Learning About Protein Localization: A Lesson for Analyzing Figures in a Scientific Publication
Editor: Scott Gehler
Published online:
Abstract
In order to function correctly, proteins must be localized to a specific subcellular location. We have designed this lesson to use data from the primary literature to teach students about the mechanisms cells use to direct proteins to the appropriate destinations and about the types of experiments that scientists use to investigate these mechanisms. Exposing undergraduate students to primary literature and experimental science in biology courses can prepare them for the demands of the job market and graduate programs. However, students can struggle when asked to analyze data from publications due to the high cognitive load involved with figure interpretation. We have designed this lesson to help students draw meaningful conclusions from figures in primary literature. To make the figure interpretation process more accessible to students, we use a combination of scaffolding to break down figure interpretation into smaller attainable steps and group work to allow students to combine their knowledge and work collaboratively. In this lesson, student groups are given a subset of figures from a scientific article along with questions that guide them through the process of decoding and interpreting these figures. The students interpret three figures that use different experimental techniques to address the subcellular localization of the TIN2 protein and one figure that determines the locations of the signal sequences in the protein that are critical for the correct localization. Taken together, this lesson helps students understand both how the eukaryotic cell localizes proteins to the correct subcellular localization and how scientists study this question.
Citation
de Waal E, McGehee AM. 2020. Learning about protein localization: A lesson for analyzing figures in a scientific publication. CourseSource. https://doi.org/10.24918/cs.2020.5
Society Learning Goals
Cell Biology
- Protein Targeting & Trafficking
- How are cellular components targeted and distributed to different regions and compartments of a cell?
- Methods & Tools of Cell Biology
- How do the methods and tools of cell biology enable and limit our understanding of the cell?
Science Process Skills
- Process of Science
- Interpret, evaluate, and draw conclusions from data
- Construct explanations and make evidence-based arguments about the natural world
- Modeling/ Developing and Using Models
- Build and evaluate models of biological systems
- Quantitative Reasoning/ Using Mathematics and Computational Thinking
- Apply the tools of graphing, statistics, and data science to analyze biological data
Lesson Learning Goals
- Students will understand how different experimental techniques can be used to assess protein localization.
- Students will understand that proteins are directed to organelles within cells using amino acid-based signal sequences.
- Students will learn how to interpret data from complex figures in scientific publications.
Lesson Learning Objectives
At the end of this activity students will be able to:
- Explain how eukaryotic cells "know" where a particular protein should be located.
- Analyze immunofluorescence data to determine protein localization within a cell.
- Analyze a western blot with subcellular fractions to determine protein localization within a cell.
- Describe an experiment that can be used to analyze the subcellular localization of a specific protein.
- Describe an experiment that can be used to determine the signal sequence of a protein.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Protein localization is an important concept in cell biology because the location of a protein within a cell provides the physiological context for protein function. Although the process of translation always initiates in the cytosol, many proteins must be translocated to organelles during or after translation to perform their specific role. The trafficking of a protein to a particular location within the cell is regulated by a signal sequence encoded within the protein. The concepts behind protein localization and signal sequences are an important part of cell and molecular biology education (1), but there are relatively few resources available for teaching these important biological phenomena to undergraduate students (2-4). We wanted to design a lesson that would help students master the concepts of protein localization and signal sequences, while also strengthening their ability to apply the process of science, one of the core competencies in Vision and Change (5).
One common strategy used in lecture courses to familiarize students with the scientific process is to expose them to primary literature. Unfortunately, reading a scientific paper is a daunting task for most undergraduate students because of unfamiliar terminology and complex figures (6,7). Data interpretation, in particular, can be overwhelming because students have little experience extracting meaningful information from figures and tables that have been derived from hypothesis-based research (8).
