Using a Primary Cell Culture Model to Study the Neural Extracellular Matrix
Editor: Scott Gehler
Published online:
Abstract
The extracellular matrix (ECM) provides structural support to cells, but also has key roles in mediating cell adhesion, cell signaling, and differentiation. While this topic is discussed in the lecture setting, it is not heavily studied in laboratory sessions. This lesson was created so that students can understand how the ECM is molecularly structured by using the ECM in the brain as a model. More specifically, the students study a particular structure in the neural ECM called the perineuronal net (PNN). In this exercise, students examine which cell type in the brain helps produce this structure and whether changes in activity affect PNN development. In this lesson students isolate and plate single cells derived from embryonic mouse brains; they then perform aspects of a standard immunofluorescence staining protocol. Lastly, the students will image the cultures using a fluorescence microscope. All of the aforementioned fundamental techniques are widely used by cell biologists. After completion, students hand in a formal lab report that is contextualized by primary literature. Students thoroughly enjoyed this exercise, and they found it challenging but rewarding and thought provoking.
Read the Essay Article about how author KAG adapted this lesson for online in "Going remote: An online adaptation to using a primary cell culture model to study the neural extracellular matrix."
Citation
Giamanco KA. 2020. Using a primary cell culture model to study the neural extracellular matrix. CourseSource. https://doi.org/10.24918/cs.2020.9
Society Learning Goals
Cell Biology
- Methods & Tools of Cell Biology
- How do the methods and tools of cell biology enable and limit our understanding of the cell?
Lesson Learning Goals
Students will:
- Appreciate the use of the mouse as a model organism.
- Understand how the neural extracellular matrix is structured.
- Understand the structural and molecular components of the perineuronal net.
- Address how the methods and tools of cell biology enable and limit our understanding of the cell.
Lesson Learning Objectives
Students will:
- Isolate single cells from dissected cerebral cortices of embryonic mice.
- Develop hypotheses and find support for these hypotheses in the literature.
- Treat primary cultures with agents to inhibit glial cell growth and increase activity levels.
- Fix and block primary cultures in addition to applying primary antibodies.
- Use fluorescence microscopes to analyze and interpret their results.
- Complete a lab report that is contextualized by primary and secondary literature.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
The American Association for the Advancement of Sciences (AAAS) has emphasized the importance of providing opportunities for undergraduate students to engage in research in the classroom laboratory setting (1). This requires students to learn how to set forth hypotheses, design experiments, and analyze their results in a manner consistent with that of research scientists. As part of this process, students become versed in performing literature searches and discussing their results in the context of a greater body of work. To address the changes set forth by the AAAS, it is essential that as instructors, we allow students the freedom to carry out these experiments, hone these skills, and become immersed in the exercise.
One of the most fundamental techniques in cell biology is mammalian cell culture, which involves the generation and maintenance of cells isolated from specific animal tissues. This procedure necessitates the use of a sterile cell culture hood and incubator, as well as a proper medium and supplements to sustain cell growth. Many undergraduate students only learn about this technique in the lecture setting and do not have opportunities to perform such experiments. Recently, Bowey-Dellinger and colleagues devised an exercise in which students learn how to trypsinize cells, assess cell viability, and generate cell counts using immortalized HeLa cells (2). Additionally, others have created exercises utilizing human embryonic kidney cells (HEK293 cells) and mouse fibroblast cells (3T3 cells) (3-4). Similarly, Catlin and colleagues generated an experiment in which students work with purchased primary cultures to study a number of cellular processes as well as the mechanisms underlying neurodegeneration (5). However, in these exercises the students do not directly learn how to isolate cells from tissue.
To address this, many undergraduate instructors have developed experiments in which the students receive hands-on experience isolating cells from a particular tissue. In exercises such as these, students employ enzymatic and mechanical methods to create a single cell suspension, plate the cells, and then use them in downstream applications, such as Western blotting, staining, or qPCR. Chicken eggs represent one such model system that can be utilized to generate primary cultures. In particular, neurons from the chick forebrain can be readily isolated, and then students have the opportunity to examine axon growth and neuronal structure (6). Additionally, fundamental pharmacological concepts can be studied through the isolation and subsequent culture of chick cardiomyocytes (7). Others have reported using chick cardiomyocytes as an exercise to demonstrate that living tissue can maintain normal structure and function even in a culture dish (8). Keratocytes have been isolated from fish scales to allow students to study the intricacies of cell migration (9). In the lesson described here, students isolate single cells from previously dissected cerebral cortices of embryonic mice to study how a particular structure within the neural extracellular matrix (ECM) is organized to address two specific questions. The two questions addressed are (1) Is aggrecan expression modulated by activity? and (2) Is aggrecan likely produced by neurons or glia?
While Cell Biology lectures primarily focus on cellular functions and processes, most courses include discussions of the ECM and more broadly, the extracellular environment. The ECM not only provides structural support to cells, but mediates cellular events, including cell migration, adhesion, differentiation, and tissue development (10). Classical ECMs like the basement membrane, that are found within epithelial, endothelial, muscle, and adipose tissues are comprised of fibrous proteins, like fibronectin, laminin, and collagen in addition to glycoproteins and proteoglycans (11). Interestingly, the neural ECM has a somewhat different composition from that of more classical ECMs (12), which demonstrates to students that there is a great degree of heterogeneity within the extracellular environments of different tissue types. This laboratory exercise would dovetail well with presentations about the ECM in the lecture setting and would stimulate discussions of the interplay between structure and function.
The perineuronal net (PNN) is a specialized substructure found within the neural ECM that plays an integral role in restricting plasticity within the brain. Structurally, the PNN is highly organized and surrounds the cell body and extensions stemming from particular neurons. In terms of molecular composition, PNNs are made up of proteoglycans and complex sugars (13). Interestingly, appropriate sensory input is needed for PNNs to develop properly (14). A complete understanding as to how these molecules link together to form PNNs still eludes us. Because of this, many investigators have turned to in vitro or cell culture models to more thoroughly analyze and manipulate this structure with the hopes of gathering information about how PNNs are organized (15-18).
In this exercise, students will answer two central questions focused on one particular component of the PNN, aggrecan: (1) Is aggrecan-based PNN formation modulated by sensory activity? and (2) Is aggrecan likely made by neurons or glia? This work will shed light onto how the PNN forms and which cell types contribute to its formation—two areas that are currently under active study. To carry this out, students will undertake a four-week experiment in which they isolate single cells from dissected cerebral cortices of embryonic mouse brains and plate the cells into culture dishes (Lab Session 1). Then they treat these cultures with agents to increase activity levels to address Question 1 and a glial cell inhibitor to address Question 2 (see above). In Lab Session 2, the students will focus on setting forth hypotheses and predictions based on the two central questions, while the cultures are incubating. Additionally, the students will have time to gather primary and secondary literature articles to support their hypotheses and predictions, as well as discuss their thoughts in small groups. To end this particular session, the discussion is opened up to the entire class to ensure that all students are able to find supporting literature. Students then fix and block their cultures and apply antibodies directed against aggrecan and glial fibrillary acidic protein (GFAP, a marker of glial cells) (Lab Session 3), and use fluorescence microscopes to analyze their results (Lab Session 4). Each lab session takes about 3 hours to complete, with one week separating each exercise.
This lesson requires the use of a cell culture hood (one that is approved for mouse work), incubator, centrifuge, and fluorescence microscopes. To maintain the cells, specific cell culture media and supplements need to be purchased, but these could easily be stored for use in multiple semesters, or even used for research purposes.
In this laboratory project, students employ techniques that they learned about in the lecture setting (use of trypsin and manual trituration to isolate single cells, fixation and blocking of primary cultures, application of primary antibodies, and use of fluorescence microscopy), all of which are widely used in scientific research. Therefore, this experiment affords students the opportunity to engage in authentic, hypothesis-driven research. While the two main questions being addressed are provided to the students as well as the experimental design, the students are responsible for delving into the literature to find support and rationale for their own hypotheses and corresponding predictions. Formative assessments are given after the completion of Lab Session 1 and Lab Session 3 to help the students prepare for the lab report, with instructor feedback provided. The lab report mimics the style of a journal article, consisting of the following sections: Introduction, Materials and Methods, Results, and Discussion. Students also need to provide images of PNNs and glial cells as well as tables documenting how they plated their cells and how the cells were treated and stained.
The students thoroughly enjoyed conducting this experiment and found it fascinating that they were able to isolate cells, stain them, and then visualize the staining under the microscope. Throughout this experiment, students worked in groups that they selected, and they established a rapport with one another. In the second session there is time to discuss the background information as well as the procedural details pertaining to the experiment. Additionally, in this session, students discuss the results not only within each group, but as a class to ensure that everyone feels comfortable interpreting and thinking about what they found.
