A CURE for Salmonella: A Laboratory Course in Pathogen Microbiology and Genomics
Editor: William Morgan
Published online:
Abstract
Rapid advances in genomics and bioinformatics, the vast amount of data generated by next-generation sequencing, and the penetration of the ‘-omics’ into many areas of biology have created a need for students with hands-on experience with computational and ‘big data’ methods. Additionally, laboratory experience in the isolation, identification, and characterization of unknown bacteria is a vital part of a microbiology student’s training. This lesson is a course-based undergraduate research experience (CURE) focusing on Salmonella enterica, a common and relatively low-virulence foodborne pathogen. In Module 1, students isolate and identify S. enterica strains from stream sediment, poultry litter, or other sources. They conduct phenotypic evaluation of antimicrobial resistance (AMR) and can search for plasmids. Isolates’ whole genomes may be sequenced by the United States FDA or public health laboratories, typically at no charge. In Module 2, students learn basic methods of genome assembly, analysis, annotation, and comparative genomics. They use easily accessible, primarily web-based tools to assemble their genomes and investigate areas of interest including serotype, AMR genes, and in silico evidence of mobile genetic elements. Either module can be used as a standalone learning experience. After course completion, students will be able to isolate and identify Salmonella from natural sources, and use computational analysis of microbial genomic data, particularly of the Enterobacteriaceae. This lesson offers undergraduate microbiologists a genuine research experience and a real-world microbiology application in genomic epidemiology, as well as a valuable mix of field, laboratory, and computational skills and experiences.
Citation
Jurgensen SK, Harsh J, Herrick JB. 2021. A CURE for Salmonella: A Laboratory Course in Pathogen Microbiology and Genomics. CourseSource. https://doi.org/10.24918/cs.2021.24
Society Learning Goals
Biochemistry and Molecular Biology
- Evolution
- What is the molecular basis of evolution?
Bioinformatics
- DNA - Information Storage [GENOMICS]
- Where are data about the genome found (e.g., nucleotide sequence, epigenomics) and how are they stored and accessed?
- How can bioinformatics tools be employed to analyze genetic information?
- Ecology and Evolution [METAGENOMICS]
- How can bioinformatics tools be employed to examine ecological niches?
- Computational Skills
- What higher-level computational skills can be used in bioinformatics research?
Ecology
- Impacts of Ecosystems on Human Health and Well-being
- How do humans depend on ecosystems for their health and well-being?
Microbiology
- Evolution
- How do human impacts on the environment influence the evolution of microorganisms (e.g., emerging diseases and the selection of antibiotic resistance)?
- Considering the immense variety of microenvironments, how have mutations and horizontal gene transfer selected for a huge diversity of microorganisms?
- Information Flow and Genetics
- How do genetic variations impact microbial functions (e.g., in biofilm formation, pathogenicity, and drug resistance)?
Lesson Learning Goals
Students will:
- Describe how whole genome sequences (WGS) of pathogens such as Salmonella can be used in genomic epidemiology to track and source outbreak strains.
- Understand how human impact on the environment can influence the evolution of microorganisms (e.g., emerging diseases and the selection of antibiotic resistance).
- Describe the processes of isolation, identification, and characterization of human pathogenic bacteria found in environments such as streams and manure.
- Gain experience collecting microorganisms in the environment.
- Learn how to properly store genomic data and other large scientific datasets in cloud systems designed for this purpose.
- Learn how to assess the quality of whole genome sequencing runs, to assemble microbial genomes, and to use WGS to serotype isolates and determine their phylogenetic relationships to other strains.
- Learn how to use various bioinformatics methods to annotate genes and to study antibiotic resistance, virulence, and mobile genetic elements in pathogens.
- Work in groups on a semester-long project and present their results.
- Gain practice in navigating scientific obstacles
- Learn about potential careers in public health microbiology and bioinformatics.
Lesson Learning Objectives
Module 1 Students will:
- Work in groups on a semester-long project and communicate research findings in oral and poster presentations. Maintain a physical lab notebook.
- Learn guidelines for safely handling Salmonella and other potential pathogens in a laboratory environment.
- Learn how to make Salmonella pre-enrichment and enrichment media.
- Keep a laboratory notebook.
- Collect and record sample metadata using a sampling probe and metadata management application.
- Collect sediment from streams aseptically.
- Use liquid and plate media to enrich and isolate Salmonella from stream sediments and other sources..
- Use biochemical tests such as the Gram stain, KOH, oxidase, and catalase tests to identify putative Salmonella.
