Mapping a Mutation to its Gene: The "Fly Lab" as a Modern Research Experience
Editor: Srebrenka Robic
Published online:
Abstract
Although genetics is an invaluable part of the undergraduate biology curriculum, it can be intimidating to students as well as instructors: Students must reduce their reliance on memorization and dive deep into quantitative analysis, and instructors must make a long, rich history of genetics experiments clear, coherent, and relevant for students. Our Lesson addresses these challenges by having students map an unknown mutation to its gene using a modern suite of genetic tools. Students receive a Drosophila melanogaster strain with a mutation that causes the normally flat wing to bend at distinct sites along its length. Although we recently mapped this mutation to its gene, here we have renamed it "crumpled wing" (cw), an example of a pseudonym that you could use in the classroom. Like many standard "fly labs" that are taught at undergraduate institutions, this Lesson reinforces classic genetics concepts: students selectively mate fly strains to determine mode of inheritance, test Mendel's Laws, and three-point map an unknown mutation relative to known markers. But here, we expand on this tradition to simulate a more modern primary research experience: we greatly increase mapping resolution with molecularly-defined transgene insertions, deletions, and duplications; then cross-examine our data with key bioinformatic resources to identify a short-list of candidate cw genes. After extensive data interpretation and integration, students have been able to map cw to a single gene. This Lesson has a flexible design to accommodate a wide range of course structures, staffing, budgets, facilities, and student experience levels.
Citation
Dean DM, Deitcher DL, Loehlin DW, Banta LM. 2020. Mapping a mutation to its gene: The “fly lab” as a modern research experience. CourseSource. https://doi.org/10.24918/cs.2020.51
Society Learning Goals
Biochemistry and Molecular Biology
- Information storage and flow are dynamic and interactive
- What is a genome?
Bioinformatics
- Computation in the life sciences
- What is the role of computation in hypothesis-driven discovery processes within the life sciences?
- DNA - Information Storage [GENOMICS]
- Where are data about the genome found (e.g., nucleotide sequence, epigenomics) and how are they stored and accessed?
- How can bioinformatics tools be employed to analyze genetic information?
Genetics
- Transmission - Patterns of Inheritance
- How can one deduce information about genes, alleles, and gene functions from analysis of genetic crosses and patterns of inheritance?
- How does the phenomenon of linkage affect the assortment of alleles during meiosis?
- What are the mechanisms by which an organism’s genome is passed on to the next generation?
- Molecular biology of gene function
- How is genetic information expressed so it affects an organism's structure and function?
- Genetic variation
- How do different types of mutations affect genes and the corresponding mRNAs and proteins?
- Methods & Tools in Genetics
- What experimental methods are commonly used to analyze gene structure and gene expression?
Science Process Skills
- Process of Science
- Locate, interpret, and evaluate scientific information and primary literature
- Pose testable questions and hypotheses to address gaps in knowledge
- Interpret, evaluate, and draw conclusions from data
- Construct explanations and make evidence-based arguments about the natural world
- Modeling/ Developing and Using Models
- Recognize the important roles that scientific models, of many different types (conceptual, mathematical, physical, etc.), play in predicting and communicating biological phenomena
- Make inferences and solve problems using models and simulations
- Build and evaluate models of biological systems
- Quantitative Reasoning/ Using Mathematics and Computational Thinking
- Apply the tools of graphing, statistics, and data science to analyze biological data
- Communication and Collaboration
- Share ideas, data, and findings with others clearly and accurately
Lesson Learning Goals
Students will understand how to map a mutation to its associated gene, just as might be done in primary research. In addition, students will know how to use their dataset to conduct several of the analyses that are taught in a standard college genetics curriculum (assessing mode of inheritance, chi-square tests, three-point mapping, complementation tests). Finally, students will apply bioinformatics to classical genetics to understand the integrative nature of modern research.
Lesson Learning Objectives
After analysis of multiple datasets and cross-examining their findings with bioinformatic resources, students will be able to map a mutation to a single gene conclusively. During this process, students will:
- handle adult Drosophila and score their phenotypes
- use Punnett squares and cross diagrams to predict outcomes of genetic crosses and compare these predictions to their data
- determine the mode of inheritance of a mutant trait
- use chi-square tests to determine whether the data from genetic crosses fit with the predictions of Mendel's First Law (Equal Segregation) and Second Law (Independent Assortment)
- three-point map an unknown mutation relative to known loci (a known gene and several transposable element insertions at known genomic locations)
- interpret complementation tests between the unknown mutation and deletions and duplications of known genomic segments
- demonstrate their ability to gather bioinformatics data from a model organism database (Flybase), genome browser (GBrowse), and search tool (BLAST)
- critically evaluate their data and hypothesize the identity of the unknown gene in a written lab report
- design experiments to further confirm or extend their findings
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
The fruit fly Drosophila melanogaster is an invaluable model system for genetics research and, perhaps even more so, for genetics education. One reason for its popularity in the undergraduate curriculum is the centrality of Drosophila throughout the past century of genetics research. During this long run, Drosophila has helped us answer fundamental questions about the sex-linkage of traits, linkage mapping, epistasis, cell-fate determination, body-patterning, behavioral genetics, and many other key biological topics (2-7).
In addition to its historical significance, Drosophila is, today, an ideal toolbox to train budding scientists in the lab. Its array of powerful but user-friendly genetic tools offers diverse opportunities for active learning (e.g., 1, 8-11). Its short life cycle, high reproduction rate, small size, and affordability make for efficient generation of large datasets and by extension, efficient troubleshooting as students develop their technique. Highly patterned external tissues such as the wing and the eye, and the disruption of their patterns by certain mutations, provide clear readouts for data analysis, as well as visual intrigue (e.g., 1,9). And in spite of its very distant relatedness to mammals, Drosophila has proven itself a credible model for a number of human diseases (7). These qualities make for an organism that is, in a sense, an excellent teacher: it offers many different learning opportunities, gives frequent and clear feedback, and is engaging and relatable.
And so, Drosophila is an extremely useful educational aide, both as a historical figure in the lecture canon and as a model system for the laboratory classroom. Despite these positive qualities, students often have difficulty connecting the ostensibly distant history of fly research to their own experiences in the lab. It is clear that student learning is improved if they feel a sense of ownership and significance in their work, and that guided- or open-inquiry activities foster this sense (12-14). A challenge, however, is to create a narrative for students that makes working with a classic genetic model seem relevant to them.
Our solution is to have our students use modern techniques to answer classic, unanswered questions. Drosophila mutant strains have been collected long before we even knew that DNA is the genetic material, and so there are a good number of Drosophila mutations that have been reported in the literature long ago, are still maintained in available strains, but the mutations have not been mapped to their genes (15,16). Recently, we worked with students to map the wavy mutation to the IP3-kinase 2 (IP3K2) gene. wavy causes the normally planar wing to buckle in a distinct pattern (Figure 1). After mapping wavy in the teaching lab, we worked with a handful of upper-level undergraduate researchers to refine, expand, and publish our findings, solving an interesting mystery that had lingered for over 80 years (1,17).

Figure 1. What you will investigate. In this Lesson, your class will map a mutation that affects Drosophila wing structure. This figure shows flies from two major types of strains that you will use in the classroom. Left: a fly from a wild-type (WT) strain showing straight wings, and also wild-type, red eyes. Right: a fly from the Mutant strain, showing the “crumpled wing” (cw) trait that you will investigate. cw mutant wings buckle at a specific location along the lateral edge (solid arrow), and often curl upwards at the distal end (dotted arrow). Note that crumpled wing is an example of a pseudonym that you could use to simulate a primary research experience for your students—wavy is the actual name of the mutant trait, and it is a mutation in the IP3K2 gene (1). Flies of the Mutant strain also carry two other mutations: white causes white eyes (seen above), and forked affects bristle length and shape (bristles not readily visible in this photo; see Supporting File S1: Mapping a Mutation-Class Preparation Guide, Figure S1-5 for additional images of all traits). You will use white, forked, w+-transgene insertions, deletions, duplications, and bioinformatics to map cw to its gene.
More recently, we have mapped other unmapped mutations in our lab class (unpublished results). However, we could not ignore the exceptional educational value of the wavy/IP3K2 exercise. This Lesson Plan stems from a tradition of "fly labs" that have given generations of students excellent hands-on experience with three-point mapping (18-22). However, previous lesson plans used genetic markers that were relatively distant from the unknown gene, and unlike this Lesson, they did not use molecularly-defined deletions, duplications, or tightly-linked transgene insertions to further narrow down the chromosomal interval where the unknown gene could be. Therefore, even if students had been trained to cross reference their results with the Drosophila genome, their list of candidate genes would have been too long to map an unknown mutation to its gene with any degree of certainty. In contrast, our Lesson provided a definite answer: students interpreted the results of at least 15 different genetic crosses and cross-examined their findings with intensive bioinformatics. This was a significant effort to be sure, but then after this hard work, they could positively identify IP3K2 as the wavy gene. Students expressed much satisfaction—at times genuine excitement—at solving the puzzle. For these reasons, we chose to develop this exercise into a Lesson Plan to share with other educators. To help you simulate the primary research experience that our students had, we suggest renaming the mutant strain with a pseudonym, which we model here by renaming wavy as "crumpled wing" (cw). Otherwise, your class will undergo the same process that we used to map wavy to IP3K2.