Intended Audience
This laboratory exercise was designed for an upper-level undergraduate course in Cell Biology in which students are either Biology or Biochemistry majors. This experiment has been tested over multiple semesters with 2-3 laboratory sections offered per semester. Each section has had a maximum of 16 students.
Presenting material in lecture detailing the various molecular constituents of the ECM before the laboratory sessions ensures that the students have a strong foundation on which to build upon.
Required Learning Time
The experiment requires a total of four laboratory sessions that are one week apart, with each session lasting about three hours. There are some steps in each of these sessions that require waiting time so you have the freedom to incorporate active learning techniques based on lecture material or allow students time to start working on the laboratory assignments and report. To ensure that the activities can be carried out during the given lab period, you will need to perform some parts of the experiment yourself. (Consult Table 1: Use of a Primary Cell Culture Model to Study the Neural Extracellular Matrix Teaching Timeline for further information).
Prerequisite Student Knowledge
Students should be aware of what the ECM does and the molecules that are found in these matrices. Additionally, prior to this exercise, my students complete an experiment where they work with immortalized cell lines, which instills in them the importance of maintaining a sterile environment. Specifically in this exercise, they wash, trypsinize, and split cells in addition to generating cell counts. Thus, when they perform the neural ECM experiment, they are able to draw upon that previous experience. They should have learned about immunofluorescence and microscopy in the lecture setting before they perform that part of the experiment.
In addition, students should have experience using PubMed, Google Scholar, JSTOR or similar resources to gather primary and secondary literature pertaining to this lab report. Since this exercise is best suited for an upper level Biology or Biochemistry major, it is highly likely that the students have either taken a course on scientific communication or writing, or at the very least have been required to write a formal lab report in another science-based class. Lab Session 2 is designed to allow for students to utilize lab time to work on the fundamentals of the lab report. You will also have the opportunity to work with the students either one-on-one or in small groups during this time.
Prerequisite Teacher Knowledge
The instructor should be able to generate primary neuronal cultures from embryonic mouse brains, maintain these cultures, instruct students how to perform staining experiments and use fluorescence microscopes. In terms of concepts, the instructor needs to have an understanding of how the neural ECM differs in molecular composition from other ECMs. Some knowledge about PNNs would help, but that information could easily be learned in enough detail to instruct the class (13-18).
SCIENTIFIC TEACHING THEMES
Active Learning
Students are provided with questions that will be answered in this exercise, and then they individually come up with specific hypotheses. They will find primary literature articles to support their hypotheses and put the results they obtained in the context of this work. The students will process the previously dissected mouse brain tissue to generate single cells. Importantly, students will count the cells and determine how many cells to plate based on a given density. In addition, the students will treat their cultures with a glial cell inhibitor as well as with potassium chloride to increase overall activity. Next, they will stain the cultures, and lastly, analyze the cultures using a fluorescence microscope.
Assessment
The main form of assessment for this activity is a lab report. Since this is a multi-week experiment and the lab report is intensive, small homework assignments can be given after the students conduct the exercise in Lab Session 1 and Lab Session 3 to assess student understanding (Supporting File S1: Studying the Neural Extracellular Matrix – Lab Session 1 assignment and Supporting File S2: Studying the Neural Extracellular Matrix – Lab Session 3 assignment). In my class, the small lab assignments are each worth approximately 1% of the final grade, while the lab report is worth 5-10% of the final grade. Weekly lab quizzes are a good idea to ensure that students are reading the handouts before completing the exercise.
Inclusive Teaching
Students are allowed to pick the groups (maximum 4 members) in which they work for the duration of the project. After working together for weeks, students become comfortable with their peers, which creates a more interactive and positive environment (19). When students select their groups, they get started on the project more quickly as the time needed to get to know one another is reduced (20), which could lead to increased productivity and even creativity. Students will employ a variety of techniques in this activity, including: generation of single cells that are then plated, cell counting, staining, and microscopy, which will engage all types of learners. Group work allows for students to teach one another, which is an effective pedagogy tool. For example, a student proficient in microscope work can help other members of the group that may be intimidated or unsure of how to use such equipment. In terms of assessments, the small homework assignments are designed to help the students gather and organize the information that needs to be incorporated into the lab report, which can set the students up for success on the formative assessment. Additionally, feedback provided to each student on the small homework assignments directly addresses each individual's background and preferences. Lastly, since the formative assessment is a written lab report, students who have anxiety around test-taking or oral presentations will have an opportunity to boost their grade in this course through this assignment.
LESSON PLAN
Execution of this lesson requires planning. It takes place over four weeks, and there are a number of preparatory steps that need to be carried out by the instructor (Table 1: Use of a Primary Cell Culture Model to Study the Neural Extracellular Matrix Teaching Timeline).
Lab Session 1: Pre-laboratory preparation
Obtaining timed-pregnant mice and embryos
The Institutional Animal Care and Use Committee (IACUC) must approve any work requiring the use of vertebrate animals. The work described here adhered to the guidelines set forth by the Western Connecticut State University IACUC. Pregnant mice can be obtained from a number of companies, including Charles River Laboratories (Wilmington, MA, USA) and Taconic Biosciences (Rensselaer, NY, USA). It is important that timed-pregnant mice be ordered because the age of the embryos matters, in terms of how easy it is to perform the dissections, isolate cells, and ultimately, generate PNNs. These experiments are performed at embryonic day 16 (E16). Usually, mice are delivered a few days prior to the experiment so that the animals can acclimate to the new environment and are not stressed on the day of the exercise.
I recommend using wild-type CD-1 mice [referred to as Crl:CD1(ICR) on Charles River's website: https://www.criver.com/products-services/find-model/cd-1r-igs-mouse?region=3611]. These mice are excellent breeders and produce on average 12 embryos per litter (22). For this lesson, I recommend that each group have about 3-4 dissected mouse brains to work with, which means that 2 timed-pregnant mice per laboratory section (with 4 groups per lab section) should suffice. This can be altered if necessary.
Preparation of cell culture materials and reagents
The first session of this exercise requires instructor preparation of cell culture materials and reagents prior to lab in order to ensure that students will be able to fully complete the experiment in the allotted time. First, load glass coverslips into the coverslip holders and then, along with forceps and Schifferdecker Staining Jars, autoclave all the equipment under a standard solid cycle. The following steps must be carried out under sterile conditions in a designated cell culture hood. After cooling, incubate the coverslips within the holders in 0.5 M hydrochloric acid for one hour at room temperature (RT) in the Schifferdecker Staining Jars. Next, rinse the coverslips in water for 15 minutes at RT. Distribute coverslips to individual wells in a 24-well plate using forceps and then wash with 1x Dulbecco's phosphate buffered saline for 10 minutes at RT. Lastly, coat coverslips with laminin (final concentration 5 μg/ml) and poly-D-lysine (final concentration 100 μg/ml) made up in Dulbecco's phosphate buffered saline. Incubate the plates overnight at 37°C with 5% CO2 in a standard cell culture incubator.
Prepare and aliquot the following solutions under sterile conditions: Hank's Buffered Saline Solution with 1x penicillin/streptomycin, 0.25% trypsin-EDTA, 10/10 trypsin solution (made up of Earle's Balanced Salt Solution, 10 mg/ml ovomucoid inhibitor, and 10 mg/ml bovine serum albumin), 1/1 trypsin solution (Earle's Balanced Salt Solution, 10% of the 10/10 solution and DNase I), and Neurobasal Medium with 3x B27 supplement, 1x GlutaMAX, and 1x Penicillin/Streptomycin. The Hank's Balanced Salt Solution and 0.25% trypsin-EDTA can be prepared and stored days before the exercise, while the other solutions should be prepared on the day of the experiment.
Mouse brain dissections
After the mouse has been euthanized according to the IACUC protocol, remove the embryos from the abdominal cavity and place them in Hank's Balanced Salt Solution with 1x Penicillin/Streptomycin. Then remove the brain from the skull and dissect the cerebral cortex (21). Supporting File S3: Studying the Neural Extracellular Matrix – Dissection Instructions, provides detailed information as to how to carry out the dissections of the mouse brains including pictures to orient anyone interested in implementing this lesson in their class. Perform these dissections prior to the lab session and place them in the supplemented Neurobasal Medium. Right before the class, place tissue from 3-4 brains (total of 6-8 cortices) into a tube of 10 mL of pre-warmed 0.25% trypsin-EDTA while working in the cell culture hood. This will create enough tissue/group; you can decide how many tubes are needed. Lastly, place the tube in a 37°C water bath, shaking at maximum speed to dissociate the tissue for 25 minutes.
