My Dog IS My Homework: Exploring Canine Genetics to Understand Genotype-Phenotype Relationships
Editor: Lisa McDonnell
Published online:
Abstract
To facilitate understanding of the fundamental genetic concept of the genotype-phenotype relationship in our introductory biology students, we designed an engaging multi-week series of related lessons about canine genetics in which students explore and answer the question, "How does the information encoded in DNA lead to physical traits in an organism?" Dogs are an excellent model organism for students since the genetic basis for complex morphological traits of various breeds is an active area of scientific research and dog DNA is easily accessible. Additionally, examination of students' pets offers a relatable, real-world, connection for students. Of the more than 19,000 genes that control canine genetics, simple genetic mutations in three genes are largely responsible for the coat variations of dogs –specifically, the genes that control hair length, curl, and the presence/absence of furnishings. In our lessons, students collect DNA samples from dogs, isolate and amplify targeted sections of DNA through polymerase chain reactions (PCR), and then sequence and analyze DNA for insertions and single nucleotide polymorphism (SNP) mutations. Utilizing gel electrophoresis and bioinformatics tools, students connect how the physical manifestation of traits is rooted in genetic sequences. Students also participate in discussions of scientific literature, group collaboration to construct a final poster, and presentation of their findings during a mock scientific poster conference. Through this module students engage in progressive exploration of genetic and molecular techniques that reveal how simple variations in a few DNA sequences in combination lead to a broad diversity of coat quality in domestic dog breeds.
Primary image. Genetic Analysis of Canine Coat Morphologies. Three dogs with differing coat morphologies analyzed by students (A, B, C), an agarose gel post-electrophoresis (D), and a chromatogram of a DNA sequence highlighting a relevant mutation (E). This collage contains original images taken by authors and course participants.
Citation
Muilenburg V, Best A, Chase L, McDonough V, Stukey J, Hoey S, Scogin SC. 2020. My dog IS my homework: Exploring canine genetics to understand genotype-phenotype relationships. CourseSource. https://doi.org/10.24918/cs.2020.24
Society Learning Goals
Genetics
- Genetic variation
- How do different types of mutations affect genes and the corresponding mRNAs and proteins?
Lesson Learning Goals
Students will:
- Understand the scientific method, including asking questions, articulating research objectives, making observations, analyzing results, and drawing conclusions.
- Learn how to read and interpret primary literature.
- Develop collaborative skills.
- Understand basic genetics including the relationship between genotype and phenotype and the consequences of genetic mutations on organismal phenotypes.
- Learn about various genetics-related procedures and how to follow strict protocols.
- Learn how to keep a scientific laboratory notebook.
- Learn about the parts of a scientific poster and how to deliver a research presentation.
Lesson Learning Objectives
Students will be able to:
- Interpret selected articles from scientific journals and synthesize relevant information related to canine genetics and coat inheritance.
- Work together as a team to conduct experiments following strict protocols that produce usable results.
- Keep a detailed laboratory notebook.
- Collaboratively create a research poster and participate in a poster session mimicking a scientific conference.
- Explain the difference between genotype and phenotype; describe the consequences of gene mutations on protein structure and function; articulate the combinatory effects of three specific genes on dog coat phenotypes.
- Perform the following skills in a laboratory setting: isolate DNA, amplify DNA using PCR, analyze DNA sample using gel electrophoresis, use basic bioinformatic tools to analyze DNA sequence data.
Note: Additional, more specific objectives and goals are included with each of the lessons.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Origin and Rationale
In our college's previous introductory biology lab curriculum, students learned the scientific process and a wide array of biological topics through instructive, research-like lessons. Although engaging, each lesson was independent from each other, and concepts and findings could not be directly related to one another as the course progressed. In the case of the genetics lessons, we believe this segmented lesson style made it difficult for students to understand the effects of multiple genes and how they simultaneously affect the phenotypes of organisms. We redesigned our curriculum into modules that contain a series of progressive lessons that aim to answer an overarching question. We present here our genetics module in which students ask, "How does the information encoded in DNA lead to physical traits of an organism?" To answer this question, we analyze the genotype of our beloved dogs at three genetic loci that control coat morphology.
In this series of four lessons, students learn genetic and molecular techniques while exploring the world of canine genetics. Prior to the first lesson, students take home a DNA sample collection kit to obtain cheek cells from their dogs (Supporting File S1. Dog Genetics – Module Introduction, pages 5-6). In lesson 1, students extract DNA from their samples and setup polymerase chain reactions (PCR) to amplify regions of the three genes (Supporting File S2. Dog Genetics – Lesson 1. DNA Extraction and PCR Sample Preparation). In lesson 2, students analyze alleles of one of the genes using gel electrophoresis (Supporting File S3. Dog Genetics – Lesson 2. Gel Electrophoresis). Meanwhile, the other two genes are sequenced off-site. In lesson 3, the DNA sequences of the two remaining genes are then analyzed for single nucleotide polymorphisms (SNPs) using bioinformatics software (Supporting File S4. Dog Genetics – Lesson 3. DNA Sequence Analysis). In the fourth lesson of the module, students collaboratively create a poster about the three genes they analyzed. In the poster, students connect the genotype to the phenotype of their dogs and present information on an additional, assigned gene that has been studied in dogs by scientists (Supporting File S5. Dog Genetics – Lesson 4. Final Genotype: Collaborative Poster). Finally, students present their posters in a mini- poster conference that mimics a scientific conference. During this module, students and instructors also engage in discussion of the scientific paper by Cadieu et al. (2009) in Science that inspired this module (1) as well as discuss fundamental concepts of genetics related to information storage and transfer, DNA replication, mutation, and artificial selection. By the end of this module, students have a mechanistic understanding of how variation in organisms' genotypes leads to variation in phenotypes.
Dogs: A Model Organism
More than 180 breeds of dogs are currently recognized by The American Kennel Club (2). Each dog breed has unique traits that are governed by one or more of the 19,000 genes that make up the canine genome (3). Given the well-documented breeding practices, scientists have been able to explore the genetic basis of many complex physical traits exemplified by specific breeds. Extensive research has explained the genetic basis for coat texture (1), coat color (4,5), body size (6), leg length (7) and skull shape (8,9), to name a few. Thus, the dog is an excellent model for undergraduates to explore the molecular basis of complex morphological traits. In our lessons, students focus on three genes that regulate the coat quality of dogs as discovered by Cadieu et al. (2009) (1). Students specifically analyze genes that encode hair length (FGF5), curl (KRT71), and the presence/absence of furnishings (RSPO2) (1). The diversity in appearance of coat quality that we observe across dog breeds today largely result from combinatory effects of simple genetic mutations in these three genes (1).