Intended Audience
For several years, we have administered this Lesson as part of our Genetics class (BIOL 202) at Williams College, a 4-year, small liberal arts institution. As is the case at many colleges and universities, Genetics is the third course in our biology department sequence. Preceding Genetics, our BIOL 101 course focuses on cell/molecular biology and our BIOL 102 course focuses on evolution and organismal biology; both 101 and 102 are comprised of lecture and lab. Therefore, virtually all of our students had experienced at least a year of college and had some familiarity with the college laboratory classroom (although we feel that most if not all of this exercise could be done by more junior students—see Teaching Discussion). Approximately two-thirds of our students were intending to or had declared a major in biology, about half of the remaining students were intending to or had declared a major in chemistry, while the rest were distributed over an array of science and non-science majors. All 65-120 students attended the same lecture section three times per week, one hour per class. For lab, students were divided into sections of 12-24 that met once per week, 1.5-3 hours per class. This Lesson was completely administered through the lab sections, but we provided some background in lecture to prime students for the lab assignments (see Pre-requisite Student Knowledge for background content, and then Table 1 for suggested timing of background relative to lab meetings).
Specifics of our Genetics class and institution aside, we intend this Lesson to be an educational and meaningful experience for a broad population. This Lesson could be administered at small liberal arts colleges, large universities, 4-year community colleges, and perhaps even in advanced high school classes. While developing this Lesson Plan, we worked to widely address the needs of students, instructors, and prep staff, even if they have not worked with flies before. To this end, our Supporting Files include a complete prep guide (Supporting File S1. Mapping a Mutation – Class Preparation Guide), a complete student lab manual (Supporting File S2. Mapping a Mutation – Fly Lab Manual), all of our teaching aides and grading keys, and a full sample dataset (Supporting File S8. Mapping a Mutation – Sample Final Dataset). The Teaching Discussion and Supporting File S1. Mapping a Mutation – Class Preparation Guide (Part F, section F5) suggest accommodations for various class sizes, class schedules, student experience levels, budgets, lab facilities, prep staffing, and instructor teaching goals.
Required Learning Time
As taught, students completed the Lesson in about 5 weeks: each lab section met once per week on Weeks 1, 3, and 4; students had to perform a <5-minute procedure independently on Week 2; and after the Week 4 class, we gave students 7-10 days to write their Final Assignments (see Table 1 for a detailed Lesson Plan Timeline). Should instructors wish to shorten or lengthen this timeline, we provide a number of suggestions and resources in the Teaching Discussion and in Supporting File S1. Mapping a Mutation – Class Preparation Guide (Part F, section F5)—indeed, you could administer a sizeable portion of this module during a single class without any lab work, or at the opposite extreme, you could expand the total timeline several more weeks to accommodate additional student lab exercises and/or a lack of 25°C incubators.
Prerequisite Student Knowledge
Our institution's earlier biology courses do not involve fly work in the lab, yet our students learned how to handle and score flies within the first class period of this Lesson. Therefore, you could administer at least a portion of this Lesson to undergraduates with little-to-no experience in a biology lab (see Teaching Discussion and Supporting File S1. Mapping a Mutation – Class Preparation Guide, Part F, section F5 for suggestions). Parts of this Lesson could even work in an advanced high school class. However, as described in "Intended Audience" above, a majority of our students had had a year of introductory biology with lab prior to the Genetics course. Their general familiarity with the lab classroom may have boosted their confidence as they worked through this project.
To complete prior biology course assignments, students had received some training with spreadsheet software (Microsoft Excel and/or Google Sheets). This experience was at times helpful but not essential to analyze the fly cross data from this Lesson. There are clear benefits to learning spreadsheet software. However, we encourage instructors to give students flexibility in how they approach their data analysis, since many students seemed to better comprehend the content by performing manual calculations, or at least by using software of their choosing. If your students do not own Excel, we recommend Google Sheets over Numbers because Google Sheets does a better job at preserving our datasheet formatting.
Regarding lecture material, students had learned about basic gene structure and the Central Dogma in our Biology 101 class, and these topics were reviewed during early lectures of our Genetics course. Otherwise, very little material from previous courses applied directly to this Lesson. However, we have found it very helpful to coordinate lecture and lab within the Genetics course itself. To best prepare students for this Lesson's assignments, we recommend that they will have experienced, through lectures and/or problem sets earlier in your Genetics course: (1) determining the mode of inheritance of a mutation, (2) chi-square tests of Mendel's Laws, (3) three-point cross analysis, and (4) complementation tests. It is not necessary to use flies as your model when covering these topics in lecture, although we do use Drosophila to lecture about three-point cross analysis. Not all of these topics need to be covered before Week 1 of this Lesson—see Table 1 for suggested timing of lecture material relative to the Lesson schedule.
Beyond this background, all the information that students will need to know is in their Fly Lab Manual (Supporting File S2. Mapping a Mutation – Fly Lab Manual), in the Pre-laboratory Lecture Slideshow (Supporting File S5. Mapping a Mutation – Pre-laboratory Lecture Slideshow), and in the instructor's demo of fly work. Students will complete Pre-laboratory Assignments prior to Weeks 1 and 3 (Supporting File S2. Mapping a Mutation – Fly Lab Manual), and these assignments will help them frame their data into a biological context. At the start of Week 1 class, before students start working, we recommend that you present the slideshow (Supporting File S5. Mapping a Mutation – Pre-laboratory Lecture Slideshow), then give students a live fly handling demo similar to that shown in the video link at the end of the slideshow. You may wish to review the key points of the lecture (Supporting File S5. Mapping a Mutation – Pre-laboratory Lecture Slideshow) and fly handling demo at the start of Week 3 class, since the students will not have examined flies closely for two weeks.
Prerequisite Teacher Knowledge
Instructors should have sufficient background in genetics to guide students through the entire student Fly Lab Manual (Supporting File S2. Mapping a Mutation – Fly Lab Manual), including the Pre-laboratory Assignments, Week 1-4 Procedures, final dataset analysis, and writing of the Final Lab Assignments. Instructors who have not worked with flies will need to spend time familiarizing themselves with certain particularities of the Drosophila genetics workflow: duplication/deficiency crosses (23-25), balancer chromosomes (16), and navigating the Flybase model organism database (26; also see FlyBase tutorial in Supporting File S2. Mapping a Mutation – Fly Lab Manual, Week 4 Procedure, pp. 41-61). For additional outside references on Drosophila genetics, see Supporting File S1. Mapping a Mutation – Class Preparation Guide, Section C2 (pp. 13-14). Even if other staff will prep the flies, we recommend that instructors read through the Prep Guide early during the prep (Supporting File S1. Mapping a Mutation – Class Preparation Guide)—besides being useful background, reading the Prep Guide will greatly facilitate coordination between instructors and technical staff. However, it is a fairly lengthy, broad-ranging document, and so to help instructors and staff find prep sections relevant to their responsibilities, p. 1 lists where to find the essential prep information (text box, middle of page), and pp. 1-2 display a complete Table of Contents.
To further assist in instructor preparation, we provide the slideshow that we have presented to students before Week 1 and sometimes before Week 3 labs (Supporting File S5. Mapping a Mutation – Pre-laboratory Lecture Slideshow). At the end of this slideshow is a link to a fly handling demo video. We provide grading keys in S4. Mapping a Mutation – Grading Keys, and in Supporting File S9. Mapping a Mutation – Answer Key for the Sample Final Dataset Calculations; these files should not be given to students, but will help instructors guide their students' work.
Regarding fly work, instructors and/or preparatory staff should be able to maintain Drosophila cultures and set up the crosses described in Supporting File S1. Mapping a Mutation – Class Preparation Guide. Instructors should also be able to demonstrate fly handling and assist students with fly scoring (Supporting File S1. Mapping a Mutation – Class Preparation Guide, Supporting File S2. Mapping a Mutation – Fly Lab Manual, and Supporting File S5. Mapping a Mutation – Pre-laboratory Lecture Slideshow). We recommend that instructors and TAs practice the exercise ahead of time to develop familiarity with the phenotypes and tools.
A significant portion of this Lesson utilizes datasheet software and bioinformatics. This in mind, instructors should be familiar with Microsoft Excel and/or Google Sheets to help students wrangle their data (other spreadsheet software such as Numbers can work too, but this software alters our datasheet formatting significantly). To assist in this effort, Supporting File S8. Mapping a Mutation – Sample Final Dataset is a complete set of sample data, and Supporting File S9. Mapping a Mutation – Answer Key for the Sample Final Dataset Calculations is an answer key to sample data calculations and mapping. Do not distribute this key to students, but to help students with their calculations, you could verbally describe the embedded formulae. In addition, instructors should know FlyBase (26), GBrowse (27), and BLAST (28) well enough to guide students through the Week 4 Procedure (Supporting File S2. Mapping a Mutation – Fly Lab Manual, and key in Supporting File S4. Mapping a Mutation – Grading Keys).