Undergraduate students often lack the foundational knowledge and reasoning resources to understand a manuscript, which can pose challenges for instructors who want to incorporate an analysis of research articles in their course (6). An alternative pedagogical approach to introduce students to the scientific process is to focus solely on data. The use of this strategy forces students to concentrate on data analysis and has been found to improve comprehension of complex data sets as well as reduce anxiety when examining primary literature (8,9). It also allows instructors to emphasize data interpretation skills in their lesson plan and not burden students with comprehending the dense material in the various sections of the manuscript. Although omitting the text of the publication substantially reduces the content of a scientific article, students can still struggle with data analysis due to the high cognitive load required to understand complex figures that are common in primary literature (10). To overcome some of the difficulties associated with interpreting authentic data, we have incorporated two pedagogical strategies into this data-centric approach: scaffolding and working in small groups.
Interpretation of data and graphs is an activity with a high cognitive load for students (7). These representations have many interconnected elements (of both the experimental design and the visual representation of the data) that must be decoded in order for students to be able to interpret data. One strategy to reduce the cognitive load of a task is to break it down into smaller elements (11).
In this lesson, we use a series of questions about each figure that are designed to help students decode the content of a figure before they attempt to interpret experimental data within the figure. This process allows students to understand the experimental design and visual representation of the data before asking them to attempt to draw conclusions through interpretation. The sequence of these questions is a critical component to successfully implementing this strategy. The first set of questions for each experiment are designed to scaffold the process of understanding the experimental design (e.g., which antibodies are being used in this figure?). We place these questions under the heading "What is the experiment?" Once students have a grasp on the basics of the figure, they can start answering questions that involve higher levels of critical thinking (e.g., are the data supportive of the notion that this protein is localized to mitochondria?). We place these questions under the heading "What does the experiment tell us?". This approach provides a framework that enables groups of students to understand a complex figure by first identifying important and relevant aspects of the figure, then using this knowledge to comprehensively analyze data and develop meaningful interpretations.
Group work is a relatively common practice in college courses because it promotes active learning and provides an opportunity for students to work cooperatively to solve a problem. Small groups of students can be particularly effective in encouraging collaborative learning because students can be heard and hear from their peers instead of just listening to what an instructor says during a lecture. These interactions allow students within the group to engage in retrieving knowledge and processing ideas, which typically improves student learning outcomes and increases positive attitudes toward learning the material (12). Group work also facilitates more confidence in completing challenging tasks because groups of students can work together to solve a problem even when no one in the group initially knows the answer (13). We have opted to employ an informal approach in this lesson whereby small groups of 3-4 students work together to analyze a subset of figures from a pre-selected publication during one class period. This strategy allows students to use peer discussions to analyze figures and increase their conceptual understanding of the scientific process. The use of group work in this context can also help reduce the anxiety and frustration that is common when examining primary literature in the classroom (8).
Overview of protein localization lesson
Assessing protein localization involves a wide variety of laboratory techniques such as subcellular fractionation, immunofluorescence, and fusion proteins. In this lesson, undergraduate students were tasked with analyzing multiple figures from a published paper (14) that investigates the trafficking of a protein, TIN2, to both the nucleus and mitochondria. TIN2 was previously known to localize to the nucleus where it interacts with telomeres, but the authors in this study wanted to determine if TIN2 was also translocated into mitochondria. To answer this question, immunofluorescence, subcellular fractionation, and western blots were employed to track the localization of TIN2 within cells using anti-TIN2 antibodies. The location of TIN2 was also assessed by using TIN2-GFP and TIN2-FLAG fusion proteins. After it was shown that TIN2 localizes to both nuclei and mitochondria using different experimental techniques, a series of TIN2-GFP fusion proteins were synthesized that had specific segments of the TIN2 protein sequence deleted. These fusion proteins allowed the authors to determine the location of mitochondrial signal sequences in the TIN2 protein, and therefore identify which segments of the protein were both necessary and sufficient for translocation into mitochondria. The figures students analyze in this lesson (Supporting File S1: Protein Localization - All Figures) have been adapted with permission from this paper (14). Copyright permission to modify and use these figures was obtained through the Copyright Clearance Center (www.copyright.com).
The paper we have chosen for this activity (14) uses a wide variety of experimental approaches, which provides an opportunity for undergraduate students to see how different lab techniques can be used to answer one scientific question. By breaking down data interpretation into a series of questions, the overall goal for this activity was to provide students a strategy or framework to draw their own conclusions from authentic data.