- Verify isolates as Salmonella enterica using Salmonella-specific PCR and gel electrophoresis or real-time PCR.
- Verify the isolates as S. enterica using Enteropluri(TM) tubes, designed for the identification of Enterobacteriace (optional).
- Use Kirby-Bauer and/or Sensititre MIC plates to test isolates for their resistance to antibiotics used to treat systemic Salmonella infections.
- Freeze isolates in cryotubes for long-term cryostorage
- Ship isolates to the U.S. FDA or other public health laboratories for Illumina whole-genome sequencing.
Module 2 Students will:
- Work in groups on a semester-long project and communicate research findings in oral and poster presentations. Maintain a group electronic bioinformatics lab notebook.
- Learn how to use GalaxyTrakr and Galaxy for the computational analysis of genomes.
- Learn the use of Open Science Framework for data storage and retrieval.
- Understand the advantages and disadvantages of short- and long-read genome sequencing.
- Learn how to name data and analysis files so they are machine- and human-readable and sortable.
- Learn how and why DNA sequence and assembly data quality is assessed. Use FastQC and Trimmomatic to analyze and improve the quality of raw reads and assemblies, respectively.
- Learn how and why microbial genomes are assembled and the limitations of short-read sequences for assembly.
- Assemble isolate genomes using SPAdes and visualize assemblies using Bandage.
- Serotype isolates using SeqSero and SISTR on GalaxyTrakr.
- Learn how to find and download reference genomes from NCBI.
- Order their assembled contigs and visualize the order using Mauve.
- Understand the purpose and process of genome annotation.
- Annotate their isolates’ genomes using Prokka and RAST and visualize annotated genomes using a genome browser.
- Determine and compare the antibiotic resistance genotypes and phenotypes of isolates.
- Learn how to find and compare mobile genetic elements – plasmid-specific genes, transposons, integrons, pathogenicity islands, prophages, etc. – in isolates.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
Introduction
Course-based undergraduate research experiences (CUREs) are becoming an increasingly valuable feature of college science teaching (1, 2) . CUREs have the potential to engage all students in authentic research practices by offsetting barriers commonly associated with traditional apprenticeship models (3). CUREs engage students in collaborative, iterative research activities as the students work to collect, analyze, and communicate novel data of broader interest to the community (4, CURENet). Though the nature of these experiences can vary widely as seen in the growing catalogue of published descriptions in bioscience education at the introductory (5–8) and upper division levels (9–11), studies on CURE participation have documented a range of cognitive, affective, behavioral, and psychosocial gains (as reviewed in (12)).
Microbial genomics is a particularly fruitful field in which to focus a CURE, especially considering CURE design features highlighted in the literature (4) as well as how the experience may help science students achieve their educational or career goals (12). Genomics, transcriptomics, and related fields are now of central importance in microbiology, and in the life sciences more broadly. The concept knowledge and technical skills developed in practice with common technologies and tools learned in studying microbial genomes are largely applicable to other organisms. In addition, students gain exposure to and training in data-driven discovery by working with large data sets, which has been deemed key to the preparation of a data-capable workforce.
Common foodborne pathogens are of great interest to the US FDA, CDC, and other public health laboratories and therefore these agencies are often eager to sequence foodborne pathogens at little or no cost to researchers and educators because the data can provide needed context for tracking future outbreaks (13). This urgency combined with the advances in whole genome sequencing (WGS) make this system ideal for the development of a CURE. We have developed a lesson that is part of an ongoing research project in our laboratory. We use Salmonella enterica as our model organism in this lesson because it is a foodborne pathogen that infects over 1.3 million Americans every year, causing approximately 420 deaths, and is one of the leading infectious causes of hospitalization in the United States (14, Centers for Disease Control). However, Salmonella are also relatively less virulent than other foodborne pathogens of interest to these labs, such as Listeria and pathogenic E. coli (15), making it more appropriate in an undergraduate lab setting. Additionally, introducing students to ongoing large-scale governmental projects, such as the epidemiological tracking of foodborne illnesses like Salmonella, gives students valuable insight into potential career paths that they may otherwise be unaware of. Thus, this lesson may be of particular interest to students interested in pursuing careers in public health or infectious disease.