We staffed each lab section with one instructor and 1-2 teaching assistants (TAs). TAs were former Genetics students who we felt would be responsible intermediaries between the instructor and our students. We had them grade the Pre-laboratory Assignments (keys in Supporting File S4. Mapping a Mutation – Grading Keys); assist students with fly scoring, FlyBase work, etc.; and bring points of student confusion to the attention of the instructor. Instructors met with their TAs the week before each lab unit for about 15-30 minutes to review the upcoming lab material and logistics.
SCIENTIFIC TEACHING THEMES
Active Learning
- Hands-on learning: Students actively learn standard Drosophila lab work: handling cultures, setting up crosses, scoring fly sexes, and scoring phenotypes of several different tissues.
- Prediction: Pre-laboratory Assignments help students predict and interpret potential outcomes from all experimental crosses, giving them a feeling of a stake in the outcome.
- Data analysis: During Week 4, students use a practice dataset and the bioinformatic sites FlyBase and GBrowse to map another mutation to its gene. Students also use BLAST to ask questions about protein function and evolution. This tutorial provides a template of an effective research strategy which students can apply in the Final Assignment.
- Integrating multiple lines of evidence: In the Final Assignment, students: (i) determine the modes of inheritance of three mutant traits, (ii) use chi-square analyses to test the fit of their three-point cross data to Mendel's First and Second Laws, (iii) determine locus order with 4-5 different three-point crosses, (iv) analyze 10-15 different complementation tests, and (v) cross-reference their findings with FlyBase and GBrowse to identify the cw gene.
Assessment
- Two problem sets: Students work through two different Pre-laboratory Assignments (due Weeks 1 and 3). These short problem sets help students understand the genetics underlying each experimental cross.
- Practice data analysis: Throughout the Week 4 Procedure, instructors and/or TAs check student answers. We do not grade student work on Week 4, and focus on helping students find the correct answers if they misunderstand the material.
- Final lab report: Students use Week 4 Procedure as a template to map cw: 10-13 days after Week 4 class, students complete and turn in a 12-18-page Final Assignment that summarizes their findings.
- Assessment materials: All student assignments are in Supporting File S2. Mapping a Mutation – Fly Lab Manual. Supporting File S4. Mapping a Mutation – Grading Keys, and Supporting File S9. Mapping a Mutation – Answer Key for the Sample Final Dataset Calculations are sample grading keys.
Inclusive Teaching
- Fostering a common goal, as well as individual learning: The class works collectively to gather a dataset, and class data is analyzed for the assignment, yet we want individuals to understand the material of this collective effort. With this in mind, we ask students to prioritize care over speed: rather than quickly scoring the requested number of flies, we prefer that they score fewer than requested and use the relaxed pace to better recognize the phenotypes and understand the cross genetics. (If you are concerned about insufficient sampling, you could supplement your class data with Supporting File S8. Mapping a Mutation – Sample Final Dataset) During the Week 1 and 3 Procedures (Supporting File S2. Mapping a Mutation – Fly Lab Manual), we encourage students to discuss anomalous/interesting observations rather than just assume that they are "wrong" and change their individual data to match group consensus.
- Encouraging student initiative: We allot class-time during Week 1 for students to work ahead on the Week 3 Pre-laboratory Assignment, asking questions as needed. Similarly, if time is available the week before the Week 1 class, instructors may wish to allot time for students to work ahead on the Week 1 Pre-laboratory Assignment.
- Welcoming a diversity of working styles: During our "study hall" style office hours between Weeks 4 and 5 (Table 1), we make every effort to accommodate different working styles: we reserve a large room with wireless access and chair arrangements that allow for small group or solitary work, we encourage students to collaborate if they wish, and they may work nearby if they prefer a quieter/less-crowded location. At least one instructor and one TA are available for questions throughout all study halls. If students cannot or would rather not attend these study halls, we are happy to meet with them at another time.
- Welcoming a diversity of thinking and writing styles: We give students explicit guidance for how to organize their Final Assignments. However, to help increase their engagement in data analysis, we ask students to prioritize content and a clear narrative over strict form (Supporting File S2. Mapping a Mutation – Fly Lab Manual, Guidelines for Final Assignment section). For example, we give them the option of writing the Results and Discussion separately or combined, since both forms are used in journal articles.
- Clear guidance while encouraging creative thinking: Finally, we encourage creative thinking as students design follow-up experiments in their Final Assignments (Supporting File S2. Mapping a Mutation – Fly Lab Manual, Guidelines for Final Assignment section; Supporting File S4. Mapping a Mutation – Grading Keys). Although we provide a clear template for mapping an unknown mutation to a gene, we welcome creative use of FlyBase and GBrowse to gather supporting evidence (Supporting File S2. Mapping a Mutation – Fly Lab Manual, Week 4 Procedure and Guidelines for Final Assignment).
LESSON PLAN
Overview
Your students will map crumpled wing (cw), an unknown locus, relative to multiple known markers. Supporting File S1. Mapping a Mutation – Class Preparation Guide (Figure S1-5) and Supporting File S5. Mapping a Mutation – Pre-laboratory Lecture Slideshow show examples of all traits that you will score during this Lesson Plan: cw, white (w), forked (f ), and Bar.
Figure 2 summarizes the purpose of each experimental cross and the ultimate goals—you may wish to absorb this figure before reading through the Classroom Preparation description (next subsection). Class will span over 4 weeks, and there are two major groups of crosses that your students will analyze to map cw: The first group, Crosses A and B (Figure 2, left) are essentially three-point crosses, and the second group, the Df and Dp Crosses (Figure 2, right) are complementation tests. Figure 2 describes the fly stocks used for each cross by their general categories: WT (wild type), Mutant, Df (deficiency), and Dp (duplication). We list the individual stocks of each category elsewhere (Supporting File S1. Mapping a Mutation – Class Preparation Guide, Table S1-4), but here, we summarize the genetics that the stocks of each category have in common:
- WT strains are cw+ f +, and although they are mutant at their endogenous w gene, they are phenotypically w+ because they carry a w+-transgene insertion. Each WT strain carries a different w+-transgene insertion at a different chromosomal location, but all of the insertions, as well as f, are closely linked to cw and have known sequence coordinates. Therefore, they are useful reference points for three-point mapping of cw.
- Conversely, the Mutant strain is mutant for w, cw, and f and does not carry any w+-transgene insertions. This strain is also used for three-point mapping of cw.
- Each Df stock is heterozygous for a small deletion of the X chromosome (23,24). We use deletions that are in the general cw region for complementation tests. Some deletions do and some do not remove the cw gene. The Bar mutation is used to mark the X chromosome that does not carry the deletion (discussed in Supporting File S2. Mapping a Mutation – Fly Lab Manual, pp. 29-30).
- Each Dp stock carries a small duplication of a wild-type X chromosome sequence on chromosome 3 (25). As with deletions, we use duplications that are in the general cw region for complementation tests. Some duplications do and some do not carry the (wild-type) cw locus.