Intended Audience
This lesson was used in an upper level cell biology course with junior and senior biology majors. This course typically has 15-20 students.
Required Learning Time
This lesson was taught in a single 50-minute class period. Additional time may be required to familiarize students with the various laboratory techniques that are used for different experiments. Moreover, this lesson is one of many group activities that are used throughout the semester in this class at Suffolk University, so students may need additional time or instructions to understand the guidelines for working in groups during class.
Prerequisite Student Knowledge
In order to successfully complete this lesson students need to have a basic understanding of the following techniques: immunofluorescence, subcellular fractionation, western blotting, and fusion proteins. Students should also be familiar with protein targeting in eukaryotic cells, signal sequences, and the biological use of the terms "necessary" and "sufficient." Some of these techniques and concepts can be introduced along with the lesson, if needed. For example, when this lesson is taught at Suffolk University, students have been previously exposed to many of these techniques and concepts, but immunofluorescence and colocalization of fluorescent probes are introduced just before students start working in groups for this lesson.
Prerequisite Instructor Knowledge
Instructors need to be familiar with the concepts that are covered in this lesson, which can be found in most cell biology textbooks [e.g., Molecular Biology of the Cell by Alberts and colleagues (15)]. Additionally, instructors should thoroughly read the Chen et al. paper (14), so they can explain the rationale of the experimental design and understand the figures the students will analyze during the lesson.
This lesson deals with a protein that has two distinct subcellular localizations: the nucleus and the mitochondria. While the lesson is focused primarily on understanding where in the cell a protein is located, it would be helpful for the instructor to understand how a single protein can function in different subcellular compartments. For a review about dual targeted nuclear and mitochondrial proteins see Monaghan and Whitmarsh (16).
SCIENTIFIC TEACHING THEMES
Active Learning
In this lesson, students are actively engaged in collaborative learning. Students work together in small groups of 3-4 students to answer a set of questions that guides them through the data interpretation process. This framework allows the students to build an understanding of the experimental set up and then draw conclusions from data produced by the experiment.
Assessment
Each group of students is tasked with answering a set of questions on worksheets that are distributed in class (Supporting Files S2-S5: Protein Localization - Figure 1, 2, 3, and 4 questions). These worksheets are used as formative assessments and are collected at the end of the lesson. Although all students receive the questions, one worksheet is graded from each group for completion, and the instructor circulates through the room throughout the lesson to ensure that all student groups are able to answer the questions. Individually assigned homework questions are used to reinforce the concepts from the lesson, and exam questions provide a summative assessment to further evaluate student learning (Supporting File S6: Protein Localization - Related Assessment Questions).
Inclusive Teaching
Analyzing and interpreting authentic data is a difficult endeavor that typically requires participation from all group members. We use instructor-assigned groups for this activity so all groups have students with a variety of prior courses and experiences to ensure that group success requires contributions from all members. The instructor also facilitates participation and group discussion by asking questions to various group members during class. Importantly, the interpretation of immunofluorescence images can be difficult for individuals who have red/green colorblindness, but the use of alternate color schemes can make this type of data more accessible (17). A set of alternate green/magenta images (Supporting File S7: Protein Localization - All Figures Magneta) has been generated for this lesson so all students can participate in the analysis of the immunofluorescence data.
LESSON PLAN
Pre-class Preparation: Instructor
Successful implementation of this lesson requires small student groups and handouts. Groups should be established before class and can be chosen by the instructor or self-selected by the students. Handouts that accompany this lesson should also be prepared before class. There should be one handout for all four figures (Supporting File S1: Protein Localization - All Figures) and separate handouts that have the questions for each figure (Supporting Files S2-S5: Protein Localization - Figure 1, 2, 3, and 4 questions). Using separate handouts for figures and questions is critical because this allows the students to see the figure while working on the questions. The figures need to be printed in color to allow for interpretation of the immunofluorescence data. The images are easiest to analyze in the red/green set of images (Supporting File S1: Protein Localization - All Figures); however, to accommodate students with red/green colorblindness, some sets of the green/magenta images (Supporting File S7: Protein Localization - All Figures Magneta) should also be printed. Since the images in the original publication use the red/green/blue color scheme, there is a minor loss of information in the green/magenta version of figures, which is why both figure sets are provided and used in class. If color printing is not available, the color figures can be projected in the classroom using a PowerPoint presentation (Supporting File S8: Protein Localization - Presentation Slides).