Other aspects of dog genetics have also been explored in undergraduate labs. In a multi-week investigation, the relationship between dog size and variation in SNPs has been examined using a combination of PCR, restriction digests, and gel electrophoresis (10). Nuclear and mitochondrial DNA from dogs have also been analyzed to solve mock forensic crimes using PCR and gel electrophoresis (11). The module of lessons we present here is unique in that the concepts and methodologies are readily accessible to first-year biology students with little to no prior knowledge of modern molecular techniques. Other strengths of this module include (a) consideration of combinatory effects of three genes on the phenotype of an organism, (b) analysis of the impact of both insertion and SNP mutations, (c) use of different methodological approaches associated with different kinds of mutations, and (d) exposure to basic bioinformatic tools (NCBI blast, DNA alignment, and protein alignment). Finally, the use of dog samples eliminates the privacy concerns associated with the use of human (often student) samples.
Gaining Fundamental Molecular Skills
One important curricular aspect of these lessons is introducing fundamental techniques of molecular biology to our students early in their (scientific) collegiate careers – techniques like DNA extraction, PCR, gel electrophoresis, and sequence analysis. Mastering basic molecular techniques and understanding their conceptual underpinnings are essential to today's graduates of biology. This view is supported in multiple publications on teaching molecular techniques in introductory and advanced courses across an array of biological topics (e.g., 12-14). PCR and/or gel electrophoresis, specifically, have been used in undergraduate classes to detect GMOs in food (15-17), to analyze how replacing traditional buffers with beverages impacts gel electrophoresis (18), to troubleshoot methodology of gel electrophoresis virtually (19), and to solve fictitious crimes via analysis of DNA (19,20). Analysis of a SNP involved in bitter-tasting ability (21) and dyslexia (22) in humans has also been integrated into the undergraduate biological curriculum at some institutions.
Intended Audience
This module of lessons was developed for students in an introductory biology class or laboratory, for biology majors.
Required Learning Time
This set of four lessons was designed as a module. Each experimental lesson within the module was designed for one, three-hour laboratory period to complement lecture material on genetics in an introductory biology course. It takes five laboratory periods to complete the entire module so that students can present their posters in a mock scientific conference.
Prerequisite Student Knowledge
No genetics knowledge is needed beyond what is written in the introductory material of each lesson. However, students are required to read the background material and procedures associated with each week's lesson before they come to class (Supporting Files S1. Dog Genetics – Module Introduction, S2. Dog Genetics – Lesson 1. DNA Extraction and PCR Sample Preparation, S3. Dog Genetics – Lesson 2. Gel Electrophoresis, S4. Dog Genetics – Lesson 3. DNA Sequence Analysis, S5. Dog Genetics – Lesson 4. Final Genotype: Collaborative Poster). This familiarizes students with basic concepts, terms, and procedures. Students are required to complete a pre-lesson assignment in their lab notebooks in which they record the title, date, research objective(s), and summarized procedures to ensure they are familiar with the lesson. They also complete pre-lesson worksheets associated with the lessons. Students learn the molecular skills needed during the lab from verbal instruction and from following the procedures written in the lessons.
Prerequisite Teacher Knowledge
In order to successfully teach these lessons, instructors must have a basic familiarity with genetics and the transfer of information from DNA to proteins. Instructors should also be familiar with common molecular techniques, including DNA extraction, polymerase chain reaction (PCR), gel electrophoresis, BLAST, and basic bioinformatic analysis. Finally, the instructor and students must also have the means to sample and extract DNA, perform PCR, perform gel electrophoresis, and sequence DNA. DNA sequencing can be done either at the home institution or off-site, depending on availability of equipment at the institution. To make posters, students need computers and access to appropriate software (e.g., PowerPoint, Google Slides), as well as the ability to print posters for the mock conference.
SCIENTIFIC TEACHING THEMES
Active Learning
In Class: In the classroom, the entire module engages students in active learning. From start to finish, students are active participants in authentic science as they collaborate on hands-on lab work and problem-solve in small groups to conduct DNA analysis. Students also engage with primary literature and, like scientists, actively maintain an accurate, up-to-date lab notebook for the duration of the module. Students also create a research poster of their results (including images of gels, chromatograms, etc.). Furthermore, students communicate like scientists as they present for one hour in a format mimicking an interactive, scientific research poster session (see Supporting File S5. Dog Genetics – Lesson 4. Final Genotype: Collaborative Poster).
Outside of Class: From the beginning of this module, students are asked to be active participants in their learning. Specifically, students are given pre-lesson reading and questions to gauge prior knowledge and build a conceptual framework for future learning. Also, students are given the materials and expertise to collect data from dogs at their home or in their community. For many, this raises engagement as they become interested in knowing specific genetic characteristics of animals that are close to them and their families. As students work on pre-lesson assignments, they model good scientific procedure by contributing to their ongoing lab notebook. Finally, a good portion of the creation of the final research poster is completed out-of-class. This is an important part of the active learning process, as students must combine content knowledge with 21st century skills to set up meetings, communicate, compromise, and ultimately develop a final product that meets the expectations of all group members.
Assessment
Assessments are embedded throughout the module and include (a) pre-and post-lesson worksheets (Supporting Files S1. Dog Genetics – Module Introduction, S2. Dog Genetics – Lesson 1. DNA Extraction and PCR Sample Preparation, S3. Dog Genetics – Lesson 2. Gel Electrophoresis, S4. Dog Genetics – Lesson 3. DNA Sequence Analysis), (b) lab notebooks (Supporting File S6. Dog Genetics – Lab Notebook Grading Rubric), and (c) student evaluation of participation of group members (Supporting File S7. Dog Genetics – Group Evaluation). We also include two summative assessments at the end of the module in the form of a quiz (Supporting File S8. Dog Genetics – Example Quiz Questions) and a collaborative poster (Supporting File S9. Dog Genetics – Poster Template). Additionally, students and instructors intermittently discuss the Cadieu et al. (2009) Science paper (1) which was the inspiration for this module during the first three lessons. Discussions occur during "down-time," e.g., when the gels are running during Lesson 2 (Supporting File S3. Dog Genetics – Lesson 2. Gel Electrophoresis). Students engage in discussion of the paper and are guided by worksheets provided by the Instructor (Supporting File S10. Dog Genetics – Cadieu et al. Discussion). For instance, discussion of the paper starts with understanding the primary objective and general methodology of the research presented and then transitions to specific methodology and results for each gene. The discussions are complemented by scaffolded homework (pre- and post-lesson worksheets) that integrates analysis of the scientific paper with understanding methodology and students' experimental results from their analyzed dogs.