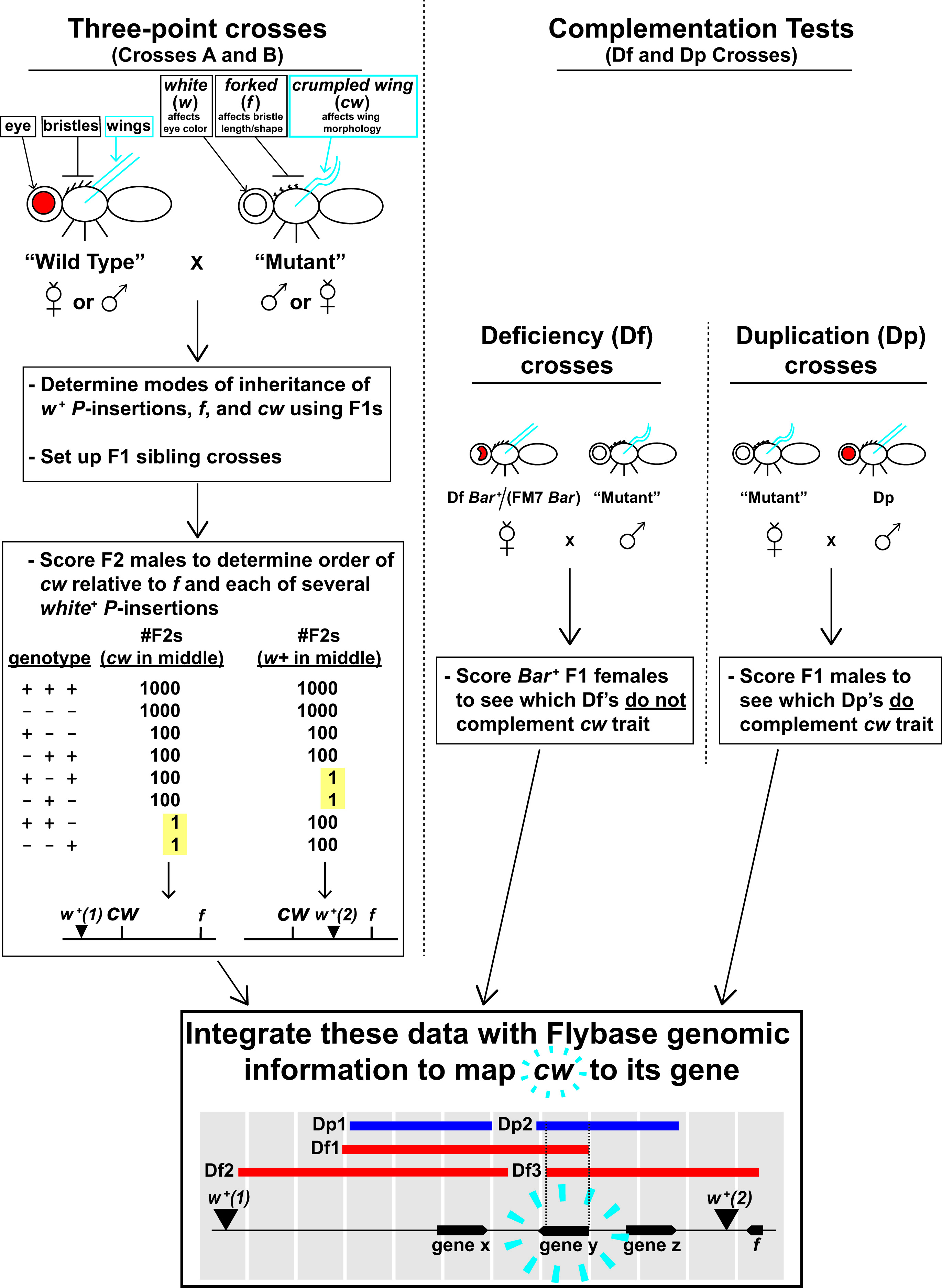
Figure 2. Overview of Fly Lab experiments. This figure may be a helpful primer before you engage in the details of your prep (Table 1 and S1. Mapping a Mutation – Class Preparation Guide). See the first subsection of the Lesson Plan (“Overview”) for additional information about the crosses and fly stocks that are listed in this figure.
Classroom Preparation
Fly Prep
Supporting File S1. Mapping a Mutation – Class Preparation Guide is a comprehensive prep guide for this Lesson, and also a general reference manual for fly work. Its full table of contents is on pp. 1-2, but in summary, the Prep Guide is comprised of the following parts:
- Part A presents a complete prep schedule and discusses schedule adjustments for various incubation temperatures.
- Part B describes how to open a Bloomington Drosophila Stock Center (BDSC) account and how to order the fly stocks for this Lesson.
- Part C describes standard fly lab equipment and compares some fly diet options. (Most standard fly diets should work fine for this Lesson's strains, but we compare and contrast three particular formulations.)
- Part D provides strategies to maintain and expand healthy fly cultures.
- Part E describes how to prepare the crosses for this Lesson. It also describes how to distinguish Drosophila sexes and wild-type vs. mutant phenotypes (supporting images provided).
- Part F describes the classroom supplies for Weeks 1-4 of the Lesson. It also discusses alternative supplies to accommodate various teaching goals, facilities, budgets, class sizes, etc.
To ensure a successful class, start fly prep early—we suggest ordering BDSC stocks 18-27 weeks before the Week 1 classes (discussed in Supporting File S1. Mapping a Mutation – Class Preparation Guide, Part A). The BDSC will only be able to send you one vial of each fly stock (Supporting File S1. Mapping a Mutation – Class Preparation Guide, Part B), and from those starter cultures, you will need to create enough cultures and crosses for all the students in your class (target numbers listed in Supporting File S1. Mapping a Mutation – Class Preparation Guide, Table S1-3). This is a long-term, but very manageable effort if you "expand" fly stocks regularly (i.e., if you regularly increase the number of vials/bottles of each fly stock). Supporting File S1. Mapping a Mutation – Class Preparation Guide, Part D describes strategies to expand stocks efficiently.
We hope that Supporting File S1. Mapping a Mutation – Class Preparation Guide will be a helpful resource for those who have not worked with Drosophila, as well as for those who have. If you are in the latter group, you might wish to skip tutorial sections and focus on key fly prep information: which fly stocks to order, when to order them, target numbers as you expand stocks, crosses to set up, when to set them up, and how many. We have compressed this information into tables:
- Table S1-3 (pp. 4-7) is a complete and detailed schedule for prep staff. Cross reference this table with Figure 2 of the Lesson Plan, which is an overview of the experimental design; and with Table 1 of the Lesson Plan, which is a generalized prep schedule alongside a detailed schedule for instructors, TAs, and students.
- Table S1-4 (pp. 10-11) lists which fly stocks to order.
- Tables S1-3 (pp. 4-7) and S1-6 (p. 25) describe which flies to collect and cross, and when.
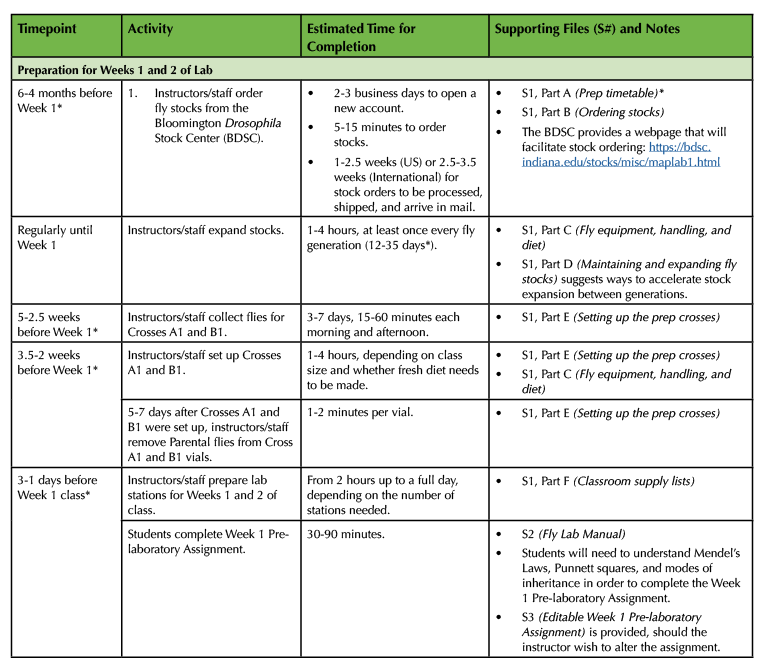
Table 1. (Part 1 of 4). Lesson Plan Timeline, prep for Weeks 1 and 2.
This four-part table lays out the relative timing between all Lesson Plan preparations, classes, and assignments. It also serves as a complete Supporting File directory, cross-referencing files alongside the activities that they support.
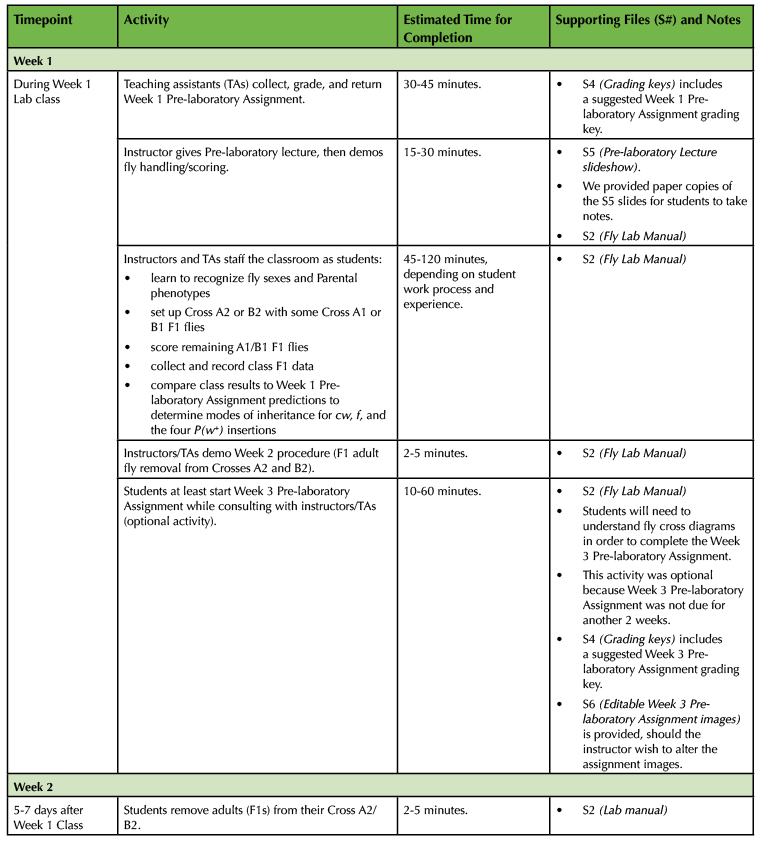
Table 1. (Part 2 of 4). Lesson Plan Timeline, Weeks 1 and 2 of class.