Beginning of Class
Ideally, students should come into the classroom and sit with their groups at the beginning of the class period.
Introduction to Methods
If any of the methods that are used in the lesson are unfamiliar to students, it can be useful to start out with a brief overview of these methods and data interpretation. Students at Suffolk have already had exposure to subcellular fractionation and western blot analysis, so they only need an overview of fluorescence imaging and what colocalization looks like. We do this using a slide that shows single color and merged color images in both the red/green and magenta/green color schemes (Supporting File S8: Protein Localization - Presentation Slides, Slide 1). For red/green images the area where the two proteins overlap looks yellow; for the green/magenta images the overlap looks white.
Group Work
Once students have been introduced to the methods, the instructor should distribute the handout that has all four figures and a set of questions that is used to analyze the first figure (Supporting File S1: Protein Localization - All Figures and Supporting File S2: Protein Localization - Figure 1 questions). Each student should receive their own copy of the figures and questions. When distributing the handouts of the figures, we give students the option of the red/green (Supporting File S1: Protein Localization - All Figures) or green/magenta (Supporting File S7: Protein Localization - All Figures Magneta) sets of images to work with (~10-15% of students choose the green/magenta set). After the students have received the figures and questions, they should individually read through the background information provided in the question handout and then work as a group to discuss and answer the questions. We have provided information below about each of the question handouts. As the students work through the questions, the instructor should circulate around the room to ask groups to explain their answers to determine if clarification or redirection is needed. When individual groups have reached the end of the questions for a particular figure, they should be given the next handout, which contains questions for the next figure. This staged approach allows the instructor to make sure the students are correctly analyzing data before proceeding to the next figure.
Figure 1
The figures and questions in this lesson are based on the Chen et al. paper (14) that investigates the mitochondrial localization of the telomeric protein TIN2. The handout associated with this figure (Supporting File S2: Protein Localization - Figure 1 questions) contains a brief overview of the TIN2 protein and the experimental question of the paper. Additionally, a terminology list is provided so that students will know the meaning of key terms they will encounter during the lesson. Students read through this background information and use the figures handout to answer the questions regarding Figure 1. The first questions (1 and 2 under the heading "What is the experiment?") are designed to help students understand what they are looking at in the figure and why it is useful. In this immunofluorescence experiment, the protein TIN2 is visualized either with antibodies to the endogenous protein, antibodies that recognize a FLAG-tagged TIN2, or through GFP fluorescence. The nuclear protein TFR2, the mitochondrial protein ATP5A1, and DNA are also indicated to accurately determine the localization of TIN2. Students must identify what is shown in each panel from Figure 1 and the experimental purpose of the reagents that are used before they move on to interpret data within the figure. These "What is the experiment?" questions are intended to be relatively straight forward to answer and ensure that students have a basic understanding of the experiment before they attempt to draw conclusions from the data. The following questions (3-6 under the heading "What does the experiment tell us?") then ask the students to interpret the figure. As the students are working through these questions, it is important that the instructor checks in with groups to see how they are interpreting the data. While most groups of students easily determine that TIN2 is localized to the nucleus (question 3), some groups have difficulty with data in Figure 1B and question 4. This figure shows that some of the TIN2 fluorescence overlaps with mitochondria. However, because the localization of TIN2 and ATP5A are not identical, students will sometimes incorrectly interpret this to mean that TIN2 is not localized to mitochondria. It is important to check that students are correctly interpreting Figure 1B at this point, so that their subsequent data interpretation is not misdirected. This can be done either by checking in with individual groups if the class is small enough or by asking individual groups to report their answers to question 4 in a class discussion.