We emphasize the value of record keeping in our introductory biology laboratory course, since scientists, health professionals, and teachers will be required to keep records of their experiments, patients, or students. Our students keep records by maintaining lab notebooks. Lab notebooks are required to include the following elements: date, experiment title, group members' names, purpose of experiment, data, sample identification numbers, calculations, figures/tables, results, and conclusions. Students also answer questions that are embedded in some of the lessons in their lab notebooks. Lab notebooks are informally scanned by instructors during the lab times to verify students are recording relevant information, data, and activities. At the end of the module, lab notebooks are formally assessed by grading selected elements from each lesson. Each student lab notebook is evaluated on the same elements that are listed on a grading rubric (Supporting File S6. Dog Genetics – Lab Notebook Grading Rubric), and each element appears at least twice on the grading rubric. This method of evaluating selected criteria is an effective grading strategy because we can give specific, informative feedback without grading the entire lab notebook.
Students are given a summative assessment at the end of the module in the form of a quiz (Supporting File S8. Dog Genetics – Example Quiz Questions). The quiz evaluates students' understanding of the first three lessons and the Cadieu et al. (2009) paper (1). In our course, students are given a quiz at the beginning of Lesson 4 and then work in their groups to create a poster summarizing their research and results from their two dogs. For most of our students, this is the first scientific poster they have made, so we discuss common mistakes and discuss expectations described in the grading rubric (Supporting File S5. Dog Genetics – Lesson 4. Final Genotype: Collaborative Poster). Students also investigate and present an additional, assigned gene that regulates a different aspect of dogs' phenotypes in a small section of their posters. We provide a modifiable PowerPoint template from which students design their posters (Supporting File S9. Dog Genetics – Poster Template).
During the final lesson, students present their results in a poster conference that models a professional conference. Students take turns asking and answering questions about their research during the conference and fill-out a questionnaire from this discussion (Supporting File S5. Dog Genetics – Lesson 4. Final Genotype: Collaborative Poster). Additionally, all participants of the conference evaluate the posters and vote to determine the best poster. The winning poster receives a "best in show" certificate, ribbon, and prize. Lastly, students self-assess their own participation in the group and the participation levels of other group members using a standard form (Supporting File S7. Dog Genetics – Group Evaluation).
Inclusive Teaching
Helping students find identity and purpose in science is an important aspect of inclusion. In this module, students are invited to have a personal investment in the project as they bring samples from their personal pets for analysis. Although this may seem trivial to some, students often respond to the module by referencing how exciting it is to analyze their own pets (see Student Reaction section).
In addition, students work in small, collaborative groups throughout the module and share expertise amongst themselves. In these situations, students of diverse backgrounds often find comfort and develop camaraderie with others as they are able to use their strengths and bolster their weaknesses in a collaborative setting. Finally, the breadth of the activities in this module promote inclusive learning through stimulation of multiple modalities. For example, students isolate and analyze DNA, conduct primary literature research, discuss/debate results, and co-create a poster presentation (which is modeled after a professional scientific poster session).
LESSON PLAN
Genetics Module Overview
The primary goal of this module is to explore the relationship between genotypes and phenotypes of organisms to answer the question, "How does the information encoded in DNA lead to physical traits in an organism?" To answer this question, students analyze three genes that are associated with the coat quality of dogs: hair length, curl, and furnishings. While exploring the genotype-phenotype relationship, students learn several fundamental techniques of genetics research, including DNA isolation, polymerase chain reaction (PCR), gel electrophoresis, and DNA sequence analysis.
Prior to the first lesson, students and instructors obtain DNA samples from dogs using a cytology brush and complete a short data sheet about the coat appearance of the sampled dog (Supporting File S1. Dog Genetics – Module Introduction). In the first lesson, students isolate DNA from the canine cheek cells they collected (Supporting File S2. Dog Genetics – Lesson 1. DNA Extraction and PCR Sample Preparation). Next, students use polymerase chain reaction (PCR) to amplify (make copies of) portions of three genes that encode hair length, curl, and furnishings. Students analyze the PCR products of the gene associated with furnishings in Lesson 2 (Supporting File S3. Dog Genetics – Lesson 2. Gel Electrophoresis). Presence of the furnishings trait (i.e., eyebrows and moustache) is a consequence of an insertion mutation in the RSPO2 gene. Dogs with furnishings have the insertion mutation in at least one of their two RSPO2 alleles. Dogs without furnishings have two RSPO2 alleles without the insertion. Students use gel electrophoresis to determine which two RSPO2 alleles are present in their dogs. Dogs with a homozygous RSPO2 genotype display a single band on the gel, whereas dogs with a heterozygous RSPO2 genotype display two bands on the gel (See Primary Image Part D). Finally, students connect the genotype revealed on their gels with the presence or absence of furnishings that they observe when looking at their dogs.