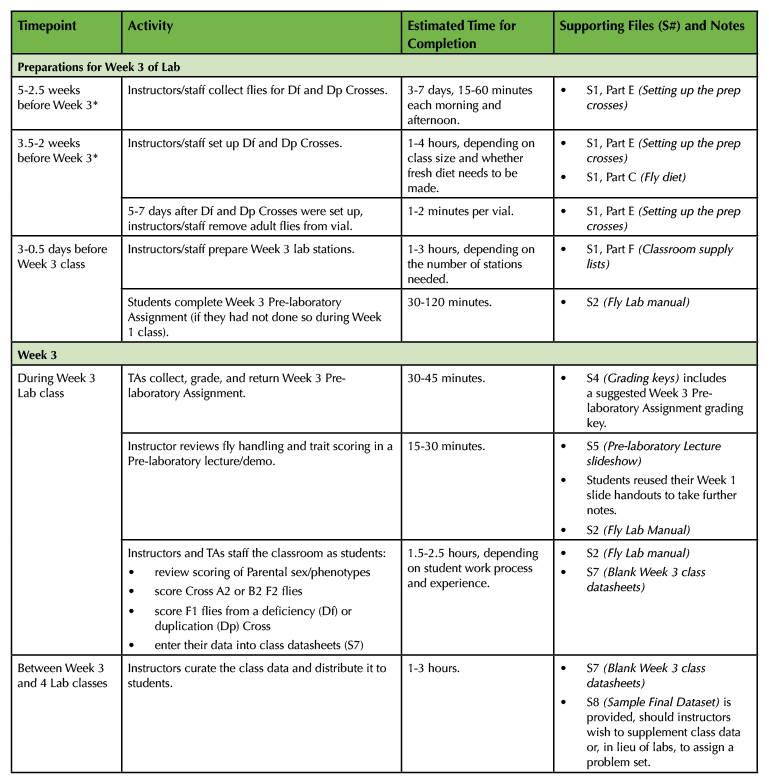
Table 1. (Part 3 of 4). Lesson Plan Timeline, Week 3 prep and class.
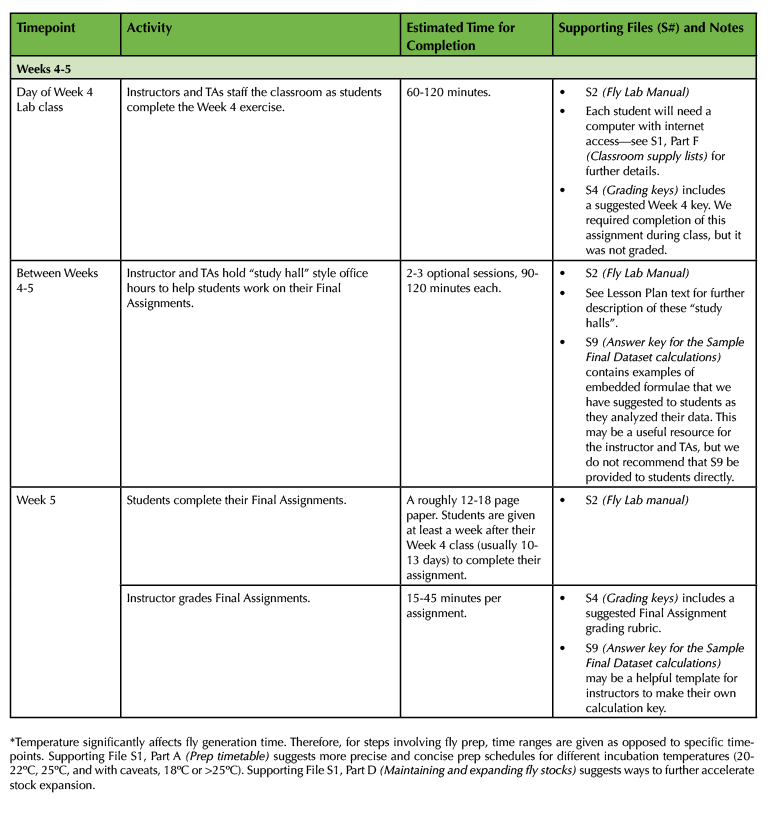
Table 1. (Part 4 of 4). Lesson Plan Timeline, Weeks 4 and 5 of class.
Classroom supplies
In addition to describing fly prep, Supporting File S1. Mapping a Mutation – Class Preparation Guide details all classroom prep:
Week 1
Part F, section F1 describes Week 1 classroom supplies (see Figure S1-7, p. 34; Table S1-7, pp. 35-36). To help you with ordering, Table S1-7 lists suppliers, catalog numbers, specs, etc.
Weeks 2-3
We swap in new fly cultures and crosses for the Week 3 class (described in Tables S1-3 and S1-6), but otherwise, we leave all the Week 1 supplies in place through Week 3 (Part F, sections F2 and F3).
Week 4
On Week 4, we remove supplies from the lab and students use the room to work through a computer-based exercise (Part F, section F4). Most students bring their own laptops, but we have 6-10 laptops available to borrow (Mac or PC, Safari or Firefox installed).
Week 5
Our preferred Week 5 office hour classroom is described in the "Scientific Teaching Themes" section under the "Inclusive Teaching" subsection.
Class-time: Instruction, Student Work, and Grading
The following section details all instructor, TA, and student work during this Lesson. We reference Supporting Files as they apply. Table 1 is a timeline of this subsection's information. Figure 3 describes all student work over the course of the Lesson—students receive this same figure in their lab manual (Supporting File S2. Mapping a Mutation – Fly Lab Manual, Figure 2).
Week 1
On the first week of the Fly Lab, students learn how to handle Drosophila, score fly sexes and phenotypes, determine modes of inheritance for mutant traits, and set up Crosses A2 and B2.
Week 1 Pre-laboratory Assignment
- At the start of Week 1 class, students turn in their completed Week 1 Pre-laboratory Assignments (Supporting File S2. Mapping a Mutation – Fly Lab Manual, pp. 14-16).
- TAs grade the Pre-laboratory Assignments (key in Supporting File S4. Mapping a Mutation – Grading Keys) and return them to students during class. If a student made significant errors on their Pre-laboratory Assignment, the instructor or a TA checks in with that student during Week 1 class to help clear up misunderstandings.
Week 1 Pre-laboratory Lecture and Fly Handling Demo
- Instructor gives a lecture at the start of lab (slideshow provided in Supporting File S5. Mapping a Mutation – Pre-laboratory Lecture Slideshow).
- After this overview, the instructor demos basic fly handling to the students—the last slide in Supporting File S5. Mapping a Mutation – Pre-laboratory Lecture Slideshow links to a video of our Week 1 demo.
Week 1 Classroom Procedure
- Students work through the Week 1 Procedure (Supporting File S2. Mapping a Mutation – Fly Lab Manual, pp. 17-24). Instructors and TAs assist students with fly handling/scoring.
- Class collects Cross A1/B1 data. (We collect class data on a dry erase board, then students record the consensus in the Supporting File S2. Mapping a Mutation – Fly Lab Manual, p. 23 table.)
- After students complete the Week 1 Procedure, either an instructor or TA demos the Week 2 Procedure to students (Supporting File S2. Mapping a Mutation – Fly Lab Manual, p. 25).
- During the rest of class-time, we give students the option to start working on the Week 3 Pre-laboratory Assignment (Supporting File S2. Mapping a Mutation – Fly Lab Manual, pp. 27-32). As students work ahead, instructors and TAs field questions (key in Supporting File S4. Mapping a Mutation – Grading Keys).
- At the end of class, staff collect Crosses that were set up by students (A2 and B2) and place them in an incubator set to 25°C (if available).
Week 2
No lab class this week, but students remove F1 adults from their Crosses A2 and B2.
- 5-7 days after their Week 1 class, but otherwise on their own time, students drop by the lab and remove adult flies from their Cross A2 or B2 (Supporting File S2. Mapping a Mutation – Fly Lab Manual, p. 25).
Week 3
During this week, students score F2s from their Crosses A2 and B2 (for three-point mapping), and score F1s from Df and Dp Crosses (for complementation analysis).
Week 3 Pre-laboratory Assignment
- At the start of Week 3 class, students turn in their completed Week 3 Pre-laboratory Assignments (Supporting File S2. Mapping a Mutation – Fly Lab Manual, pp. 27-32).
- TAs grade the Pre-laboratory Assignments (key in Supporting File S4. Mapping a Mutation – Grading Keys) and return them to students during class. If a student made significant errors on their Pre-laboratory Assignment, the instructor or a TA checks in with that student during Week 3 class to help clear up misunderstandings.
Week 3 Pre-laboratory Lecture and Demo
- Given the two-week break, instructors may wish to review the lecture slides and/or fly handling (Supporting File S5. Mapping a Mutation – Pre-laboratory Lecture Slideshow). Otherwise, the Pre-laboratory lecture should summarize the Week 3 procedure and place it within the context of the entire Lesson.