Figure 2
The second set of questions (Supporting File S3: Protein Localization - Figure 2 questions) refers to Figure 2. The experiment shown in this figure also addresses the localization of TIN2, but uses subcellular fractionation and western blotting instead of immunofluorescence. The "What is the experiment?" questions that accompany Figure 2 are intended to ensure that students understand the subcellular fractionation (Question 7) and the use of the various antibodies that are chosen for the western blot (Question 8). The "What does the experiment tell us?" questions ask the students to interpret the data in the figure to determine whether the fractionation worked correctly (Question 9) and where in the cell TIN2 is localized (Question 10). When looking at this figure, some students may notice the multiple bands in the TIN2 western blot and the fact that these bands show different subcellular localization. The lower two bands represent mitochondrially processed forms of TIN2 (14). While this finding is not explicitly used in this lesson, it is addressed in the original paper, and instructors should be familiar with the article and prepared to explain this finding to students who notice it.
Figure 3
The third set of questions (Supporting File S5: Protein Localization - Figure 3 questions) concern Figure 3. In this experiment the authors are addressing the question of where in the mitochondrion TIN2 is localized (i.e., does it associate with the outer surface of the mitochondrion, or is it actually translocated into the inner compartments of the mitochondrion?). They address this question by isolating mitochondria and then digesting accessible proteins with the enzyme ProteinaseK. The "What is the experiment?" questions help the students to understand the experimental setup, how the three reactions that were analyzed were treated differently (Question 11), and that Trition-X 100 is included in order to disrupt membranes and allow the ProteinaseK to access proteins that reside inside the mitochondrion (Question 12). The "What does the experiment tell us?" questions lead students through the interpretation of this experiment by determining whether the proteins were intact at the end of each reaction (Question 13), why these results are obtained for the controls (Question 14), and then asking students to draw a conclusion about localization of TIN2 on the basis of this experiment (Question 15).
Figure 4
The fourth set of questions (Supporting File S5: Protein Localization - Figure 4 questions) concern Figure 4. The purpose of the experiments shown in this figure is to determine where the mitochondrial signal sequences are located in the TIN2 protein. There are no "What is the experiment?" questions in this set because the experimental set up of Figure 4 is very similar to that of Figure 1D. A major focus for this set of "What does the experiment tell us?" questions is for students to understand how experiments try to establish biological causality. Students are asked to interpret this data to determine whether particular regions of the TIN2 protein are necessary and/or sufficient for mitochondrial targeting (Questions 16-19). Instructors can also use "indispensable" for necessary and "induces" or "promotes" for sufficient if they feel that is more appropriate (18). This analysis is somewhat complicated by the fact that TIN2 contains two localization sequences, either of which is sufficient for mitochondrial targeting, and students are asked to explain how the data supports this conclusion (Question 20).
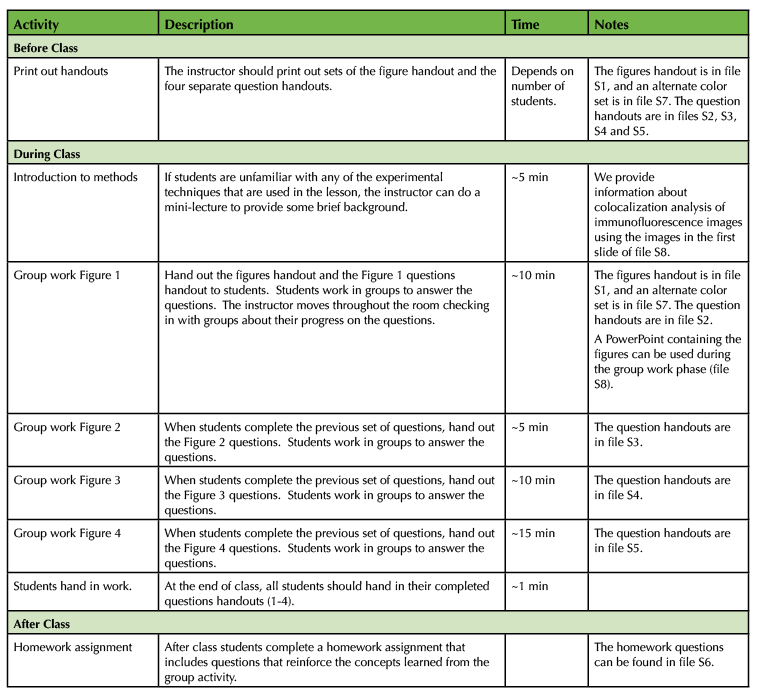
Table 1. Protein Localization - Lesson Timeline
TEACHING DISCUSSION
The overall goals of this lesson are to teach students about protein localization and the experimental techniques used by scientists to study protein localization as well as to develop skills that will enable students to critically analyze experimental data. In order to assess whether student learning objectives are being achieved, both formative and summative assessments are used.
