus gene expression -Introduction to Xenopus PowerPoint .pptx(PPTX | 485 KB)
S9. Xenopus gene expression Introduction to in situ hybridization PowerPoint.pptx(PPTX | 783 KB)
- License terms

Comments
Comments
There are no comments on this resource.