Week 3 Classroom Procedure
- Students score their Cross A2 or B2 (Supporting File S2. Mapping a Mutation – Fly Lab Manual, pp. 33-38).
- Instructor gives each student a Df or Dp Cross vial. Each student scores their Df/Dp Cross (pp. 38-39).
- Each student enters their A2/B2 and Df/Dp Cross data into a class data file (Supporting File S7. Mapping a Mutation – Blank Week 3 Class Datasheets).
- Supporting File S8. Mapping a Mutation – Sample Final Dataset is a full set of Week 3 data. If needed, use it to supplement your own class data.
Week 4
On this week, we help students work through a practice data analysis session: Using genetic mapping data similar to those of the Fly Lab, but for a different (hypothetical) unknown that we call dark ruby (dr), students use the bioinformatic resources FlyBase and GBrowse to identify candidate dr genes.
- Students bring their own laptop to class or borrow from instructor. PCs or Macs should both work fine. We recommend Safari or Firefox as a web browser and not Chrome (Chrome appeared to have issues interacting with GBrowse graphics).
- Students work through the Week 4 Procedure (Supporting File S2. Mapping a Mutation – Fly Lab Manual, pp. 41-61).
- Instructors and TAs check students' work as they proceed. This exercise is not graded, and we help students find correct answers if they encounter difficulties (key in Supporting File S4. Mapping a Mutation – Grading Keys).
Week 5
On this final week, instructors hold extra office hours and students prepare their Final Assignments.
Week 5 Office Hours
- Instructors schedule optional "study hall" style office hours (2-3 sessions, 90-120 minutes per session). We have preferred evening sessions after dinner to increase student turnout. We reserve a room with internet access and with chair/desk arrangements that enable small group collaborations, as well as solitary work.
- At these office hours or on their own, students follow Final Assignment Guidelines (Supporting File S2. Mapping a Mutation – Fly Lab Manual, pp. 63-68) to analyze class data and write their reports.
- Instructors and TAs staff office hours to provide strategic guidance, review Pre-laboratory Assignments, answer questions about Final Assignment content, assist with GBrowse navigation, etc.. Supporting File S4. Mapping a Mutation – Grading Keys contains a qualitative key for the Final Assignment, Supporting File S8. Mapping a Mutation – Sample Final Dataset is a complete sample dataset, and Supporting File S9. Mapping a Mutation – Answer Key for the Sample Final Dataset Calculations is a key for the sample data calculations. To help students with their calculations, you could verbally share the embedded formulae in Supporting File S9. Mapping a Mutation – Answer Key for the Sample Final Dataset Calculations. However, do not distribute this key to students, because with it in hand, a student could complete all calculations without understanding them conceptually.
Week 5 Final Assignment
- Students turn in their Final Assignments 7-10 days after their Week 4 class (guidelines in Supporting File S2. Mapping a Mutation – Fly Lab Manual, pp. 63-68).
- Instructors grade Final Assignments (keys in Supporting File S4. Mapping a Mutation – Grading Keys and Supporting File S9: Mapping a Mutation – Answer Key for the Sample Final Dataset Calculations).
TEACHING DISCUSSION
Student performance and feedback
Most of our students performed very well on their Fly Lab assignments. When this Lesson was implemented in its final form, average scores were: Week 1 Prelab 97.4 ± 8.9% SD, Week 3 Prelab 93.0 ± 11.7%, and Final Assignment 92.7 ± 4.7% (n=163). Virtually 100% of our students successfully completed the Week 4 Procedure. The vast majority (90-95%) of our students successfully mapped cw (wavy) to IP3K2, and their writing suggested that they understood how to integrate their cross data with bioinformatics. All students who failed to map cw to IP3K2 came close, but then either implicated an adjacent gene or were not able to narrow down the short-list to one gene. Jafrac1 is a gene adjacent to IP3K2 that also overlaps with the non-complementing deficiencies and complementing duplications (1), so it is definitely reasonable—in fact astute—for students to bring up this gene as a cw candidate. However, only IP3K2 has complete transcripts duplicated by both complementing duplications [Dp(1;3)DC267 and Dp(1;3)DC268], and Dp(1;3)DC267 does not duplicate anywhere near the entire coding sequence of Jafrac1. Students can see these alignments for themselves by using GBrowse to visualize the IP3K2 and Jafrac1 transcripts alongside the "Deleted segment" and "Transgene Duplication" tracks (Supporting File S2. Mapping a Mutation – Fly Lab Manual, Week 4 Procedure; also see Figure 3 of reference 1 for a visual of the IP3K2/Jafrac1 chromosomal region).
Otherwise, when students lost points, it was usually for minor clerical/calculation errors, or for not fully explaining the reasoning behind a particular conclusion (see prompts in Supporting File S2. Mapping a Mutation – Fly Lab Manual, Final Assignment Guidelines, pp. 63-68).
Fostering discovery, future directions
Instructors and TAs should be well-versed in the procedure (Supporting File S2. Mapping a Mutation – Fly Lab Manual), overall experimental design (Figure 2), and expected results (Supporting File S4. Mapping a Mutation – Grading Keys, Supporting File S9. Mapping a Mutation – Answer Key for the Sample Final Dataset Calculations). With this knowledge, they can gently guide students through their fly scoring, calculations, and data analysis. On the other hand, students learn by deducing most of the experimental design and expected results for themselves. To be sure, we give students very clear, step-by-step guidance through procedures and assignments, and we want them to know, from the outset, what activities they will do and generally why they will do them (Figure 3; Supporting File S2. Mapping a Mutation – Fly Lab Manual, and Supporting File S5. Mapping a Mutation – Pre-laboratory Lecture Slideshow). However, we want them to discover how these activities will map cw. Perhaps the best illustration of this point is in the differences between Figures 2 and 3 of this Lesson Plan: Figure 2 (intended for instructors) is a visual, detailed representation of the expected results for every experiment, while Figure 3 (intended for students) is a simple list of student activities and their purposes. To provide a specific example of this comparison, Figure 2, as it describes the Df Crosses, indicates that Bar+ F1 females with a cw mutant phenotype will positively locate cw. Figure 3 omits this information, only describing the Df Cross scheme and its general purpose. We believe that students will best understand how the Df Cross maps cw if they derive the answer for themselves (Week 3 Pre-laboratory Assignment in Supporting File S2. Mapping a Mutation – Fly Lab Manual), and we feel similarly about Crosses A and B, the Dp Crosses, and FlyBase/GBrowse work.
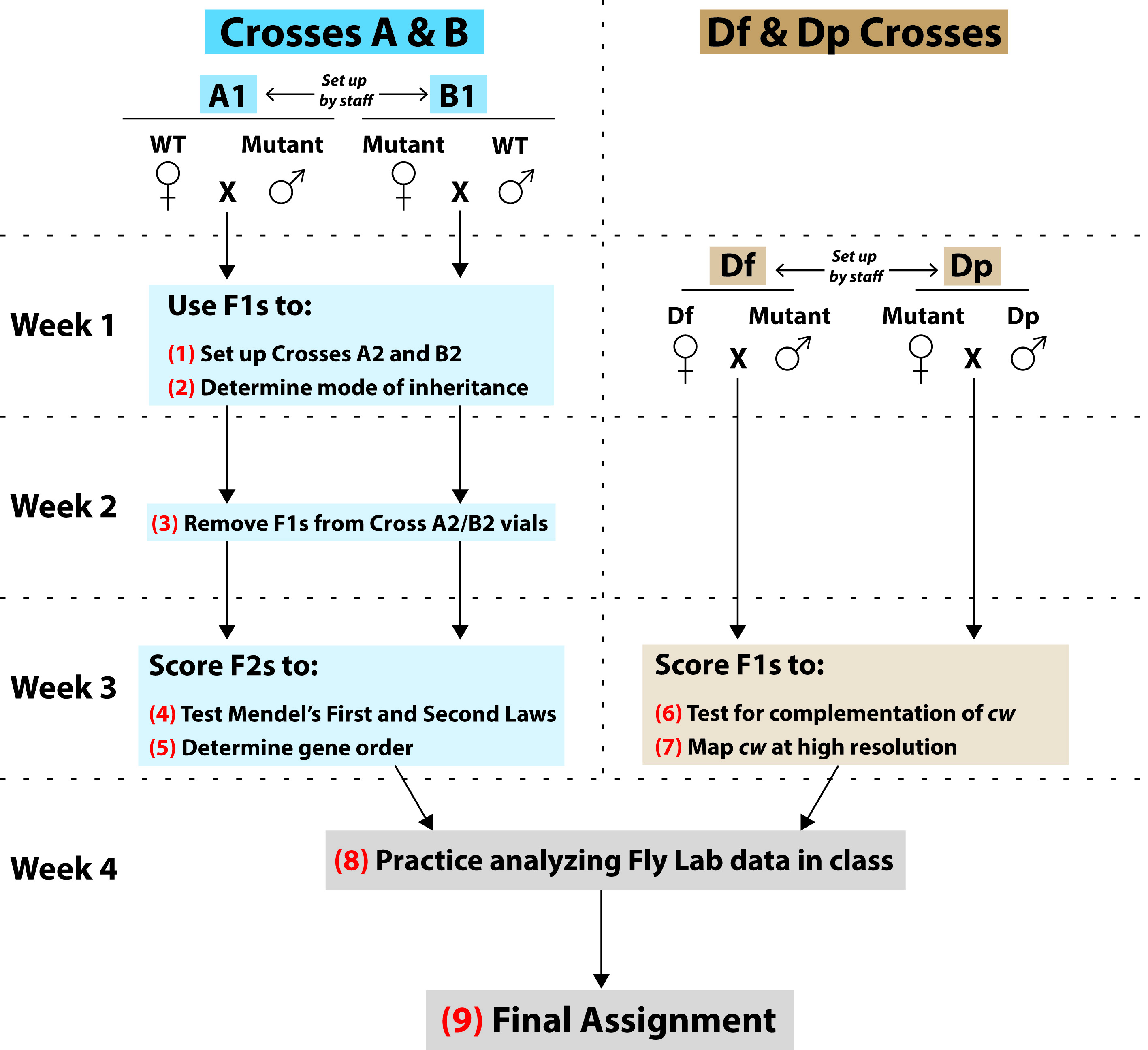
Figure 3. Summary of student work during the Fly Lab. This figure describes each experimental cross and summarizes the content and goals of each major student activity [numbered in red (1) through (9)]. See “Overview” at the start of the Lesson Plan section for further information about each type of cross and fly stock that is displayed in this figure.