While its pathogenicity has made Salmonella a commonly studied organism, its occurrence in natural environments has not been extensively investigated (16, 17). Pathogens have traditionally been studied in the context of human infection and food, with less regard to their potential environmental reservoirs. These potential reservoirs include reptiles, fresh waters sources and manure (18). We typically sample sediment from agriculturally impacted streams because it potentially harbors a more stable microbial community than water (15). For this CURE, we also worked with several local small-scale and industrial poultry farmers who provided poultry litter from their farms, as Salmonella is typically a commensal organism in turkeys and chickens.
Recently-developed high throughput (or “next generation”) sequencing methods have made it possible to sequence entire bacterial genomes quickly and affordably (19). As a result, a vast amount of microbial DNA sequence data is being generated. In particular, an abundance of DNA sequence data is being generated by the U.S. FDA, CDC, and state public health laboratories as part of their recent thrust to use whole genome sequences for epidemiological tracking of pathogens. The CDC has successfully used WGS to identify the source of dozens of foodborne illness outbreaks since its initial implementation in 2013, as well as to uncover new or unknown sources of infection. For example, recently the CDC traced an outbreak of Salmonella Javiana to cut fruit produced in New Jersey by using WGS to show that the ill people were infected with genetically similar Salmonella, which suggested a single infection source. These high throughput methods allow for higher resolution in typing, distinguishing, and characterizing outbreak strains at the subspecies level because small genetic differences (such as single-nucleotide polymorphisms or SNPs) can be identified. The datasets also constitute a valuable and under-utilized resource for genomic and bioinformatics lessons such as this.
Intended Audience
This two-part CURE can be directed to different student populations and levels. We use the experience in its entirety in an upper-division course at a large master’s degree-granting university with biology majors concentrating in microbiology. The course (previously Bacterial Discovery, now known as Laboratory in Bacterial Pathogenomics) has been offered nearly every term since Spring 2018, and is taught by a faculty member assisted by an upper division undergraduate research student with previous experience in the course. Both modules provide students with experiences and skills similar to those gained from working in a research laboratory, so this course is particularly useful for students who are unable to work in a one-on-one mentored research setting. As the first module – on Salmonella isolation and identification – requires a biological safety level (BSL) 2 laboratory space, not every institution may be able to incorporate it into their course. However, the bioinformatics module of the lesson can be adapted and implemented in biology and biotechnology courses for introductory through graduate level students. In order for the material to be accessible to novices in bioinformatics, this module exclusively uses freely available online tools that do not require computer programming experience or the use of the command line.
Required Learning Time
This laboratory lesson is divided into two modules, each taking roughly 8 weeks of a 16-week semester. We taught the course in twice weekly 90-minute lab periods as a standalone laboratory course (with no required lectures). Our microbiology laboratory spaces can accommodate up to 24 students, and we currently offer one section of this course per semester. The Teaching Timeline (Table 1) includes the approximate time required for each laboratory activity, set up, and out of class time for preparation (as necessary).
Table 1A. Recommended course activity timeline for A CURE for Salmonella Module 1: Wet Lab. “Labs” in both modules, refers to distinct activities to be completed, not to laboratory days. Multiple labs could be done on one day (and the reverse may occur, as well: one lab could be extended over multiple days).
|
Activity |
Description |
Estimated Time |
Notes |
|
Lab 1 |
|||
|
Course overview, lab safety, class safety and pre-course survey |
|
90 min |
|
|
Lab 2 |
|||
|
Preparation for field sampling |
|
90 min |
|
|
Lab 3 |
|||
|
Sample collection and processing |
|
90 min |
|
|
Lab 4 |
|||
|
Inoculation of enrichment media |
Inoculate enrichments |
15-30 min |
|
|
Lab 5 |
|||
|
Plating from enrichments |
Streak plate onto CHROMagar Plus and XLT-4 plates |
15 min |
|
|
Lab 6-8 |
|||
|
Isolation and characterization of isolates using classical microbiological techniques |
|
Varies, typically 3 or more 60-90 minute lab periods depending on the number of isolates |
|
|
Lab 9 |
|||
|
Enteropluri tube identification (optional) |
Inoculate and read Enteropluri tubes |
5-15 min (inoculation), 45-60 min (reading) |
|
|
Lab 10 |
|||
|
PCR |
PCR using invA primers |
60-90 min |
|
|
Lab 11 |
|||
|
PCR agarose gel electrophoresis |
|
60-90 min |
|
|
Lab 12 |
|||
|
Prep strains for shipping, freezing |
|
15-20 min |
S10. CURE for Salmonella – Preparing isolates for shipping and freezing |
|
Lab 13 |
|||
|
Kirby-Bauer testing |
Typical Kirby-Bauer protocol on Mueller-Hinton plates |
20-30 min (should be read in 16-24 hrs) |
|
Table 1B. Recommended course activity timeline for A CURE for Salmonella Module 2: Genomics. “Labs” in both module 1 and 2, refers to distinct activities to be completed, not to laboratory days. Multiple labs could be done on one day (and the reverse may occur, as well: one lab could be extended over multiple days).