Lab Session 1
Since the protocol for the first session takes up the entire laboratory period, I recommend starting the exercise (Supporting File S4: Studying the Neural Extracellular Matrix – Lab Session 1 Handout) right after the quiz. Pre-warm all solutions that will be used prior to the start of the exercise. When students enter the lab, the samples will already be incubating in 0.25% trypsin-EDTA to ensure separation of single cells. At this point, students will break into small groups and receive their own conical tube with tissue in it. All of the following steps, with the exception of cell counting, must be executed in a cell culture hood. Students will manually triturate the tissue, by vigorously pipetting up and down to isolate single cells. They should avoid cell clumps, as this can affect the health of the cultures and skew the total cell number. Next, spin samples in a centrifuge that is maintained at RT to remove the trypsin. All centrifugation steps are carried out at RT for 7 minutes at 400 x g or 1,389 RPM. Remove the supernatant, leaving the cells within the pellet. To inhibit the trypsin, resuspend the pellet in the 1/1 solution containing ovomucoid inhibitor and bovine serum albumin. At this point, some of the pellet will be insoluble, likely requiring filtering the sample with a 70 μm cell strainer. Next, layer the 10/10 solution beneath the sample. The easiest way to do this is to place the pipette filled with the 10/10 solution at the bottom of the tube and dispense the liquid in full before moving the tip upwards. After another centrifugation step, wash the pellet in the supplemented Neurobasal medium. Lastly, centrifuge the samples and then resuspend the resulting pellet in an appropriate amount of media (for samples of this size, it is usually between 500 μl and 1 mL). Students then dilute their samples 1:10 in Neurobasal medium to count the cells using a hemocytometer. It is a good idea to have students practice how to count cells and plate them at desired densities prior to this exercise, but if not, work closely with the students to ensure they are counting properly and performing the calculations correctly. Depending on whether time allows, students can plate the cells, or you can plate the cells yourself. The amount of cells each group generates will determine how many wells their samples will occupy in a 24-well plate. The desired plated density is 1.4 x 106 cells/mL, with 500 μl distributed into each well. Thus, each well will contain a total of 700,000 cells. Once this is completed for all groups, immediately place the plate in the cell culture incubator. This is considered to be 0 days in vitro (0 DIV).
It is imperative that all work be conducted in a cell culture hood. If there are multiple hoods available for use, then move between the hoods to ensure the students are properly guided. If there is only one cell culture hood to work with, then have students rotate to complete their work. Those that are not actively working in the cell culture hood can engage in another activity or assignment. You will likely need to demonstrate particularly difficult steps in the protocol (ex: layering the 10/10 trypsin inhibitor solution beneath the cell suspension). Additionally, pay special attention to the sterile technique displayed by each student to ensure that the cultures are not contaminated. Set up biohazard bags so that waste is disposed of according to the IACUC protocol.
Students enjoy this session because they are able to utilize tools that we have talked about in lecture. For example, they learn first-hand what trypsin-EDTA does and why we use it to create a single cell suspension. They are fascinated by the fact that they can count the cells under the microscope and that they are able to generate so many cells from the small pieces of mouse brain tissue.
Care of cultures between Lab Session 1 and Lab Session 3
The cultures need to be incubated until 14 DIV in order to observe PNN development and formation. The day after plating (1 DIV), require the students to come to the laboratory and divide their cells into four categories: untreated control, potassium chloride (KCl), cytosine-β-D-arabinofuranoside (AraC), and then lastly, KCl + AraC. For example, if 8 coverslips were generated/group, then each group would have 2 coverslips for each of the four groups listed above (Figure 1). Have them treat the cells in the KCl group with 20 mM KCl (final concentration is 25 mM because there is 5 mM in the base media), treat the AraC group with 5 μM AraC, and treat a group of cells with both KCl and AraC. You may wish to be responsible for any media changes moving forward yourself, or you can require students to perform these steps. On 3 DIV, all of the media within the wells must be removed and replaced with fresh pre-warmed supplemented Neurobasal media. The subset of cells that have been exposed to KCl are continuously maintained in media with KCl. Therefore, there will be a set of cells that are exposed to normal Neurobasal media (untreated control cells and cells that were exposed to AraC) and elevated KCl levels (KCl group and those cells treated with KCl and AraC). On 6 DIV, 9 DIV and 12 DIV, perform a half media exchange, where half of the media in the well is removed and replaced with either normal Neurobasal media or Neurobasal media with elevated KCl levels. Carry out all of the aforementioned steps in a cell culture incubator under sterile conditions.
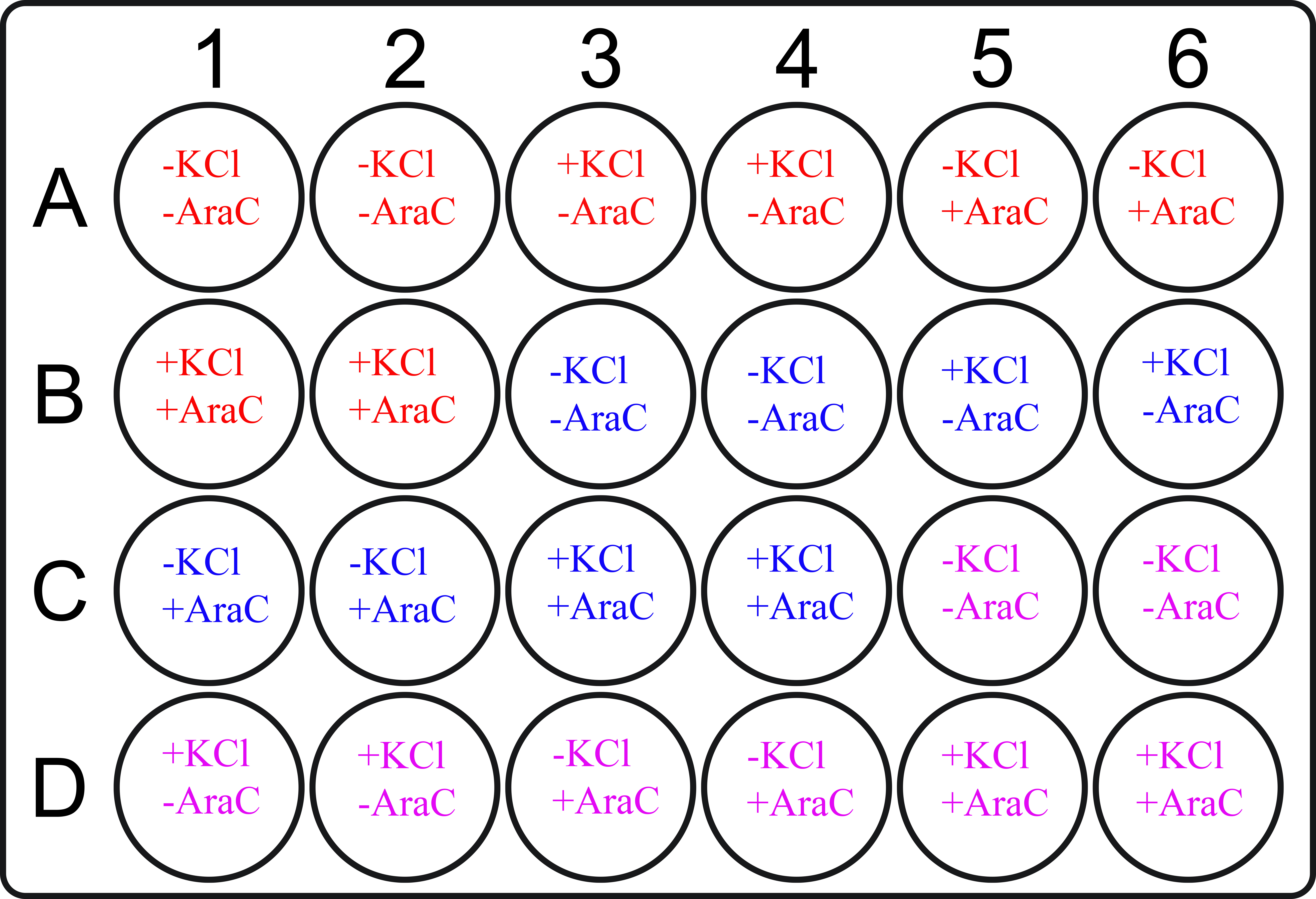
Figure 1. Treatment paradigm. This is an example of a multi-well plate depicting how to divide the 24 wells amongst the four treatments groups (-KCl -AraC, +KCl -AraC, -KCl +AraC, and +KCl +AraC) and three groups of students. Each group of students will be able to work with two coverslips/treatment. This can be modified to accommodate the number of students and number of student groups. Each color corresponds to a different group of students. Schematic of the multi-well plate was taken from: https://en.wikipedia.org/wiki/File:24-well-plate.svg. Author: Volker-Morath. License information: https://creativecommons.org/licenses/by-sa/4.0/deed.en. Text was added to the file in order to convey information about treatment groups and student groups.