Starting at top left and following arrows down through blue boxes: In one set of experimental crosses, students will conduct three-point mapping of cw. On Week 1 of the Fly Lab, each student will receive a vial of either Cross A1 or Cross B1, both of which were set up by staff. These were reciprocal crosses between the WT and Mutant strains. Staff will have emptied parental flies before the F1s emerge, and so by classtime, the adult flies in each A1/B1 vial will all be F1s. (1) Each student will anesthetize their A1/B1 F1 flies and brush some into a fresh culture vial. This Cross A2 or B2 will produce an F2 generation of flies that they will score on Week 3. (2) Students will then score remaining F1s to determine the mode of inheritance of cw (i.e. is the mutant trait X-linked or autosomal, and is it dominant or recessive to the wild type trait). They will also score F1s for two other traits—forked (f ) and white (w)—that will be used as reference points to map cw. Note that students will not map cw relative to the endogenous white gene, but rather relative to several different white+-transgene insertions that are more closely linked to cw. The Week 1 Pre-laboratory Assignment will help students determine expected F1 phenotypes for each possible mode of inheritance, and they will use this assignment’s predictions to analyze their F1 data (Supporting File S2: Mapping a Mutation-Fly Lab Manual, pp. 14-16). (3) You will not hold class on Week 2, but 5-7 days after your Week 1 lab, each student will empty F1 flies from their Cross A2/B2 vial. This will ensure that all the flies that they will score on Week 3 will be of the F2 generation. (4) On Week 3, students will score F2s for cw, f, and w. They will use these data to test if the cw alleles segregated equally (Mendel’s First Law), and also if alleles for cw and f assorted independently of each other (Mendel’s Second Law). (5) In addition, students will use the F2 data for three-point cross analysis. This will determine the order of cw relative to forked and several different white+-transgene insertions—forked and the insertions have known chromosomal locations that closely surround cw, and students will learn how to look up their locations. Therefore, this process will define the location of cw down to a small, contiguous chromosomal interval. The Week 3 Pre-laboratory Assignment (Supporting File S2: Mapping a Mutation-Fly Lab Manual, pp. 27-32, Question 1) will help students understand the non-recombinant and recombinant classes that they will see in their three-point cross data.
Starting at top right and following arrows down through brown boxes: In a parallel set of experimental crosses, your class will perform complementation analysis of cw. Geneticists have produced strains of flies with chromosomal deletions (i.e. deficiencies, abbreviated as “Df”) and duplications (“Dp”) of precisely defined sequences (references 23-25). Each Df and Dp will overlap only a small number of genes. Staff will have set up Df Crosses (Df females x Mutant males) and Dp Crosses (Mutant females x Dp males) and removed the parental flies before Week 3 class. (6) On Week 3, students will score Df and Dp Cross F1s to see if the cw mutant phenotype was complemented or not by various deletions and duplications. The Week 3 Pre-laboratory Assignment (Supporting File S2: Mapping a Mutation-Fly Lab Manual, pp. 27-32, Questions 3a and 3b) will help students understand how these crosses work. (7) Students will eventually learn how to look up the precise sequence boundaries of each deletion and duplication, then use their complementation data to map cw to an even smaller chromosomal interval than what was defined by the three-point crosses.
Gray boxes at bottom: (8) As evident in this figure, there are several lines of evidence that your students will integrate to map cw. The Week 4 Procedure will navigate your students through this type of multifaceted data analysis: They will receive a practice dataset and learn how to use Flybase, GBrowse, and other important online tools to map a different mutation to its gene, then gather information about that gene’s function to support their hypothesis. (9) After this guidance, students will use similar approaches to map cw in the Final Assignment (Supporting File S2: Mapping a Mutation-Fly Lab Manual, pp. 63-68).
We did not collect course evaluation data for this module specifically, but anecdotally, many students expressed appreciation for making a genuine discovery. We have already mapped wavy to the IP3K2 gene (1), and so a pseudonym is necessary to simulate a primary research experience. As an example, we used the pseudonym crumpled wing (cw) throughout this manuscript. We prefer to focus on student learning as opposed to discipline, but note that students could search the internet and locate this article. This may not concern you—students taking this initiative must still absorb a good deal of material from this manuscript, then defend their logic using your class data. Nevertheless, you may wish to replace the pseudonym in our files, swap in different fly stocks, etc. especially if you plan to offer this Lesson over multiple terms. In part to accommodate such needs, we provide editable versions of all files that students will receive; see next subsection, "Potential Lesson Plan Modifications", for further information.
To provide other opportunities for novel discovery in the classroom, we are curating many other unmapped Drosophila mutations. We plan to share our data and recommendations with the broader education community (Derek Dean and David Deitcher, in preparation).
Potential Lesson Plan Modifications
This Lesson can accommodate a wide variety of course structures, student experience levels, class sizes, available prep time, facilities, budgets, etc. We describe the Lesson as we taught it, but there are many ways to modify this module with minimal sacrifice of learning goals. For example, if you do not wish to prep all of the recommended stocks (Table S1-4) or your course does not have a lab component, Supporting File S8. Mapping a Mutation – Sample Final Dataset is a complete Week 3 dataset that can be used to supplement or even fully stand in as class data. For a more advanced genetics class, you could have students collect for their own crosses, or potentially conduct additional genetic analyses of cw (e.g., sequencing of the mutation, RNAi, or genetic rescue; see reference 1). To facilitate communication between instructors and prep staff, we discuss an array of potential Lesson modifications in our Prep Guide (Supporting File S1. Mapping a Mutation – Class Preparation Guide, Part F, section F5).
Should you wish to modify the text, figures, and/or assignments of this Lesson Plan, we have provided fully editable Supporting Files:
- The prep guide (Supporting File S1. Mapping a Mutation – Class Preparation Guide) and lab manual (Supporting File S2. Mapping a Mutation – Fly Lab Manual) are provided as DOCX files.
- S3. Mapping a Mutation – Editable Week 1 Pre-laboratory Assignment is an XLSX file of the Week 1 Pre-laboratory Assignment Punnett squares.
- Supporting File S6. Mapping a Mutation – Editable Week 3 Pre-laboratory Assignment Images is a compressed folder of all three Week 3 Pre-laboratory Assignment images (EPS format). After downloading, unzip this folder to view and edit each image separately.
- All other Supporting Files are editable as well.
- We are happy to provide further materials to facilitate your implementation of the Lesson. For example, we could send you lab manual figures, or even their individual panels. Please contact the corresponding author with requests (Derek Dean, ddean@williams.edu).
- Finally, all DOCX files were typed on a Mac with Microsoft Word (Version 16.21). We realize that page breaks, margins, figure grouping, and other formatting can shift between versions of Word. To preserve our original formatting, we also provide PDF versions of all three of the DOCX Supporting Files: Supporting File S10. Mapping a Mutation – PDF of Class Preparation Guide (S1); S11. Mapping a Mutation – PDF of Fly Lab Manual (S2); and S12. Mapping a Mutation – PDF of Grading Keys (S4).
SUPPORTING MATERIALS
- S1. Mapping a Mutation – Class Preparation Guide. Complete guide for staff to prepare all fly cultures, crosses, and lab supplies for this Lesson. Also includes basic information about fly work for those who have not worked with Drosophila.
- S2. Mapping a Mutation – Fly Lab Manual. Student lab manual for entire Lesson.
- S3. Mapping a Mutation – Editable Week 1 Pre-laboratory Assignment. Original XLSX file used to produce the Punnett squares in the assignment.