|
Activity |
Description |
Estimated Time |
Notes |
|
Lab 1 |
|||
|
Introduction to whole-genome sequencing & assembly, using GalaxyTrakr |
|
45 min |
|
|
Lab 2 |
|||
|
In silico serotyping of Salmonella |
|
10 min |
|
|
Lab 3 |
|||
|
Assessing assembly quality, filtering and trimming reads using FastQC and Trimmomatic on GalaxyTrakr |
|
30-60 min |
|
|
Lab 4 |
|||
|
Naming files, genome assembly using SPAdes, assessing assembly quality using QUAST and Bandage. |
|
45-60 min (not including run time) |
|
|
Lab 5 |
|||
|
Ordering contigs to a reference, read mapping, using Mauve |
|
60 min |
|
|
Lab 6 |
|||
|
Annotation 1: Whole genome using Prokka and RAST; visualization using Artemis |
|
30-45 min |
|
|
Lab 7 |
|||
|
Annotation 2: Antibiotic resistance genes |
|
30-45 min |
|
|
Lab 8 |
|||
|
Annotation 3: Miscellaneous genes (genomic islands, virulence genes, mobile elements, etc.) |
Background info (e.g., a mini-lecture) on virulence factors, Salmonella pathogenicity and other islands, and mobile elements (plasmids, phages, transposons, integrons) as needed |
30 min to ? |
|
|
Lab 9 |
|||
|
Poster presentation, oral presentation, scientific communication, group work |
|
Multiple days |
|
Prerequisite Student Knowledge
Module 1 - Salmonella Isolation, Isolation, and Identification
For this first module, students should have taken a general or introductory microbiology course with a laboratory to develop sufficient skills in culture maintenance, basic diagnostic biochemical tests, and common isolation methods. However, if module 2 is not being used, there would be sufficient time to teach these concepts before Salmonella isolation. Thus, this module can be implemented alone in an introductory microbiology laboratory course with the laboratory skills discussed above taught before beginning the module. BSL-2 safety training for all students is required for the implementation of this module since the target organism (Salmonella) is a human pathogen, and there is also the possibility that other pathogens (e.g., Klebsiella pneumoniae or pathogenic E. coli) could be isolated.
Module 2 Bioinformatics
For the second module, students should have a basic knowledge of the characteristics of the bacterial genome and foundational genetic concepts such as the Central Dogma and horizontal gene transfer. No prerequisite knowledge of bioinformatics is required, though many of our students have a basic introduction to bioinformatics in JMU’s CURE-model first-year biology curriculum (5). All tools used in this lesson are freely available online, so only basic computer expertise is required to complete this module.
Prerequisite Teacher Knowledge
Module 1
The instructor should have significant training/experience in microbiology as well as some familiarity with field sampling and working with environmental samples in the laboratory before implementing this lesson. We recommend that the lesson be piloted before full implementation so the instructor can become familiar with sampling sources that reliably yield Salmonella and be able to recognize Salmonella on the selective media. Familiarity with BSL-2 safety protocols is a necessity.
Module 2
This lesson assumes that the instructor has some basic knowledge of microbial genomics. For those who are not familiar with bioinformatics and are interested in incorporating this module into their course, we recommend the excellent paper by Edwards and Holt (20) with its accompanying tutorial. GalaxyTrakr and especially Galaxy provide a variety of tutorials to introduce users to the interface (21,24).
Scientific Teaching Themes
This lesson was designed and implemented as a semester-long CURE (4, 22) comprising two modules (Supporting File S25. CURE for Salmonella – Course Overview). It is a hands-on introduction to laboratory and bioinformatics techniques that encourages students to work in teams to produce and analyze genomic and other data with real-world applications. The main goal of this lesson is to produce students who are knowledgeable and confident in their abilities to work at the bench and with a computer on a project that they initiate, carry out, and conclude within the timescale of the course.