Lab session 2
Prepare a lecture to go over all the necessary material to understand the project as well as discuss the experimental details from the first session. Typically, this lecture should take about one hour to one hour and a half of lab time (Supporting File S5: Studying the Neural Extracellular Matrix – Lab Session 2 Pre-Lab Lecture). With the remaining time, students will work in small groups to develop hypotheses and predictions based on the two central questions that are being asked in this exercise: (1) Is aggrecan expression modulated by activity? and (2) Is aggrecan likely produced by neurons or glia? Circulate the room to ensure that students are able to ask questions and articulate their hypotheses. Next, they will work on gathering primary and secondary literature articles to use in the lab report, allowing them the opportunity to ask questions about finding suitable literature articles and discuss the experiment in groups (Supporting File S6: Studying the Neural Extracellular Matrix – Lab Session 2 Handout). Additionally, this will afford the students more time to work on the actual writing of the lab report. You might find it helpful to hold this session in a computer lab where students have access to workstations, or alternatively, students could bring their own devices to the laboratory.
Lab Session 3: Pre-laboratory preparation
Preparation of solutions
On the day of the exercise, prepare the following solutions: 4% paraformaldehyde in 1x phosphate buffered saline, and the blocking medium: Dulbecco's Modified Eagle's Medium, 5% fetal bovine serum, 0.5% Triton X-100, and 0.2% sodium azide. Store both reagents at 4°C. Additionally, aliquot the primary antibodies: anti-aggrecan and anti-glial fibrillary acidic protein (GFAP), and store aliquots at -20°C until needed.
Prepare a brief pre-lab lecture that explains why cells need to be fixed and blocked prior to staining (Supporting File S7: Studying the Neural Extracellular Matrix – Lab Session 3 Pre-Lab Lecture).
Lab Session 3
Fixing and blocking cells; Incubation in primary antibodies
When the students come to class, the cells will be in the cell culture incubator. While working in a designated fume hood, they will fix cultures in 4% paraformaldehyde in 1x phosphate buffered saline. To do this, the students will first remove all the cell culture media from the well and dispose of it in a waste container. Next, they add 500 μl of the fixative and incubate cultures for 20 minutes at RT. After this period, wash the cultures 3 times for 5 minutes each in 1x phosphate buffered saline. Once the last wash is complete, remove all of the buffer, and block cells to prevent non-specific binding of the primary antibodies. The blocking medium is made up of Dulbecco's Modified Eagle's Medium, 5% fetal bovine serum, 0.5% Triton X-100, and 0.2% sodium azide. At this point, incubate the plate at RT on a shaker for one hour. After the requisite time, wash the cultures again (3 times for 5 minutes each) and then apply primary antibodies. Incubate the cells overnight at 4°C in primary antibodies that are diluted in the blocking medium. The primary antibodies used are anti-aggrecan and anti-GFAP, both at a dilution of 1:2000 (Supporting File S8: Studying the Neural Extracellular Matrix – Lab Session 3 Handout).
During the blocking step, while cells are incubating, the students can assemble a list of reagents that were used in the exercise that need to be included in the lab report. You can provide the name of the company and hold the students responsible for including the company location. If there is any remaining laboratory time at the end, answer questions about the lab report and touch base with each group of students about their hypotheses and predictions. Students can also work on their small homework assignments and get feedback on them.
This session allows for students to understand how immunofluorescence works and reinforces the relationship between an epitope within the protein, the primary antibody, and the secondary antibody with an associated fluorophore. This exercise further highlights why antibodies need to be specific in terms of what they recognize.
After Lab Session 3: Instructor steps
Incubation in secondary antibodies; Hoechst staining and coverslipping
The day after Lab Session 3, carry out the remaining steps: incubation in secondary antibodies, Hoechst staining, and coverslipping. After the primary antibody incubation, wash the cells as above and incubate in appropriate Alexa Fluor secondary antibodies that are diluted at 1:800 in the aforementioned blocking medium. Cover plates in foil and subject them to slight agitation for 2 hours at RT. Once the two-hour period has lapsed, wash the cells as previously described. Next, to stain all nuclei, incubate cells in Hoechst or 4',6-diamidino-2-phenylindole (DAPI) at 1:50,000 in 1x phosphate buffered saline for 30 minutes at RT while covered in aluminium foil with slight agitation (Figure 2). After another series of washes as above, mount the coverslips onto glass slides. Using forceps, remove the coverslips from each well, rinse the coverslip in milliQ water, and then mount the coverslip onto a glass slide containing a drop of ProLong Antifade Reagent, which has been prepared according to the manufacturer's instructions. Store slides at room temperature until analysis.
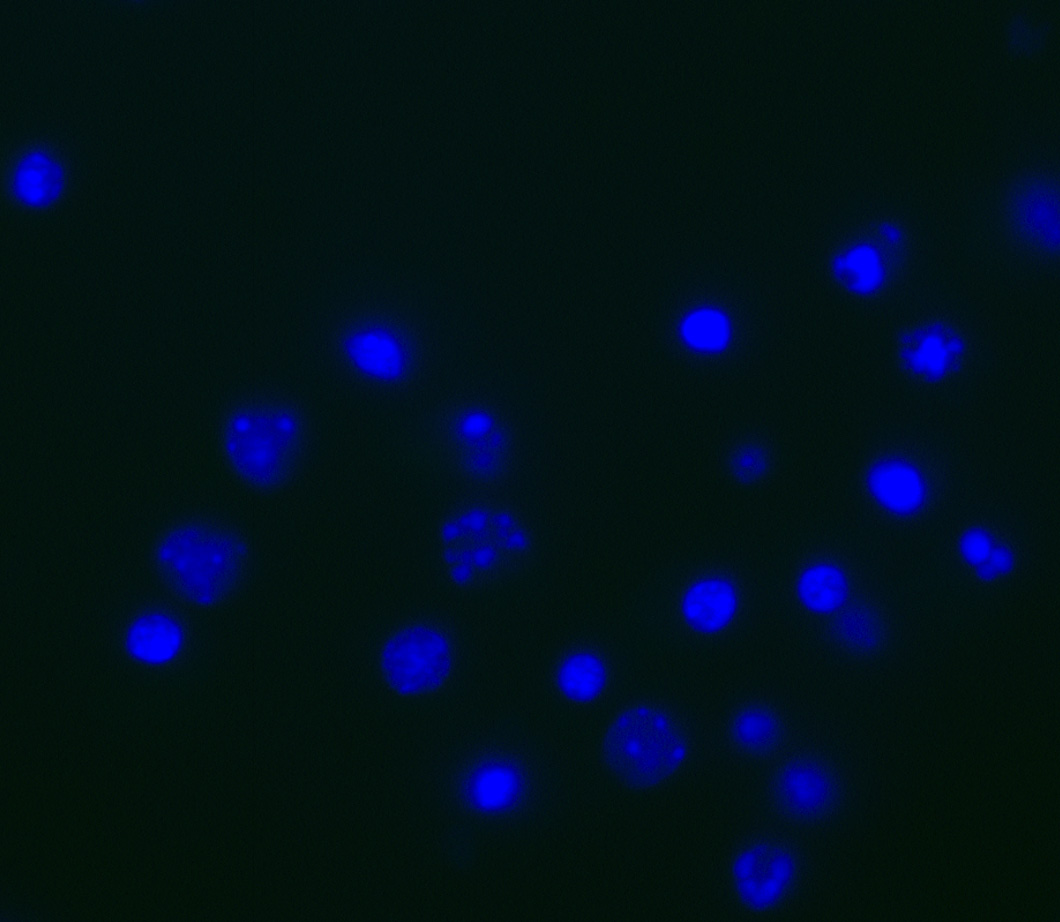
Figure 2. Hoechst staining of cortical cultures. In order to visualize nuclei, students apply the fluorescent Hoechst 33342 stain (blue) to cultures after the secondary antibody incubation. This was included to ensure that students were analyzing cellular staining and not any staining artifacts.
Lab Session 4: Pre-laboratory preparation
Set out fluorescence microscopes and prepare a brief pre-laboratory lecture about how students will analyze their data and how to use the microscopes (Supporting File S9: Studying the Neural Extracellular Matrix – Lab Session 4 Pre-Lab Lecture and Supporting File S10: Studying the Neural Extracellular Matrix – Lab Session 4 Handout).
Lab Session 4
Student groups will use the fluorescence microscopes in groups to analyze their cells, which fosters student discussion. Remind the students how immunofluorescence works and which fluorophores are associated with which antibodies. Students should record all their observations and can talk to other groups. If possible, students can obtain images to use for the lab report. At the end of the session, hold a discussion with the entire class to assess what each group saw. In the case of this experiment, they will be looking at expression of aggrecan-based PNNs (Figure 3) and GFAP (Figure 4) in four groups: Control, +KCl, +AraC, and +KCl +AraC.