For hair length (FGF5) and curl (KRT71) genes, instructors and teaching assistants remove impurities from the PCR products between lessons 1 and 2 using commercially available biological kits and protocols. Students could also complete this purification step in an additional laboratory period, but we do not have time in our current curriculum. Purified products are sequenced off-site using Sanger Sequencing (cost $3.87 per sample in 2019). Analysis of the sequence data for hair length and curl genes allows students to investigate the consequences of point mutations (Supporting File S4. Dog Genetics – Lesson 3. DNA Sequence Analysis). A change in the identity of a nucleotide from G to T at position 284 in exon 1 of the FGF5 gene alters one amino acid in the translated protein (Cys95Phe), resulting in the long hair phenotype of dogs. Similarly, the curly hair phenotype results from a change in nucleotide C to T at position 451 in exon 2 of KRT71, which also alters the identity of one amino acid of the translated protein (Arg151Trp). Students analyze the nucleotide sequences and amino acid sequences using a variety of free software programs, including NCBI's Blast, ExPASy, and sequencing chromatogram viewing software. To determine the identity of their dog's nucleotides at position 284 on FGF5 and 451 on KRT71, students align their dog's DNA sequence to a wolf DNA sequence because wolves express the ancestral phenotype. Students then use the chromatogram viewing software to identify if their dog has a homozygous genotype (which appears as a single chromatogram peak at the SNP position of interest – see Primary Image Part E) or a heterozygous genotype (which appears as two overlapping chromatogram peaks at the SNP position of interest). Ultimately, students link the DNA sequence, amino acid sequence, and the phenotype of their dogs by analyzing their DNA sequence, identifying the SNPs at the positions of interest, and translating codons into amino acids (Figure 1).
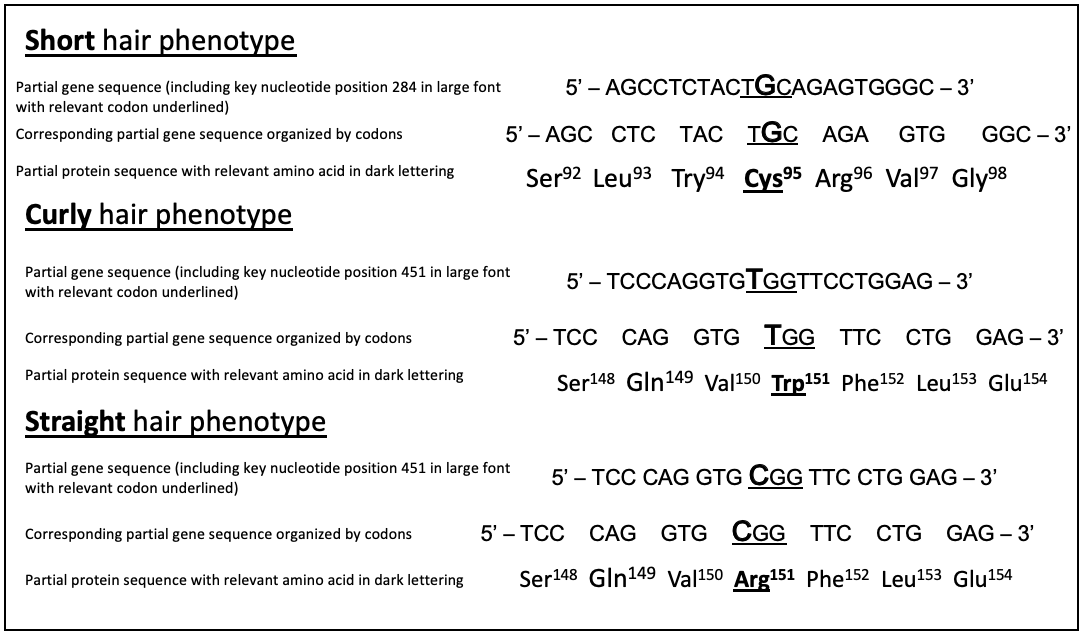
Figure 1. A model student figure presenting the link between the genotype and phenotype of an experimental dog that has only one allele for FGF5 (short hair phenotype) and both the curly and straight alleles for KRT71 (curly hair phenotype).
Student groups collaboratively create a scientific poster to share their research on the genotype-phenotype analysis of their two dogs in a mock scientific conference. In the poster, students provide background material, predictions, research objectives, methodology, and results of their RSPO2, FGF5, and KRT71 analyses. Students also present the genotype-phenotype relationship of an additionally assigned gene that scientists have studied to explore more about canine genetics (Supporting File S5. Dog Genetics – Lesson 4. Final Genotype: Collaborative Poster). Every group is assigned a different canine gene to present that has been found to affect a particular phenotype in dogs. By participating in the poster conference, students learn about different combinations of the three genes analyzed in the lessons that were not expressed by their group's two dogs, as well as about the assigned genes. Students gain practice at communicating scientific information to colleagues and the general public. Additionally, students gain exposure to a variety of poster designs and have the opportunity to reflect on effective and ineffective approaches in disseminating information as they vote for the best poster.
Over two years in our course, our students have sampled DNA from 198 dogs representing over 80 breeds or mixed breeds that exhibit variable phenotypes (Figure 2). Based on the observed phenotype, the ancestral form of no furnishings and straight hair were more common than the variant form of furnishings and curly hair, respectively. However, the variant form of long hair has been slightly more common than the ancestral form of short hair (Figure 2).
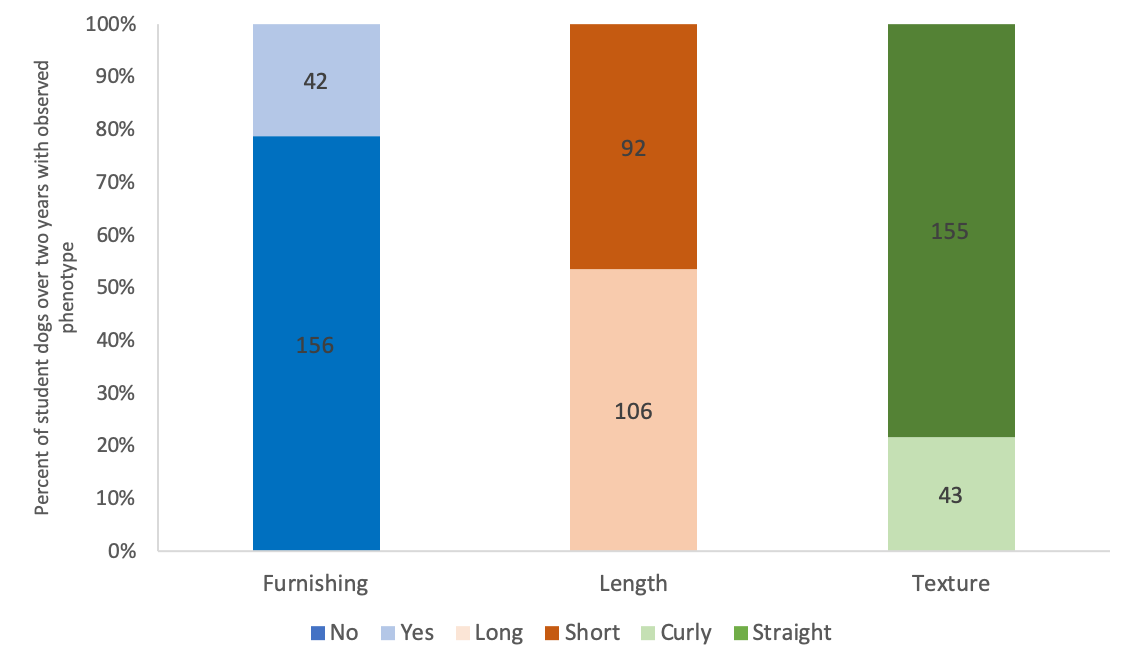
Figure 2. Percent of student dogs over two years with observed phenotype. For each trait, the ancestral version is indicated as a dark color and the variant version is indicated as a light color. The total numbers of dogs observed for each version of each trait are indicated within bars.