Formative assessment questions (Supporting Files S2-S5: Protein Localization - Figure 1, 2, 3, and 4 questions) are answered by groups of students during class. The student answers are graded for completion and are checked for accuracy by the instructor. This approach lets the instructor determine whether students understand the concepts covered in the activity and provides an opportunity to address inaccuracies either through written feedback to the students or additional instruction in later class periods. Answers to these formative assessment questions suggest that the learning goals are being met, as students were able to answer all questions from all four figures the last time this activity was used in a cell biology course at Suffolk University (Fall 2019).
Students also answer summative assessment questions in homework assignments and exams to determine their comprehension of concepts that were taught in this lesson. We use questions about protein targeting from a textbook question bank (15) as well as questions we have written ourselves (Supporting File S6: Protein Localization - Related Assessment Questions). Student performance on these questions varied, but in every case a majority of the students (>60%) were able to correctly answer the assessment question. Thus, the combination of these formative and summative assessments allows us to assess all of the learning goals for this lesson and has provided us with evidence that this lesson is effective in achieving the learning goals.
Beyond the learning objectives of this lesson, we are interested in helping our students to achieve the goal of learning how to interpret data from complex figures in scientific publications. Reading an entire manuscript and trying to understand the figures at the same time can be discouraging to undergraduate students because at this point in their academic career they are still developing the ability to think like a scientist. To overcome the knowledge barriers and frustration that is often experienced with this endeavor, this lesson uses a data-centric approach and scaffolding that enables small groups of students to interpret authentic data about protein localization. By scaffolding the data interpretation process, we are allowing students to gain a first-hand experience in how trained scientists extract meaningful conclusions from authentic data so the same process can be applied to novel data sets encountered in the future. Competently analyzing authentic data does not come from one activity or even one college course, and undergraduate students that repeatedly examine figures from scientific publications can improve their ability to comprehend complex graphical data representations and draw accurate conclusions on their own (9). By implementing strategies that reduce apprehension and increase confidence when analyzing data from primary literature, students can eventually develop the skills needed to engage with the investigative process of scientific research.
Another important aspect of this lesson is the coupling of understanding protein localization with understanding how to analyze data from the primary literature. To answer the formative questions associated with this lesson, students must rely on content knowledge from earlier in the course and use science process skills to interpret the data. For example, students must understand that different proteins reside in different organelles in order to be able to interpret the data in Figures 1-2 (Supporting Files S2 and S3: Protein Localization - Figures 1 and 2). Additionally, the interpretation of the data presented in this lesson serves to reinforce students' content knowledge. For example, when students interpret the data in Figure 4 and see that removing particular stretches of amino acids from the TIN2 protein alters its subcellular localization, this reinforces the concept, taught earlier in the course, that it is sequences of amino acids in proteins that direct proteins to particular subcellular locations (Supporting File S5: Protein Localization - Figure 4). The ability to make connections between content knowledge and science process skills will help these students meet the demands of the job market and graduate programs.
Student Reactions
In general, this lesson is well received by the students. They do have some initial difficulty with the interpretation of the immunofluorescence data, but this is frequently a point of discussion for the groups and they work together to discuss how to understand and interpret this data. One of our goals in designing this and other similar activities that are used in the cell biology course is to teach students how to analyze data from the primary literature. By scaffolding the data interpretation and breaking it down with guided questions under the headings "What is the experiment?" and "What does the experiment tell us?" we hope that students will learn to do these steps on their own when confronted with novel experimental data. Students frequently mention the data interpretation activities as one of the most useful parts of the cell biology course in the student evaluation surveys at the end of the semester.