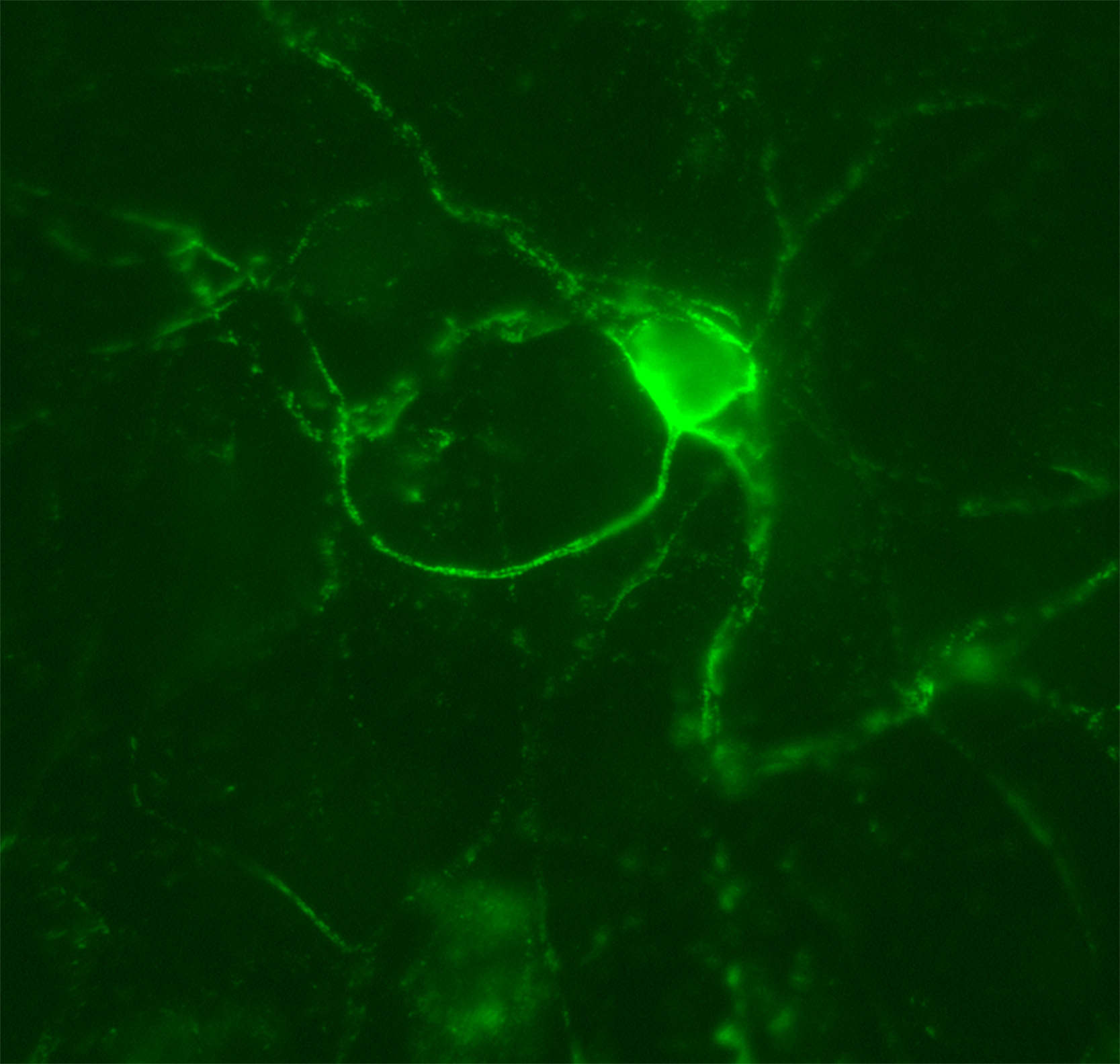
Figure 3. Aggrecan-based PNN. In this experiment, cortical cultures were stained with anti-aggrecan antibodies to label PNNs (green). This is a representative image of an aggrecan-based PNN that the students should visualize in this lesson by using a fluorescence microscope. The PNN surrounds the cell body and extensions of a particular neuron within the culture.
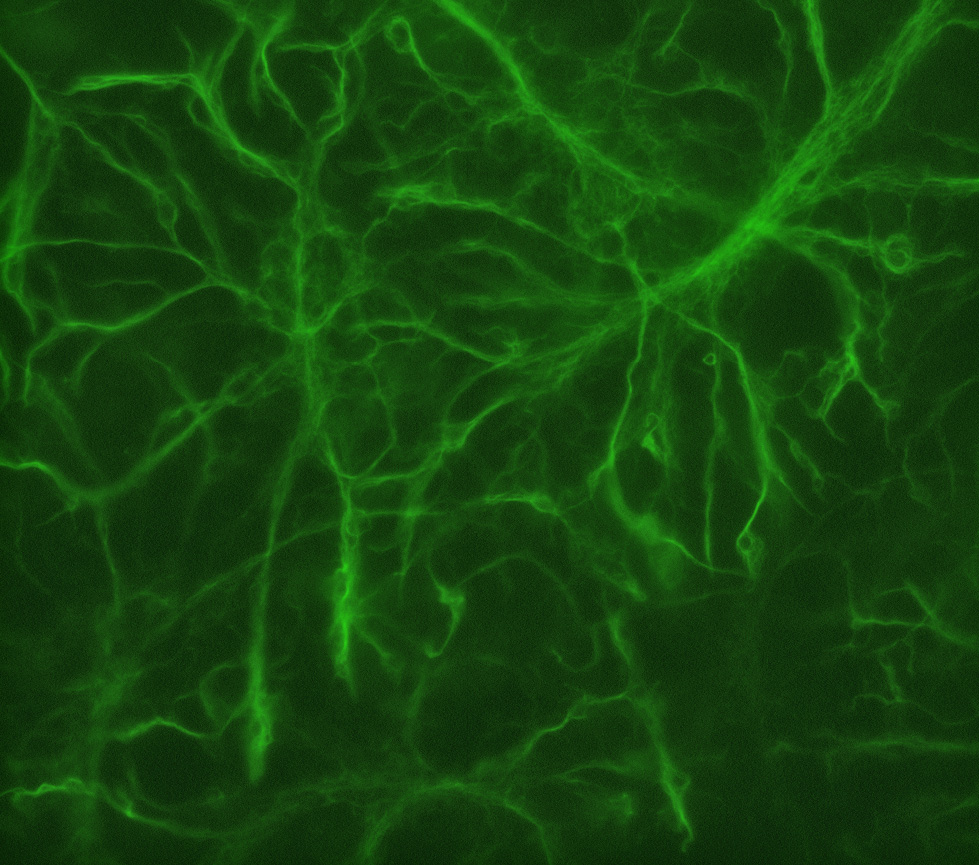
Figure 4. GFAP staining. In this lesson, students will stain primary cultures with antibodies against glial fibrillary acidic protein (GFAP) to label glial cells (green). This is a representative image of what GFAP-positive glial cells should look like.
Students find this session fascinating as they get to see the direct results of their hard work and get to answer the two main questions that were set forth in the beginning of the experiment. If you want to adopt this exercise, consult Supporting File S10: Studying the Neural Extracellular Matrix – Instructor Notes.
Assessment
Data analysis is typically qualitative rather than quantitative, but this exercise could be adapted to fit a more quantitative approach, if desired. The main form of assessment is a formal lab report containing an Introduction, Materials and Methods, Results, and Discussion sections along with figures and tables and associated legends (Supporting File S12: Studying the Neural Extracellular Matrix – Lab Report Rubric). Small formative assessments for Lab Sessions 1 and 3 would help the students understand the overall goal of the experiment and allow them the opportunity to brainstorm and collect information for their lab report.