Of the sampled 198 dogs, our students have genotyped 108 dogs. We are not able to analyze all the dog DNA samples obtained by students because of cost constraints, so we analyze two dog DNA samples per student group. Of the 108 dogs chosen for genotype analysis, students were successful in identifying the RSPO2 genotype that encodes furnishings via gel electrophoresis for 77% of their dogs. When students do not observe bands in in their gels, they repeat the analysis in an open lab at the end of the week. If the repeated analysis of RSPO2 is also unsuccessful, students report the expected genotype based on the observed phenotype.
Students analyze DNA sequences for FGF5 which encodes hair length and KRT71 which encodes hair texture. Of the 108 genotyped dogs, students successfully identified the FGF5 genotype of 68% of their dogs and successfully identified the KRT71 genotype of 89% of their dogs. We suspect that the lower success rate of the FGF5 genotype analysis compared to KRT71 was due to poorly optimized sequencing conditions during one year of analysis. When students do not get successful sequence data for a locus, we assign and provide model sequences that correspond with the phenotype of their experimental dog so they can still complete the lesson and learn the material.
For the vast majority of our dogs, the experimentally determined genotypes for RSPO2, FGF5, and KRT71 are consistent with what students predict based on the observed phenotypes. The predominant cause of inconsistent relationships between the phenotypes and genotypes is a student error in correctly identifying the observed phenotype of their dogs. When inconsistencies occur, we ask to see a picture of the student's dog and then inquire about the grooming practices. For example, dogs that get their coats trimmed have a phenotype of long hair but it is trimmed to appear short! However, in some instances the genotype truly does not correspond to the phenotype, and our students have observed this in some dogs. This can occur because of gene interactions that impact gene expression. For example, dogs can only express the curly hair genotype if they also have long hair (1). Additionally, Yorkshire terriers, silky terriers, and Afghan hounds have unidentified genes that contribute to their observed phenotype for the loci of interest (1).
Overcoming Challenges: DNA Samples, Sample Identification Numbers, and Course Spreadsheet
Logistically, one potential barrier to successful implementation of this module is getting enough high-quality dog DNA samples, ideally 2-4 dog samples per student group before DNA extraction in Lesson 1 (for an overview of Lesson 1, see Supporting File S2. Dog Genetics – Lesson 1. DNA Extraction and PCR Sample Preparation). Our students sample dogs over winter break, often their own pets when they go home, but sometimes other dogs from the community. We schedule the genetics module to begin directly after break at our institution. We also entice students with extra credit points for bringing in at least one dog DNA sample, which for our students is strong incentive. Our students gather two DNA samples per dog and use both samples during DNA extraction with kit components and protocols to increase the probability of getting a sufficient quantity of high-quality DNA. Additionally, we give sampling bags to faculty and staff in the department to supplement student samples. As a last resort, instructors can gather dog DNA samples from a local dog rescue.
To successfully execute this module, instructors also need a plan to keep track of the identities of dog DNA samples, the PCR products from the three genes, and the DNA sequence numbers. We use an identification scheme that includes dog identification numbers, PCR identification numbers, and sequencing identification numbers. Dog identification numbers are designated by the instructor and written on each DNA sampling bag and associated data sheet and typically range from 1-200. These numbers are used by students on all of their tubes during the DNA extraction process in Lesson 1 (Supporting File S2. Dog Genetics – Lesson 1. DNA Extraction and PCR Sample Preparation). Next, instructors assign each dog three PCR identification numbers, one for each of the genes that students analyze (RSPO2, FGF5, KRT71). Finally, the PCR products from the FGF5 and KRT71 genes are assigned a sequencing identification number that are generated by our off-site sequencing center. All numbers are recorded on the Sample Identification Datasheet (Supporting File S11. Dog Genetics – PCR Sample Identification DataSheet) and the electronic spreadsheet (Supporting File S12. Dog Genetics – Electronic Spreadsheet). Undergraduate students typically lack experience in using numbering schemes for experimental and record-keeping purposes, so we find it valuable to explain why there are different numbers associated with the different kinds of samples.
We use a course spreadsheet (i.e., Google Sheet) to keep track of dog identification numbers, phenotypes of dogs, and results from analysis of each gene (Supporting File S12. Dog Genetics – Electronic Spreadsheet). Students enter their experimental information in the spreadsheet and identification numbers so all samples and associated identification numbers stay organized. This spreadsheet is also helpful for instructors to review at the end of the module to analyze how successful students are at getting quality data from the three loci of the experimental dogs and to determine if any improvements or changes need to be made in the future.