Potential Alternatives
This lesson was designed to be used in an upper level cell biology course in which students have some familiarity with the techniques that are used in this paper and with this type of group-based data interpretation activity. If students are not familiar with the experimental techniques used here or with group work, then additional time beyond a 50-minute class period may be needed to complete the lesson.
We teach this lesson in a class that typically has around 15 students. This small size enables the instructor to move around the room speaking with each group individually, guiding the group discussion, and giving students the question handouts as they progress through the lesson. While this structure works very well for this lesson, it would likely be prohibitive if the class size is substantially greater than 20 students. For a larger course, all of the question handouts can be given at the beginning of the activity along with the figures handout. Students can work in groups and the instructor can keep the class on track by calling the entire class back together periodically and facilitating a class discussion to go over the questions for each figure. This can be done by asking different groups to share the answers with the class and addressing inconsistencies and questions that arise with the entire class rather than with each individual group.
This lesson is easily modifiable based on the goals of the instructor. The lesson has four figures and corresponding questions that accompany each figure. This lesson could be shortened and adapted by using only a subset of these figures and questions. Depending on student prior knowledge and instructor goals, it would be possible to adapt this lesson to focus on one of two learning goals: (1) illustrating how different experimental techniques can be used to assess protein localization (Figures 1-3) or (2) understanding that proteins are directed to organelles within cells using amino acid-based signal sequences (Figures 1 and 4). For goal 1, Figures 1-3 show students that both immunofluorescence and subcellular fractionation followed by western blotting can be used to assess protein localization. For goal 2, analyzing Figures 1 and 4 lets students see how manipulating the amino acid sequence of a protein can provide important mechanistic insights into protein targeting.
Additionally, the modular nature of this lesson allows the instructor to adapt the lesson in real time for particular classes that seem to be progressing slowly through the figures. If there is a class, or even a group of students who are moving more slowly through data interpretation than expected, these students can be instructed to skip some of the figures. If our class gets off to a slow start, Figure 3 is usually skipped so there is more time to focus on Figures 1, 2 and 4.
SUPPORTING MATERIALS
- S1. Protein Localization - All Figures
- S2. Protein Localization - Figure 1 questions
- S3. Protein Localization - Figure 2 questions
- S4. Protein Localization - Figure 3 questions
- S5. Protein Localization - Figure 4 questions
- S6. Protein Localization - Related Assessment Questions
- S7. Protein Localization - All Figures Magenta
- S8. Protein Localization - Presentation Slides
ACKNOWLEDGMENTS
We thank Arup Dey and Atosa Ahmadi for reviewing the manuscript, Maghnus O'Seaghdha for providing the immunofluorescence images that are used to illustrate colocalization, and the students in Bio403 at Suffolk University for participating in this lesson and providing valuable feedback.
The figures in the supporting files S1 and S7 are edited from a scientific publication (Chen et al. (2012). Mol Cell 47, 4271-4285). We have written permission to use these figures. Copyright permission was obtained through the Copyright Clearance Center (copyright.com). Documentation of the copyright permission can be provided if necessary.
References
- Tang BL, Teng FYH. 2005. Concepts of protein sorting or targeting signals and membrane topology in undergraduate teaching. Biochem Mol Bio Educ. 33: 188-193. doi: 10.1002/bmb.2005.494033032448.193.
- Vallen E. 2002. Analysis of protein localization and secretory pathway function using the yeast Saccharomyces cerevisiae. Cell Biol Educ. 1: 173-192. doi: 10.1187/cbe.02-08-0027.
- Shuster MI. 2019. Protein targeting gone awry: The importance of proper localization. http://sciencecases.lib.buffalo.edu/cs/collection/detail.asp?case_id=961&id=961. Accessed August 28(th), 2019.