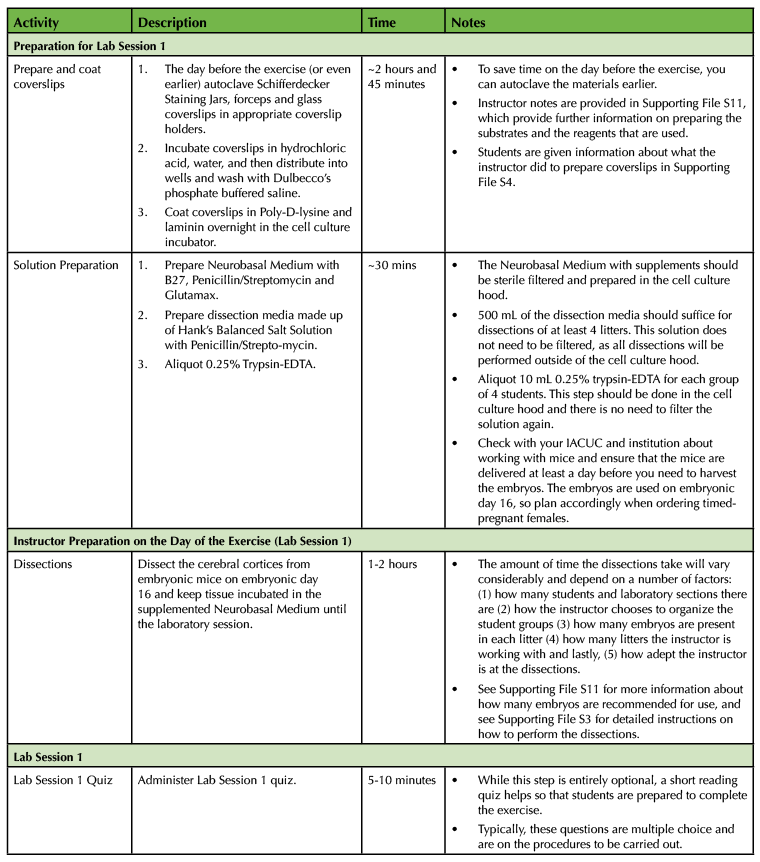
Table 1. Using a Primary Cell Culture Model to Study the Neural Extracellular Matrix Teaching Timeline (1).
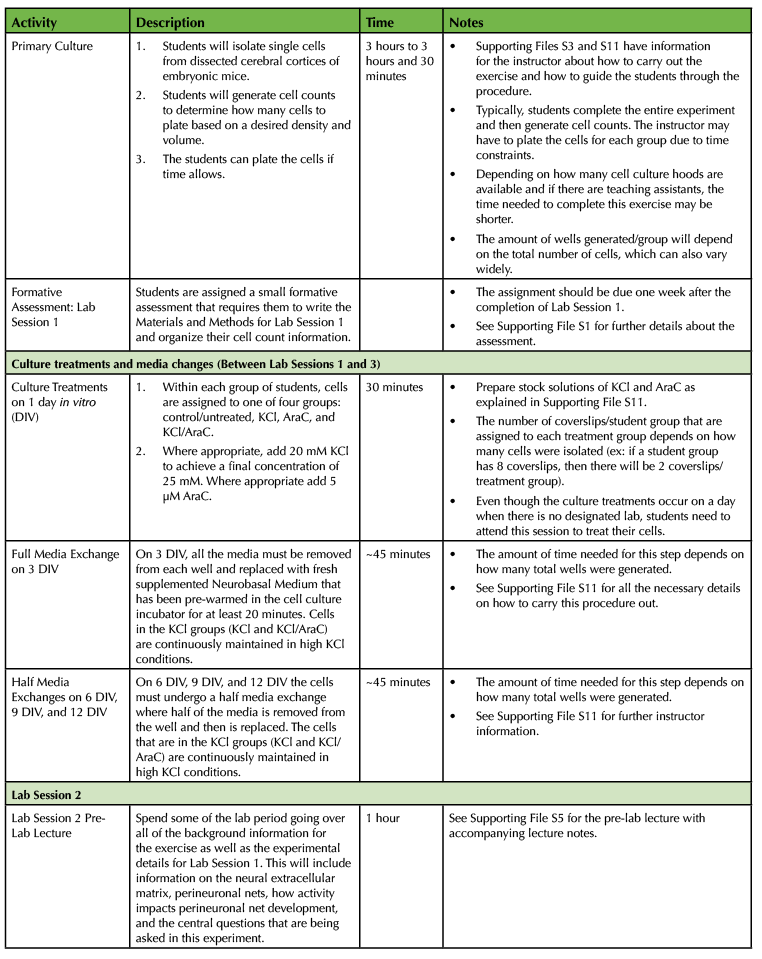
Table 1. Using a Primary Cell Culture Model to Study the Neural Extracellular Matrix Teaching Timeline (2).
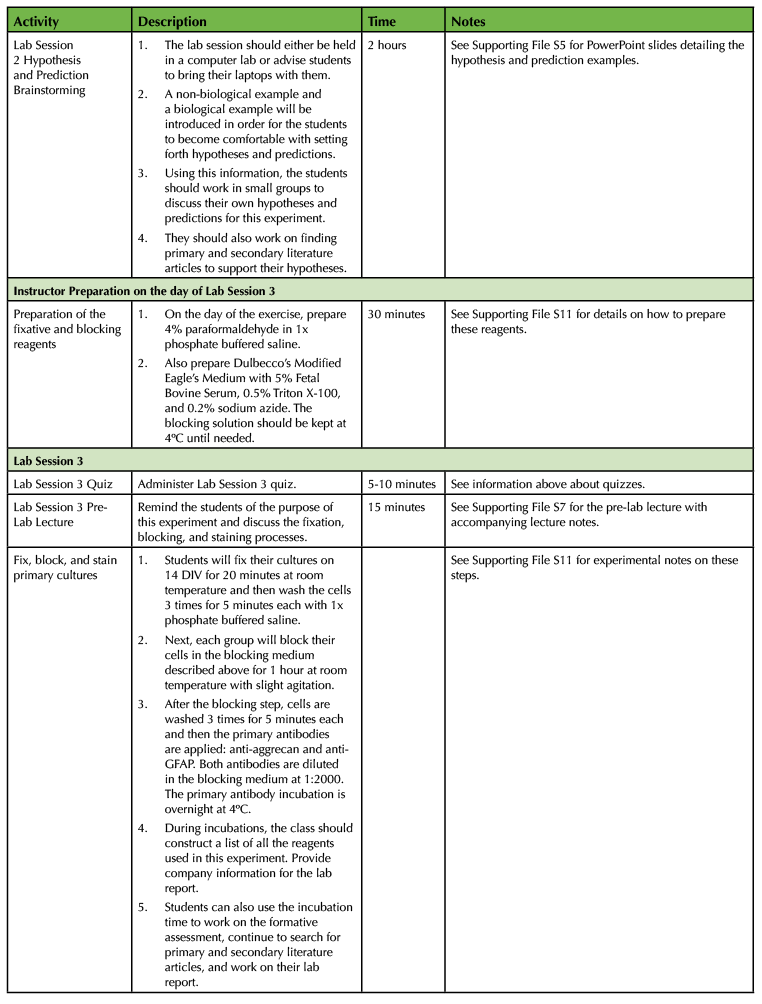
Table 1. Using a Primary Cell Culture Model to Study the Neural Extracellular Matrix Teaching Timeline (3).
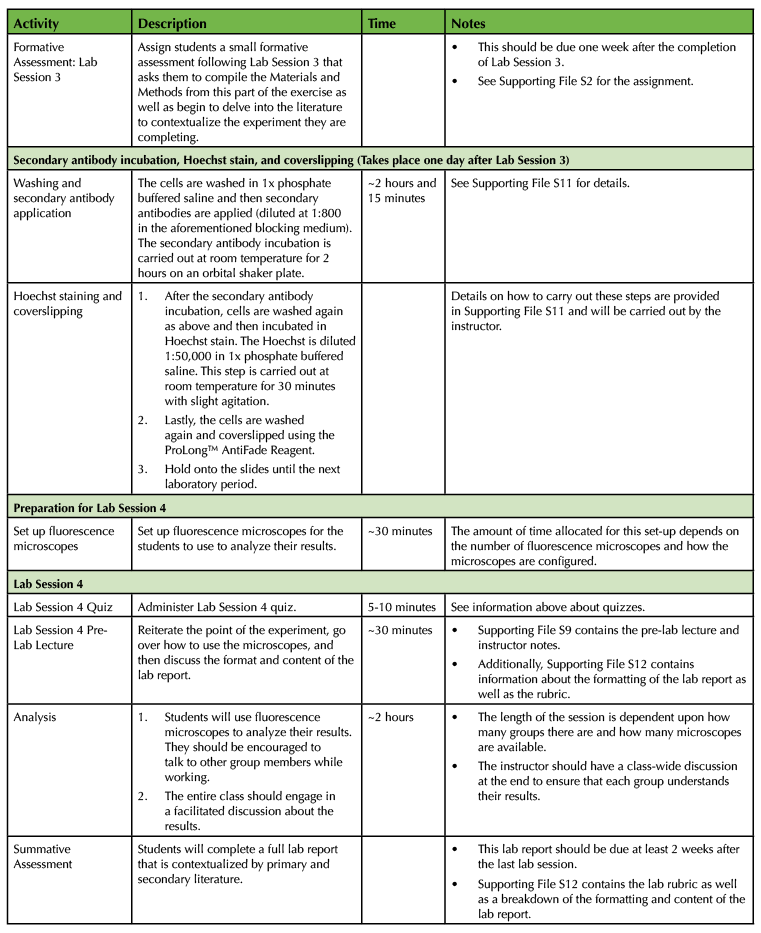
Table 1. Using a Primary Cell Culture Model to Study the Neural Extracellular Matrix Teaching Timeline (4).