- S4. Mapping a Mutation – Grading Keys. Answers and suggested grading for the Week 1 and 3 Pre-laboratory Assignments, the Week 4 Procedure (checked but not graded), and the Final Assignment.
- S5. Mapping a Mutation – Pre-laboratory Lecture Slideshow. Slides for the lecture at the start of Week 1 lab. We sometimes reviewed these slides at the start of Week 3 lab as well.
- S6. Mapping a Mutation – Editable Week 3 Pre-laboratory Assignment Images. Compressed set of three EPS files of the assignment cross diagrams.
- S7. Mapping a Mutation – Blank Week 3 Class Datasheets. During the Week 3 Procedure, students enter A2, B2, Df, and Dp Cross data into these datasheets. Use tabs at bottom left to navigate between crosses. We formatted each datasheet to calculate class totals as data are tallied.
- S8. Mapping a Mutation – Sample Final Dataset. Complete sample dataset for all the crosses scored on Week 3.
- S9. Mapping a Mutation – Answer Key for the Sample Final Dataset Calculations. Using data from Supporting File S8. Mapping a Mutation – Sample Final Dataset, this is a key for chi-square tests, three-point cross locus order, complementation analysis, and genomic location of cw. Contains embedded formulae that could be used for alternative data.
- S10. Mapping a Mutation – PDF of Class Preparation Guide (S1). Preserves the original page formatting of S1. Mapping a Mutation – Class Preparation Guide. Use this file if you do not plan to make significant edits to the guide.
- S11. Mapping a Mutation – PDF of Fly Lab Manual (S2). Preserves the original page formatting of S2. Mapping a Mutation – Fly Lab Manual. Use this file if you do not plan to make significant edits to the manual.
- S12. Mapping a Mutation – PDF of Grading Keys (S4). Preserves the original page formatting of S4. Mapping a Mutation – Grading Keys. Use this file if you do not plan to make significant edits to the keys.
ACKNOWLEDGMENTS
The authors would like to thank Audrey Werner and Debra Rogers-Gillig for their excellent technical support. Luana Maroja and Janis Bravo of Williams College, and Dawn Carone of Swarthmore College provided helpful advice for development of the lab manual. David Smith of Williams College had helpful suggestions for some key figures in the prep guide. Eric Spana of Duke University kindly provided fly stocks and insight during the early phases of this project.
This manuscript is dedicated with gratitude to Stephanie Mitchell Dean.
References
- Dean DM, Maroja L, Bomkamp B, Cottrill S, Westervelt K, Deitcher DL. 2015. The wavy mutation maps to the Inositol 1,4,5-trisphosphate 3-kinase 2 (IP3K2) gene of Drosophila and interacts with IP3R to affect wing development. G3 (Bethesda). 6(2):299-310. doi: 10.1534/g3.115.024307.
- Hartl DL, Cochrane BJ. 2017. Genetics: Analysis of Genes and Genomes (9(th) edition). Burlington, MA: Jones & Barlett Learning.
- Pierce BA. 2017. Genetics, a Conceptual Approach (6(th) edition). New York, NY: W.H. Freeman/Macmillan Learning.
- Bate M, Arias AM, editors. 1993. The Development of Drosophila melanogaster. Cold Spring Harbor, NY: CSH Press.
- Lawrence, PA. 1992. The Making of a Fly. Hoboken, NJ: Wiley Press.
- Dubnau, J. 2014. Behavioral Genetics of the Fly (Drosophila melanogaster). Cambridge, England: Cambridge University Press.
- Ugur B, Chen K, Bellen HJ. 2016. Drosophila tools and assays for the study of human diseases. Dis Model Mech 9: 235-244; doi: 10.1242/dmm.0237621.
- Marcus JM, Hughes TM. 2009. Drosophila transposon insertions as unknowns for structured inquiry recombination mapping exercises in an undergraduate genetics course. Genetics 182: 417-422; doi: 10.1534/genetics.109.101774.
- Call GB, Olson JM, Chen J, Villarasa, N, Ngo KT, et al. 2007. Genomewide clonal analysis of lethal mutations in the Drosophila melanogaster eye: comparison of the X chromosome and autosomes. Genetics 177: 689-697; doi: 10.1534/genetics.107.077735.
- Chen J, Call GB, Beyer E, Bui C, Cespedes A, et al. 2005. Discovery-based science education: functional genomic dissection in Drosophila by undergraduate researchers. PLoS Biol. 3(2): e59.
- Olson JM, Evans CJ, Ngo KT, Kim HJ, Nguyen JD, et al. . 2019. Expression-based cell lineage analysis in Drosophila through a course-based research experience for early undergraduates. G3 (Bethesda). 5; 9(11): 3791-3800. doi: 10.1534/g3.119.400541.
- Wright R. 2014. It's not about you: A simple proposition for improving biology education. Genetics 198(2): 429-430; doi: 10.1534/genetics.114.169136.
- Smith MK, and Wood WB. 2016. Teaching genetics: Past, present, and future. Genetics 204(1): 5-10; doi: 10.1534/genetics.116.187138.
- Beck C, Butler A, Burke da Silva K. 2014. Promoting inquiry-based teaching in laboratory courses: Are we meeting the grade? CBE Life Sci Educ 13: 444-452; doi: 10.1187/cbe.13-12-0245.
- Morgan TH, Bridges CB. 1916. Sex-linked inheritance in Drosophila. Publs Carnegie Instn 237: 1-88.
- Lindsley DL, Zimm GG. 1992. The Genome of Drosophila melanogaster. San Diego, CA: Academic Press.
- Nachtsheim H. 1928. Beitrag zur Topographie des X-Chromosoms von Drosophila melanogaster. Z. indukt. Abstamm.- u. VererbLehre 48: 245-258.
- MacIntyre R. 1974. Phenotypically identical but genotypically unique "unknown" stocks for genetics laboratory courses. Dros. Inf. Serv. 51: 158 (reprinted 1999. Dros. Inf. Serv. 82: 130).
- Pye Q. 1980. New white-eyed Drosophila "unknown" stocks for genetics laboratory courses. Dros. Inf. Serv. 55: 171 (reprinted 1999. Dros. Inf. Serv. 82: 137).
- Waddle FA. 1991. Some stocks for the teaching lab. Dros. Inf. Serv. 70: 262 (reprinted 1999. Dros. Inf. Serv. 82: 125).
- Ho MCW, Venema DR, Drewell RA. 2008. A concise Drosophila laboratory module to introduce the central concepts of genetics. Dros. Inf. Serv. 91: 164-168.
- Morcillo P, Tuttle R, MacIntyre RJ. 1996. The use of transposable P-elements of Drosophila melanogaster for introductory genetics laboratory courses. J Hered. 87 (5): 399-403. doi: 10.1093/oxfordjournals.jhered.a023024.
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, et al. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288-292.
- Cook RK, Christensen SJ, Deal JA, Coburn RA, Deal ME, et al. 2012. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 13: R21.
- Venken KJT, Popodi E, Holtzman SL, Schulze KL, Park S, et al. 2010. A molecularly defined duplication set for the X chromosome of Drosophila melanogaster. Genetics 186: 1111-1125.
- Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, et al. 2019. FlyBase 2.0: the next generation. Nucleic Acids Res 47: D759-D765.
- St Pierre SE, Ponting L, Stefancsik R, McQuilton P, and Fly Consortium. 2014. FlyBase 102-advanced approaches to interrogating FlyBase. Nucleic Acids Res 42: D780-D788.
- Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, et al. 2013. BLAST: a more efficient report with usability improvements. Nucleic Acids Res 41: W29-W33.
Article Files
Login to access supporting documents
Mapping a Mutation to its Gene: The "Fly Lab" as a Modern Research Experience(PDF | 1 MB)
S1-Mapping a Mutation-Class Preparation Guide.docx(DOCX | 49 MB)
S2-Mapping a Mutation-Fly Lab Manual_3.docx(DOCX | 65 MB)
S3-Mapping a Mutation-Editable Week 1 Prelaboratory Assignment.xlsx(XLSX | 18 KB)
S4-Mapping a Mutation-Grading Keys.docx(DOCX | 7 MB)
S5-Mapping a Mutation-Prelaboratory Lecture Slideshow.pptx(PPTX | 28 MB)
S6-Mapping a Mutation-Editable Week 3 Prelaboratory Images.zip(ZIP | 3 MB)
S7-Mapping a Mutation-Blank Week 3 Class Datasheets.xlsx(XLSX | 57 KB)
S8-Mapping a Mutation-Sample Final Dataset.xlsx(XLSX | 59 KB)
S9-Mapping a Mutation-Answer Key for the Sample Final Dataset Calculations.xlsx(XLSX | 68 KB)
S10-Mapping a Mutation-PDF of Class Preparation Guide S1.pdf(PDF | 42 MB)
S11-Mapping a Mutation-PDF of Fly Lab Manual S2.pdf(PDF | 67 MB)
S12-Mapping a Mutation-PDF of Grading Keys S4.pdf(PDF | 5 MB)
- License terms

Comments
Comments
There are no comments on this resource.