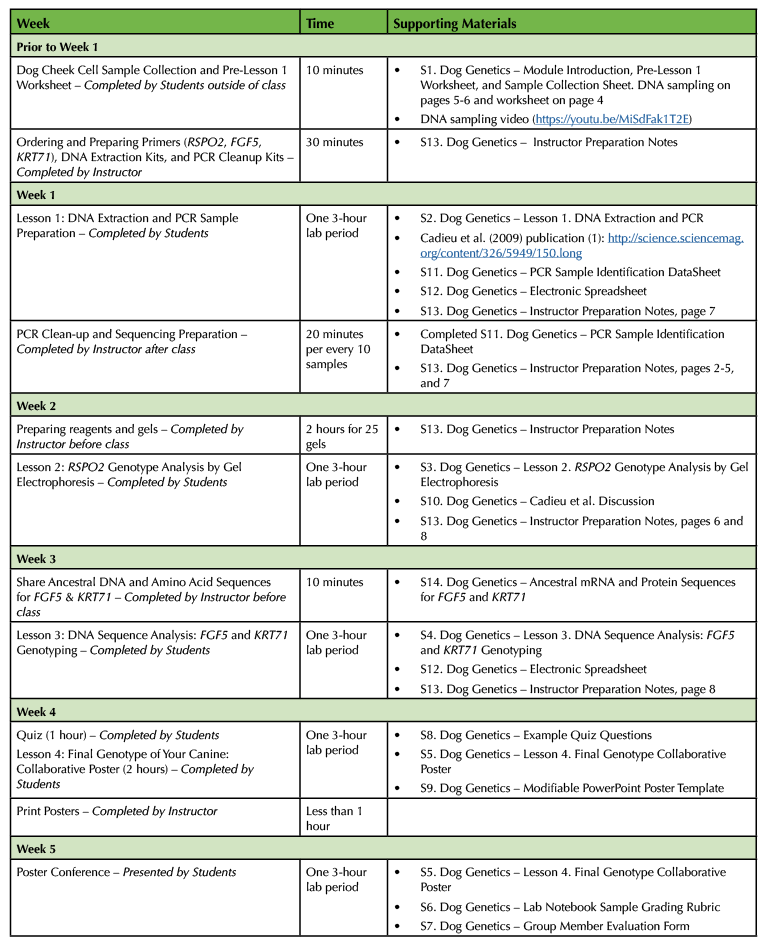
Table 1. Dog Genetics lesson teaching timeline.
TEACHING DISCUSSION
Adaptability of Lessons
The experimental components of this module could be completed in three weeks (Lessons 1-3) if instructors do not want to include a quiz or poster conference. However, our students have informally communicated to us that they do not fully understand their results until after they study for the quiz and/or create the poster. Thus, it appears that a synthetic assessment at the end of the module is important to student comprehension, but the assessment would not have to occur during class.
The sequence of Lessons 1-3 is not modifiable. Both the gel electrophoresis lesson (Supporting File S3. Dog Genetics – Lesson 2. Gel Electrophoresis) and sequence analysis lesson (Supporting File S4. Dog Genetics – Lesson 3. DNA Sequence Analysis) cannot be completed without first completing Lesson 1, where students extract DNA and prepare samples for amplification using PCR (Supporting File S2. Dog Genetics – Lesson 1. DNA Extraction and PCR Sample Preparation). Additionally, instructors must consider the time it takes to sequence DNA on- or off-site before students can complete Lesson 3. If desired and time allowed, instructors could have students complete the PCR cleanup and be involved in the sequencing on-site during class.
Students could analyze other traits using their dog DNA samples, since the genetic basis of an array of morphological traits are understood in dogs (1,4-9). We can envision investigation of an additional gene after completion of the module presented here as a great way to integrate student-directed, independent projects in an upper-level course.
This module is designed for an introductory biology lab course that is populated by biology majors, pre-professional, and education students. However, this module would also be appropriate for a genetics course or could be adapted for a general (non-majors) scientific lab course if the pace of the presented content and methods is reduced. Also, analysis of the Cadieu et al. (2009) paper (1) by students could also be removed from the module to make the lessons more accessible to non-science majors.
Effectiveness in Achieving Learning Goals and Objectives
Previous research indicates that students struggle to understand biological concepts related to molecular procedures when presented with a step-by-step list of instructions (12). Thus, we begin each lesson with a basic conceptual explanation of the procedures. Additionally, students are challenged with understanding the biological concepts related to the techniques prior to lessons as they work through pre-lesson assignments. The sequential nature of this laboratory-based module presents difficult concepts related to protocols over the course of several weeks, so students have time to grasp one idea before being presented with another too quickly. Finally, the personal nature of the lessons serves to motivate student engagement with concepts and completion of lengthy protocols.
This module additionally promotes development of 21st century skills including student collaboration, communication, interpersonal skills, and time management. Students also gain experience with common molecular laboratory equipment, software, and skills.
Student Reactions
A survey of 90 students indicated that most students enjoyed the module (Figure 3). Students felt that both the topic of phenotypes and genotypes, as well as the personal connection of using DNA of a known, and often loved canine, made the lab relatable and interesting. One student reported, "I liked how personal it was. We actually genotyped dogs that were ours." Students also mentioned how being able to visualize mutations in chromatograms to see "that a change [mutation] in a single nucleotide can change everything" raised their interest in the lab. In sum, students enjoyed being able to relate the genotypes to the phenotypes of their own dogs. One student stated, "It was helpful to study something that we can see (phenotype) and understand what is the cause behind this phenotype (genotype)."
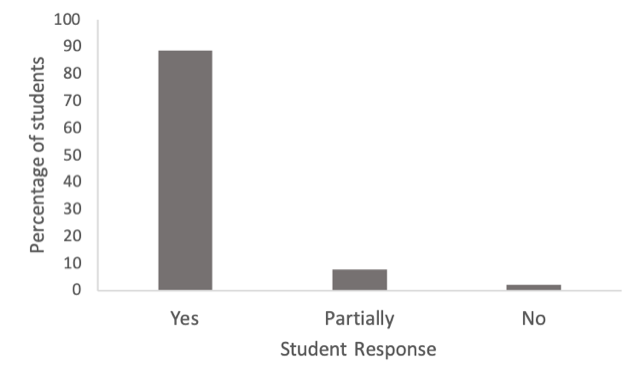
Figure 3. Student survey reactions to “Did you enjoy the genetics module?” As shown, the majority of students enjoyed the module, although a handful of students did not. Over 90% of students in the class of approximately 100 students participated in the survey.
In addition to reporting that they enjoyed the module, students responded to an open-ended question about what they learned during the module (Figure 4). Their responses revealed two primary things. First, 84% of the students referenced at least one specific genetics concept that they learned. For example, one student mentioned that, "Despite being different breeds, the same [version of] genes code for the same phenotypes." Second, over 54% of the students discussed specific lab skills that they learned including "how to analyze DNA using gel electrophoresis" and "how to analyze DNA by using a chromatogram." Finally, in addition to concepts and lab skills, some students reported that they learned how to formally present their research in a poster presentation.