- Stanton JD, Dye KM. 2017. Investigating the function of a transport protein: Where is ABC6 located in human cells? CourseSource. doi: 10.24918/cs.2017.19.
- American Association for the Advancement of Science. 2011. Vision and change in undergraduate biology education: A call to action, Washington, D.C.
- Bowen GM, Roth W, McGinn MK. 1999. Interpretations of graphs by university biology students and practicing scientists: Toward a social practice view of scientific representation practices. J Res Sci Teach 36: 1020-1043.
- Offerdahl EG, Arneson JB, Byrne N. 2017. Lighten the load: Scaffolding visual literacy in biochemistry and molecular biology. CBE Life Sci Educ 16: 1- 11. doi: 10.1187/cbe.1606-0193.
- Zogallo P, Meddleton S, Bolger MS. 2016. Teaching Real data Interpretation with Models (TRIM): Analysis of student dialogue in a large-enrollment cell and developmental biology course. CBE Life Sci Educ 15: 1-18. doi: 10.1187/cbe.15-11-0239.
- Round JE, Campbell AM. 2013. Figure facts: Encouraging undergraduates to take a data-centered approach to reading primary literature. CBE Life Sci Educ 12: 39-46. doi: 10.1187/cbe.11-07-0057.
- Kirby CK, Fleming-Davies A, White PJT. 2019. Figure of the Day: A classroom activity to improve students' figure creation skills in biology. The American Biology Teacher. 81: 317-325. doi: 10.1525/abt.2019.81.5.317.
- Sweller J. 1994. Cognitive load theory, learning difficulty, and instructional design. Learning and Instruction. 4: 295-312.
- Springer L, Stanne ME, Donovan SS. 1999. Effects of small-group learning on undergraduates in science, mathematics, engineering, and technology: A meta-analysis. Review of Educational Research. 69: 21-51.
- Hodges LC. 2018. Contemporary issues in group learning in undergraduate science classrooms: A perspective from student engagement. CBE Life Sci Educ. 17: es3. doi:10.1187/cbe.17-11-0239.
- Chen LY, Zhang Y, Zhang Q, Li H, Luo Z, Fang H, Kim SH, Qin L, Yotunda P, Xu J, Tu BP, Bai Y, Songyang Z. 2012. Mitochondrial localization of telomeric protein TIN2 links telomere regulation to metabolic control. Mol Cell. 47: 4271-4285. doi: 10.1016/j.molcel.2012.07.002.
- Alberts B, Johnson A, Lewis J, Morgan D, Raff M, Roberts K, Walter P. 2015. Molecular Biology of the Cell 6(th) Ed. New York: Garland Science.
- Mognahan RM, Whitmarsh AJ. 2015. Mitochondrial proteins moonlighting in the nucleus. Trends Biochem Sci. 40:728-735. doi: 10.1016/j.tibs.2015.10.003.
- Wong B. 2011. Color blindness. Nat Methods 8: 441.
- Yoshihara M, Yoshihara M. 2018. Necessary and sufficient in biology is not necessarily necessary - confusions and erroneous conclusions resulting from misapplied logic in the field of biology, especially neuroscience. J Neurogenet. 32: 53-64. doi: 1080/01677063.2018.1468443.
Article Files
Login to access supporting documents
Learning About Protein Localization: A Lesson for Analyzing Figures in a Scientific Publication(PDF | 159 KB)
S1 Protein Localization All Figures.docx(DOCX | 3 MB)
S2 Protein Localization Figure 1 questions.docx(DOCX | 32 KB)
S3 Protein Localization Figure 2 questions.docx(DOCX | 13 KB)
S4 Protein Localization Figure 3 questions.docx(DOCX | 13 KB)
S5 Protein Localization Figure 4 questions.docx(DOCX | 13 KB)
S6 Protein Localization Related Assessment Questions.docx(DOCX | 15 KB)
S7 Protein Localization All Figures Magenta.docx(DOCX | 2 MB)
S8 Protein Localization Presentation Slides.pptx(PPTX | 4 MB)
- License terms

Comments
Comments
There are no comments on this resource.