TEACHING DISCUSSION
The purpose of this laboratory exercise is to provide a hands-on, research-based experience for upper level Biology majors. While the premise of the experiment was given to the students, they came up with specific hypotheses and predictions based on information they gathered from the primary literature. While there were small formative assessments during this four-week experiment (lab quizzes and small assignments based on the activities carried out in Lab Sessions 1 and 3), the main form of assessment was the formal lab report. In this lab report, students delve into the literature to find support for their specific hypotheses and predictions as well as put their results in context of the greater body of work.
In the lab report, students have to explain how the neural ECM is structured and define the molecules that make up the PNN. They also need to find work that discusses the function of PNNs as well as dissect how activity plays a role in overall formation of this structure. This information allows the students to hone in on their hypotheses and explain the rationale for the experiment. Before writing the lab report, they received instructor feedback on the materials and methods section as well as their hypotheses. Additionally, in Lab Session 2, students have the opportunity to gather suitable literature articles and get one-on-one time with the instructor to discuss their hypotheses and predictions. Students will either predict that an elevation of activity levels in the cultures will cause an increase or decrease in PNN expression. In terms of the second question, students will either hypothesize that aggrecan, a component of the PNN, is made by neurons or glia. When evaluating these lab reports, I am looking for hypotheses and predictions that are supported by the literature and students must discuss how their own results fit in with a greater body of work. One of the primary ways that scientific information is conveyed is through written communication, thus, this assessment provides students with the practice in how to articulate their findings. Students are graded individually, so they are responsible for their own work. The detailed rubric breaks down exactly how the students will be graded. Moreover, you will not only be able assess student understanding and learning through the lab report, but also through the small formative assignments, as well as in conversation throughout the lab sessions.
Students generate primary cultures from embryonic mouse brains, stain the cultures, and then image them over the course of this exercise. They additionally learn sterile technique and how to generate cell counts. Importantly, many of the concepts discussed in class are put to use in this exercise, namely, cell culture technique, immunofluorescence and microscopy. Students find this laboratory intriguing and are excited when they are able to examine the stained cultures under the microscope.
To some students the lab report and delving into the associated literature might seem daunting; so you might choose to use one or more laboratory sessions throughout the semester on how to read and synthesize a journal article. One semester, I used a session to discuss a PNN paper so that students had a starting point for their own research. This dedicated session allowed us to spend more time talking about the field and students were able to freely ask questions.
This laboratory exercise requires use of the following equipment: cell culture hood, designated cell culture incubator, designated cell culture centrifuge, microscopes to perform cell counts using a hemocytometer, and fluorescence microscopes. If you only have access to one cell culture hood, then I recommend having groups rotate and providing the students with another assignment while they are not actively working in the cell culture hood. If there are additional cell culture hoods available, you need to ensure that you can monitor all the students in the hoods at a given time. This technique requires precision and sterile technique, so unless the class is well trained in this, you will need to determine how many groups you can oversee at a given time. The amount of space in the incubator is minimal, as 24-well plates can easily be stacked, so if you have to share the space with a colleague, that would work well. I recommend setting up multiple microscopes to count cells on. A simple inverted light microscope would suffice. Have multiple hemocytometers available, so more than one group of students can count at the same time. Both the cell culture centrifuge and incubator should be designated as cell culture equipment to prevent any contamination. If the centrifuge needs to be shared, it is best to ensure that bacteria are not used in the centrifuge, as any spills could contaminate the cultures. In terms of fluorescence microscopes, it would be best to have at least one microscope/group, but in the past, I have had students rotate using the microscopes because there were not enough available.
The first session might run longer than the 2 hour 50 minute time period that I typically am allotted. To prevent this, decide how many groups to have and how many people to assign to each group. I recommend no more than 4 groups and no more than 4 students/group. Another option that might work for other classes of varying sizes would be to have two groups come to the first half of lab so that they could finish the experiment in its entirety and then have the other two groups come to the latter half of lab. This could cut down on the overall time and prevent students from having to rotate in and out of the culture hood. Moreover, this adjustment would be useful for teaching larger classes. If teaching a large number of sections, then I would recommend that you establish a sign-up sheet to schedule appointments to use the fluorescence microscopes. If needed, this could be outside of class hours to accommodate the students and ensure that everyone has an opportunity to view their specimens.
Keep in mind that the embryos need to be a particular age upon harvest, so plan accordingly if there are multiple sections. Ideally, the mice would arrive about one to two days prior to the dissection. If two sections are on the same day, the embryos can be initially dissected together and then the brains can be separated for each section. Performing the exercise on a different day (other than at E16) would not be advised, as this developmental stage was selected based on previous work (17-18). Briefly, dissections are quite easy to perform at this stage and it is easier to isolate a plethora of single cells at this stage.
To make the lab a bit simpler (perhaps for a more introductory Cell Biology course at the sophomore level), you might opt to just study endogenous PNN formation—in other words, omit the use of KCl and AraC. This would eliminate a discussion of how changes in activity impact PNNs, as well as the cellular source of the molecules that comprise the PNN. However, this would require that the cells remain in culture for a longer period of time (approximately 3 weeks). If you are comfortable with a three-week gap between the beginning of the experiment and the staining, then that would be suitable. This would still provide students with the same fundamental techniques, but would make the lab report more straightforward. Alternatively, another form of assessment can be assigned instead of the lab report.
In the past, I have not had students hand in drafts of the lab report, but in my experience, some students would benefit from this. Depending on the class and the student body, you could assign this lab earlier in the semester to allow time for a first submission and for the students to get feedback. I have also thought about potentially having a workshop where students get feedback from their peers on the lab report. This allows for peer-teaching opportunities. Additionally, students could create posters instead and present their findings to the class or even the department. This would help the students hone their presentation skills.
While I have not had students analyze their results quantitatively, that can be incorporated into the exercise as well. Students can count the number of PNNs/group by picking 5-10 fields of view using the 20x objective and recording their results. They could use a one-way ANOVA to analyze their results if they are well versed in the use of statistics and have access to statistical programs. Data from multiple groups could be pooled together for the purposes of analysis or the students could be provided with a data set.
Depending on the outline of the course, you may wish to change the spacing between the sessions. As written, there is one week between each session, with the experiment taking four weeks. It would be possible for someone wishing to adopt this exercise to have more than a week between Lab Session 3 and Lab Session 4, where the students analyze their results.
This exercise provided students with an opportunity to learn fundamental techniques that are used by Cell Biologists and reinforce concepts from lecture while answering specific research-based questions on the neural ECM. Students enjoyed this exercise and found this experience rewarding and exciting.
SUPPORTING MATERIALS
- Supporting File S1: Studying the Neural Extracellular Matrix – Lab Session 1 Assignment. This file contains the formative assessment given to students after completion of the Lab Session 1 exercise. In this assignment, students need to write the Materials and Methods section as well as organize their cell count, cell density, and plating information.
- Supporting File S2: Studying the Neural Extracellular Matrix – Lab Session 3 Assignment. In the Lab Session 3 formative assessment students will organize the methods employed in Lab Session 3 of the experiment and will also find primary or secondary literature articles to address two main points. The first question asks students to think about what PNNs are, where they are found, and the molecules that are found in these structures. Secondly, students need to set forth hypotheses to answer the two central pointed questions that form the basis of the lab report. They will also need to find primary literature articles to support their hypotheses.
- Supporting File S3: Studying the Neural Extracellular Matrix – Dissection Information. This Word document contains detailed information, including pictures showing how to dissect the cerebral cortices of embryonic mouse brains.
- Supporting File S4: Studying the Neural Extracellular Matrix – Lab Session 1 Handout. This handout details the procedure that students will follow in Lab Session 1 of the experiment where they generate single cells from dissected embryonic mouse brains, count the cells, and then plate them. Please note that, where applicable, review articles were referenced in order to reduce the number of total sources as well as to encourage the students to find suitable primary sources for their lab reports.
- Supporting File S5: Studying the Neural Extracellular Matrix – Lab Session 2 Pre-Lab Lecture. This pre-lab lecture goes over the background information necessary for the students to understand the two central questions that will be answered in this experiment. Additionally, the methods that were employed in Lab Session 1 are reviewed.
- Supporting File S6: Studying the Neural Extracellular Matrix – Lab Session 2 Handout. This handout contains the instructions for Lab Session 2 in which students work on formulating hypotheses and predictions for this experiment as well as searching for applicable primary and secondary literature articles.