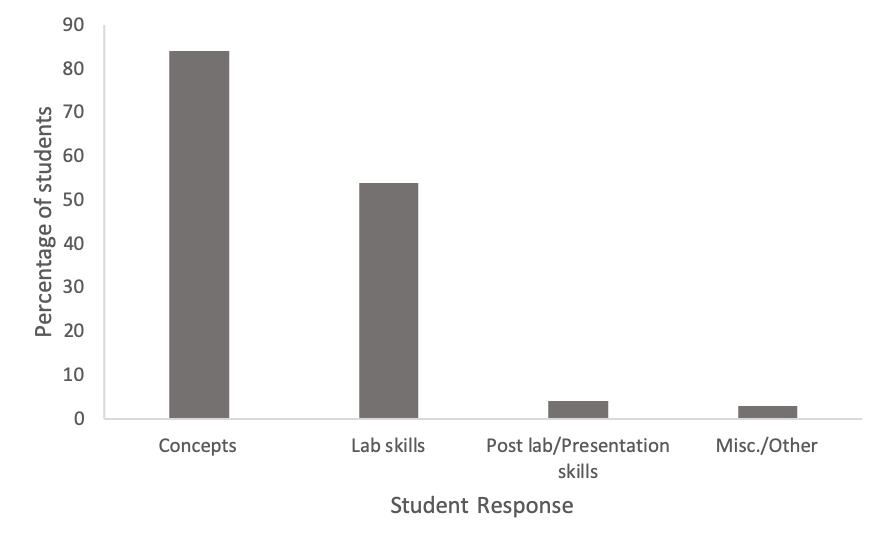
Figure 4. Student survey reactions to “what did you learn through the genetics module?” Most students reported learning concepts related to genetics or gaining laboratory skills. Over 90% of students in a class of approximately 100 students participated in the survey.
However, not all feedback was positive in nature. For example, many students disliked and/or reported difficulty in understanding the primary scientific article (1). Although the introduction and lessons in this module provided advice and support for understanding a primary research article (see Supporting Files S1. Dog Genetics – Module Introduction, S2. Dog Genetics – Lesson 1. DNA Extraction and PCR Sample Preparation, S3. Dog Genetics – Lesson 2. Gel Electrophoresis, S4. Dog Genetics – Lesson 3. DNA Sequence Analysis), instructors often observed that students did not read the background material before each lesson unless an assignment was associated with the material. Nevertheless, after completion of the module, students performed very well (82% average) on the quiz which focused on interpreting the scientific paper. This performance suggests that by the end of the module, most students were able to understand much of the paper.
In addition to not enjoying the article, the bioinformatics analysis in Lesson 3 (Supporting File S4. Dog Genetics – Lesson 3. DNA Sequence Analysis) was identified by students as dull, difficult, or hard to connect with during the lesson. However, after studying for the quiz and creating the group poster, many students responded that although it was difficult at the time, they now understood the connections among the DNA sequence, amino acid sequence, and phenotype and "why we were doing what we were doing on the computer."
We suspect that students expressed frustration with some parts of the module because their past scientific experiences may have used a more "cookie cutter" approach where expected and positive results were almost always obtained. In authentic scientific investigations, methodologies are sometimes revised and experiments repeated when they fail. As this may be a new way of approaching science for some students, we suggest that instructors prepare students for failure and build in an open lab where students can repeat failed tests such as the analysis of RSPO2 via gel electrophoresis. Student frustration associated with absence of DNA bands from their dog samples using gel electrophoresis has been minimized in our course since initiating this change. Unfortunately, some of the bigger tasks such as resequencing DNA is not possible in our course because of time constraints. However, students can still work through the DNA sequence analysis lesson and create figures for their poster using model DNA sequences from dogs with the same coat phenotype as the experimental dog that is missing data. The lack of data for some samples for any of the genes could be turned into a learning opportunity as students can be reminded that in real life, scientists commonly repeat and troubleshoot procedures until successful.
SUPPORTING MATERIALS
- S1. Dog Genetics – Module Introduction, Pre-Lesson 1 Worksheet, and Sample Collection Sheet
- S2. Dog Genetics – Lesson 1. DNA Extraction and PCR Sample Preparation
- S3. Dog Genetics – Lesson 2. RSPO2 Genotype Analysis by Gel Electrophoresis
- S4. Dog Genetics – Lesson 3. DNA Sequence Analysis: FGF5 and KRT71 Genotyping
- S5. Dog Genetics – Lesson 4. Final Genotype: Collaborative Poster, Grading Rubric, and Questionnaire
- S6. Dog Genetics – Lab Notebook Sample Grading Rubric
- S7. Dog Genetics – Group Member Evaluation Form
- S8. Dog Genetics – Example Quiz Questions
- S9. Dog Genetics – Modifiable PowerPoint Poster Template
- S10. Dog Genetics – Cadieu et al. Discussion
- S11. Dog Genetics – PCR Sample Identification DataSheet
- S12. Dog Genetics – Electronic Spreadsheet
- S13. Dog Genetics – Instructor Preparation Notes: Primers, Gels, PCR Clean-up, Supplies, Certificate
- S14. Dog Genetics – Ancestral mRNA and Protein Sequences for FGF5 and KRT71
ACKNOWLEDGMENTS
We thank our introductory biology students for their participation and feedback. Work presented in this paper is exempt from formal IRB review under the following sections of the Federal Common Rule: "45 CFR 46.101(b)(1) Research conducted in established or commonly accepted educational settings, involving normal education practices."
References
- Cadieu E, Neff MW, Quignon P, Walsh K, Chase K, Parker HG, Ostrander EA. 2009. Coat variation in the domestic dog is governed by variants in three genes. Science 326:150-153.
- American Kennel Club. 2017. Breeds by year recognized. URL http://www.akc.org/press-center/facts-stats/page-3/ Accessed 08/05/2019.
- Parker HG, Ostrander EA. 2005. Canine genomics and genetics: running with the pack. PLoS Genet 1(5):e58.
- Candille SI, Kaelin CB, Cattanach BM, Yu B, Thompson DA, Nix MA, ... & Barsh GS. 2007. A β-defensin mutation causes black coat color in domestic dogs. Science 318:1418-1423.
- Kerns J, Cargill E, Clark LA, Candille S, Berryere T, Olivier M, ... & Barsh G. 2007. Linkage and segregation analysis of black and brindle coat color in domestic dogs. Genetics 176:1679-89.
- Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, Zhu L, Padhukasahasram B, Karlins E, Davis S, Jones PG, Quignon P, Johnson GS, Parker HG, Fretwell N, Mosher DS, Lawler DF, Satyaraj E, Nordborg M, Lark KG, Wayne RK, Ostrander EA. 2007. A single IGF1 allele is a major determinant of small size in dogs. Science 316:112-5.
- Parker HG, vonHoldt BM, Quignon P, Margulies EH, Shao S, Mosher DS, Spady TC, Elkahloun A, Cargill M, Jones PG, Maslen CL, Acland GM, Sutter NB, Kuroki K, Bustamante CD, Wayne RK, Ostrander EA. 2009. An expressed Fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science 325:995-998.
- Schoenebeck JJ, Hutchinson SA, Byers A, Beale HC, Carrington B, Faden DL, Rimbault M, Decker B, Kidd JM, Sood R, Boyko AR, Fondon JW 3rd, Wayne RK, Bustamante CD, Ciruna B, Ostrander EA. 2012. Variation of BMP3 contributes to dog breed skull diversity. PLoS Genet. 8:e1002849.
- Schoenebeck JJ, Ostrander EA. 2013. The genetics of canine skull shape variation. Genetics 193:317-325.
- Hultman K, Mellgren, E. 2014. Fetching SNPs: a dog genotyping laboratory for undergraduate biology. Genetics Society of America Peer-Reviewed Education Portal (GSA PREP) doi:10.1534/gsaprep.2014.001.
- Carson TM, Bradley SQ, Fekete BL, Millard JT, LaRiviere FJ. 2009. Forensic analysis of canine DNA samples in the undergraduate biochemistry laboratory. J. Chem. Educ 86:376-378.
- Eberhardt ES, Hansen J, Riservato L, Cole M, Smaglo B, Szaniawski P. 2003. Preparing undergraduates to participate in the post-genome era: A capstone laboratory experience in proteomics. Biochem. Mol. Biol. Educ 31:402-409.
- Dymond JS, Scheifele LZ, Richardson S, Lee P, Chandrasegaran S, Bader JS, Boeke JD. 2009. Teaching synthetic biology, bioinformatics, and engineering to undergraduates: The interdisciplinary build-a-genome course. Genetics 181:13-21.
- Hancock D, Funnel A, Jack B, Johnston J. 2010. Introducing undergraduate students to real-time PCR. Biochem. Mol. Biol. Educ 38:309-316.
- Taylor A, Sajan S. 2005. Testing for genetically modified foods using PCR. J. Chem. Educ 82:597.
- Phillips AR, Robertson AL, Batzli J, Harris M, Miller S. 2008. Aligning goals, assessments, and activities: An approach to teaching PCR and gel electrophoresis. CBE Life Sci. Educ 7:96-106.
- Chao TC, Bhattacharya S, Ros A. 2012. Microfluidic gel electrophoresis in the undergraduate laboratory applied to food analysis. J. Chem. Educ 89:125-129.
- Cunningham SC, McNear B, Pearlman RS, Kern SE. 2006. Beverage-agarose gel electrophoresis: An inquiry-based laboratory exercise with virtual adaptation. CBE Life Sci. Educ 5:281-286.
- Jackson DD, Abbey CS, Nugent D. 2006. DNA profiling of the D1S80 locus: A forensic analysis for the undergraduate biochemistry laboratory. J. Chem. Educ 83:774.
- Childs-Disney JL, Kauffmann AD, Poplawski SG, Lysiak DR, Stewart RJ, Arcadi JK, Dinan FJ. 2010. Forensic analysis of canine DNA samples in the undergraduate biochemistry laboratory. J. Chem. Educ 87:1110-1112.
- Dolan DNA Learning Center, Cold Spring Harbor Laboratory. 2006. Using a single-nucleotide polymorphism to predict bitter-tasting ability. URL http://bioinformatics.dnalc.org/ptc/animation/pdf/ptc.pdf. Accessed 05/05/2019.
- Yezerski A. 2011. Genotyping SNPs associated with dyslexia. p 225-236. In McMahon K (Ed), Tested studies for laboratory teaching, Volume 32. Proceedings of the Association for Biology Laboratory Education (ABLE), 445 pages.
Article Files
Login to access supporting documents
My Dog IS My Homework: Exploring Canine Genetics to Understand Genotype-Phenotype Relationships(PDF | 322 KB)
S1. Dog Genetics- Module Introduction Pre-Lesson 1 Worksheet and Sample Collection Sheet.docx(DOCX | 2 MB)
S14 Ancestral mRNA and Protein Sequences for FGF5 and KRT71.docx(DOCX | 14 KB)
S2. Dog Genetics- Lesson 1 DNA Extraction and PCR Sample Preparation.docx(DOCX | 54 KB)
S3. Dog Genetics- Lesson 2 RSPO2 Genotype Analysis by Gel Electrophoresis.docx(DOCX | 225 KB)
S4. Dog Genetics- Lesson 3 DNA Sequence Analysis - FGF5 and KRT71 Genotyping.docx(DOCX | 208 KB)
S5. Dog Genetics- Lesson 4 Final Genotype - Collaborative Poster Grading Rubric and Questionnaire.docx(DOCX | 28 KB)
S6. Dog Genetics- Lab Notebook Sample Grading Rubric.docx(DOCX | 19 KB)
S7. Dog Genetics- Group Member Evaluation Form.doc(DOC | 27 KB)
S8. Dog Genetics- Example Quiz Questions.docx(DOCX | 861 KB)
S9. Dog Genetics- Modifiable PowerPoint Poster Template.ppt(PPT | 149 KB)
S10. Dog Genetics- Cadieu et al. Discussion.docx(DOCX | 20 KB)
S11. Dog Genetics- PCR Sample Identification DataSheet.docx(DOCX | 17 KB)
S12. Dog Genetics- Electronic Spreadsheet.xlsx(XLSX | 18 KB)
S13. Dog Genetics- Instructor Preperation Notes - Primers Gels PCR Clean-Up Supplies Certificate.docx(DOCX | 72 KB)
- License terms

Comments
Comments
There are no comments on this resource.