- Supporting File S7: Studying the Neural Extracellular Matrix – Lab Session 3 Pre-Lab Lecture. This pre-lab lecture briefly discusses the importance of fixing and blocking cultures prior to applying primary and secondary antibodies.
- Supporting File S8: Studying the Neural Extracellular Matrix – Lab Session 3 Handout. This handout details the steps that students will undertake in Lab Session 3 to fix and block their cultures as well as apply primary antibodies.
- Supporting File S9: Studying the Neural Extracellular Matrix – Lab Session 4 Pre-Lab Lecture. This pre-lab lecture describes how to use the fluorescence microscopes and analyze the results. It also explains all the components of the lab report. Lastly, it provides example images of what aggrecan-based PNNs and glial cells look like, which will open up a broader discussion of what has previously been done in the field.
- Supporting File S10: Studying the Neural Extracellular Matrix – Lab Session 4 Handout. The Lab Session 4 handout provides the necessary instructions for students to use the fluorescence microscopes and analyze their results. There is also a chart that the students can use to organize their observations as they view their slides.
- Supporting File S11: Studying the Neural Extracellular Matrix – Instructor Notes. This document provides all the necessary information for this exercise, including catalog and company information for reagents and more specific details about how to prepare the reagents that are needed for each session.
- Supporting File S12: Studying the Neural Extracellular Matrix – Lab Report and Rubric Information. This document contains a breakdown of what should be included in the lab report as well as the associated rubric.
ACKNOWLEDGMENTS
I would like to thank Dr. Russell T. Matthews for allowing me to work on PNNs when I was a graduate student in his lab. I also would like to acknowledge all the hard-working Cell Biology students at Western Connecticut State University (Spring 2017, 2018, and 2019) who performed this exercise. I also want to thank the students who helped me prepare for these experiments (Phoebe Ermert, Brittany Schappach, and Karen Velez) and those that cared for the mice (Darcy Curillo, Niko Frascone, Molly Gallagher, Jake Lipinski, Daniel Monahan, and Heather Rosenblatt).
References
- Woodin T, Carter VC, Fletcher L. 2010. Vision and Change in Biology Undergraduate Education, A Call for Action - Initial Responses. CBE Life Sci. Educ. 9:71-73. doi: 10.1187/cbe.10-03-0044.
- Bowey-Dellinger K, Dixon L, Ackerman K, Vigueira C, Suh, YK, Lyda T, Sapp K, Grider M, Crater D, Russell T, Elias M, Coffield VM, Segarra VA. 2017. Introducing Mammalian Cell Culture and Cell Viability Techniques in the Undergraduate Biology Laboratory. J. Microbiol. Biol. Educ. 18:18.2.38. doi: 10.1128/jmbe.v18i2.1264.
- Mozdziak PE, Petitte JN, Carson SD. 2004. An introductory undergraduate course covering animal cell culture techniques. Biochem. Mol. Biol. Educ. 32:319-322. doi: 10.1002/bmb.2004.494032050381.
- Marion RE, Gardner GE, Parks LD. 2012. Multiweek cell culture project for use in upper-level biology laboratories. Adv. Physiol. Educ. 36:154-167. doi: 10.1152/advan.00080.2011.
- Catlin R, Taylor A, Ditchek L, Burnett S, Khan S, Todd O, Adams M, Touhey E, Wynkoop A, Ryan J. 2016. Using Cultured Mammalian Neurons to Study Cellular Processes and Neurodegeneration: A Suite of Undergraduate Lab Exercises. J. Undergrad. Neurosci. Educ. 14:A132-A137.
- Heidemann SR, Reynolds M, Ngo K, Lamoureux P. 2003. The culture of chick forebrain neurons. Methods Cell Biol. 71:51-65.
- Freestone NS, Sam CL. 2017. Classical and novel pharmacological insights offered by the simple chick cardiomyocyte cell culture model: a valuable teaching aid and a primer for "real" research. Adv. Physiol. Educ. 41:163-169. doi: 10.1152/advan.00178.2015.
- Weaver D. 2007. Cardiac Cells Beating in Culture: A Laboratory Exercise. The American Biology Teacher. 69:407-410. Doi: 10.1662/0002-7685(2007)69[407:CCBICA]2.0.CO;2.
- Prieto D, Aparicio G, Sotelo-Silveira JR. 2017. Cell migration analysis: A low-cost laboratory experiment for cell and developmental biology courses using keratocytes from fish scales. Biochemistry and Molecular Biology Education. 45:475-482. doi: 10.1002/bmb.21071.
- Frantz C, Stewart KM, Weaver VM. 2010. The extracellular matrix at a glance. J. Cell Sci. 123:4195-4200. doi: 10.1242/jcs.023820.
- Jayadev R, Sherwood DR. 2017. Basement membranes. Curr. Biol. 27:R199-R217. doi: 10.1016/j.cub.2017.02.006.
- Bandtlow CE, Zimmermann DR. 2000. Proteoglycans in the developing brain: new conceptual insights for old proteins. Physiol. Rev. 80:1267-1290. doi: 10.1152/physrev.2000.80.4.1267.
- Celio MR, Spreafico R, De Biasi S, Vitellaro-Zuccarello L. 1998. Perineuronal nets: past and present. Trends Neurosci. 21:510-515.
- Kwok JC, Dick G, Wang D, Fawcett JW. 2011. Extracellular matrix and perineuronal nets in CNS repair. Dev. Neurobiol. 71:1073-1089. doi: 10.1002/dneu.20974.
- Brückner G, Grosche J. 2001. Perineuronal nets show intrinsic patterns of extracellular matrix differentiation in organotypic slice cultures. Exp. Brain Res. 137:83-93.
- Dityatev A, Brückner G, Dityateva G, Grosche J, Kleene R, Schachner M. 2007. Activity-dependent formation and functions of chondroitin sulfate-rich extracellular matrix of perineuronal nets. Dev. Neurobiol. 67:570-588. doi: 10.1002/dneu.20361.
- Giamanco KA, Morawski M, Matthews RT. 2010. Perineuronal net formation and structure in aggrecan knockout mice. Neuroscience. 170:1314-1327. doi: 10.1016/j.neuroscience.2010.08.032.
- Giamanco KA, Matthews RT. 2012. Deconstructing the perineuronal net: cellular contributions and molecular composition of the neuronal extracellular matrix. Neuroscience. 218:367-384. doi: 10.1016/j.neuroscience.2012.05.055.
- Chapman KJ, Meuter M, Toy D, Wright L. 2006. Can't We Pick our Own Groups? The Influence of Group Selection Method on Group Dynamics and Outcomes. J. Manag. Educ. 30:557-569. doi: 10.1177/1052562905284872.
- Hilton S, Phillips F. Instructor-Assigned and Student-Selected Groups: A View from Inside. 2010. Issues in Accounting Education. 25:15-33. doi: 10.2308/iace.2010.25.1.15.
- Pacifici M, Peruzzi F. 2012. Isolation and culture of rat embryonic neural cells: a quick protocol. J. Vis. Exp. 24:e3965. doi: 10.3791/3965.
- Tanaka T. 1998. Effects of litter size on behavioral development in mice. Reprod. Toxicol. 12:613-617. doi: 10.1016/s0890-6238(98)00045-8.
Article Files
Login to access supporting documents
Using a Primary Cell Culture Model to Study the Neural Extracellular Matrix(PDF | 910 KB)
S1. Studying the Neural Extracellular Matrix-Lab Session 1 Assignment.docx(DOCX | 14 KB)
S2. Studying the Neural Extracellular Matrix-Lab Session 3 Assignment.docx(DOCX | 14 KB)
S3. Studying the Neural Extracellular Matrix-Dissection Instructions.docx(DOCX | 511 KB)
S4. Studying the Neural Extracellular Matrix-Lab Session 1 Handout.docx(DOCX | 1 MB)
S5. Studying the Neural Extracellular Matrix-Lab Session 2 PPT.pptx(PPTX | 2 MB)
S6. Studying the Neural Extracellular Matrix-Lab Session 2 Handout.docx(DOCX | 100 KB)
S7. Studying the Neural Extracellular Matrix-Lab Session 3 PPT.pptx(PPTX | 159 KB)
S8. Studying the Neural Extracellular Matrix-Lab Session 3 Handout.docx(DOCX | 217 KB)
S9. Studying the Neural Extracellular Matrix-Lab Session 4 PPT.pptx(PPTX | 262 KB)
S10. Studying the Neural Extracellular Matrix-Lab Session 4 Handout.docx(DOCX | 125 KB)
S11. Studying the Neural Extracellular Matrix-Instructor Notes.docx(DOCX | 26 KB)
S12. Studying the Neural Extracellular Matrix-Lab Report and Rubric Information.docx(DOCX | 24 KB)
- License terms

Comments
Comments
There are no comments on this resource.