Active Learning
This laboratory lesson uses multiple approaches to engage students in active learning. Most activities are carried out in teams, and students work in small groups to plan and implement their approach to each lab period. We occasionally assign review and other summary readings as pre-class homework, followed by instructor-led group discussions. These whole class discussions also aid in troubleshooting, which are necessary due to the inherent unpredictability of research, as well as student errors. Because students follow the research process from sample collection through isolation of target organism to genomic data analysis, there is significant project ownership inherent in the lesson. In Module 1, students must make real-time decisions about the outcomes of each procedure and determine their next steps as a group. In Module 2, students follow developed tutorials at their own pace and decide as a group which advanced analyses to pursue based on their interests. Both poster and oral presentations are designed, presented, and evaluated (by the instructor) as a group.
Assessment
Using a backward design approach (23), formative and summative assessments – as well as learning activities – were mapped to course learning objectives (Table 1).
Formative assessments include observations of research-related skills (e.g., molecular techniques, communication), in-class pre-lesson quizzes on the protocols to be completed to ensure that protocols were understood before implementation, short homework assignments to help students gain experience using bioinformatics programs and pipelines, in-class activities, discussions, and maintenance of physical (for Module 1) and electronic (for Module 2) laboratory notebooks.
Summative assessments include homework assignments, group oral and poster presentations evaluated using rubrics focused on understanding of the material (S22. CURE for Salmonella – Presentation evaluation rubric and S23. CURE for Salmonella – Poster evaluation rubric), and rubric-scored physical and electronic lab notebooks (S4. CURE for Salmonella – Lab notebook grading rubric, S12. CURE for Salmonella – Bioinformatics lab notebook grading rubric). Detailed information on these assessments is included in the outlines of each weekly lesson below.
Inclusive Teaching
By their general nature, CUREs increase access to authentic research experiences for all students independent of the personal and institutional hurdles that they may face in engaging in an independent research program (3). Students work collaboratively throughout the semester as they use authentic microbiology and genomics techniques that closely align with epidemiological investigations conducted by the FDA, CDC, and other public health laboratories. Student research has the potential for discovery and to make contributions to the field (33 and E. Gline, E. Gross, B. Puma, R. Zoldork, M. Thinnes, E. Seracino, and J. B. Herrick. Presented at the Annual Meeting of the American Society for Microbiology, Virginia Branch, 8 to 9 November 2019). Through active engagement in authentic practices via wet lab and/or computer-based activities, students gain an understanding of the nature of “real world” scientific endeavors in the field, and also experience autonomy, project ownership, and how to navigate challenges in a research setting.
Other design features of the CURE lend well to inclusive teaching practices. In studying the socially relevant topics of foodborne pathogens and antibiotic resistance in their own community, this lesson would be broadly meaningful to students of varying backgrounds and professional interests (e.g., medicine, epidemiology, bioinformatics, infectious disease microbiology). Pedagogical approaches implemented in this lesson accommodate a variety of learning styles and ability through demonstrations, mini-lectures, videos, hands-on laboratory and computer-based activities, worksheets, discussions, and optional additional tutorials. Similarly, varying forms of formative and summative assessments are used to measure student learning trajectories and guide instruction.
Lesson Plan
Module 1
One of the most time-intensive aspects of preparing to incorporate this module into a course may be identifying likely sources for Salmonella. We use agriculturally-impacted local streams because we previously found that they were a source for Salmonella in our area (33). We also use poultry litter because Salmonella is typically a commensal organism in the fowl gut. Other environmental sources could include amphibians, reptiles, food, or other manures. Relative proximity to your institution should of course also be a factor when choosing sampling sites.
We use a YSI Professional Plus multiparameter instrument (SKU 6050000) to collect water temperature, pressure, salinity, and conductivity data at stream sample sites (see S3. CURE for Salmonella – Field sampling protocol) and we store this and other metadata (date, time, latitude and longitude) using EpiCollect5 which is available as both a mobile application and website. When culturing bacteria (labs 3-5, S5. CURE for Salmonella – Pre-enrichment and enrichment inoculation protocol and S7. CURE for Salmonella – Plating and purification protocol), all procedures must be conducted in a BSL-2 laboratory space. Safety documentation for students can be found in the supporting file (S1. CURE for Salmonella – Laboratory safety contract).
In this module, students prepare their own enrichment and isolation media (S2. CURE for Salmonella – Pre-enrichment and enrichment media preparation, S6. CURE for Salmonella – Plate media preparation, and S26. CURE for Salmonella – Media, Isolation, Sampling), although the media can of course simply be provided. We provided trypticase soy agar (TSA) and broth (TSB) for students to maintain cultures and to grow liquid cultures to prepare for labs 6-8 (S8. CURE for Salmonella – Miscellaneous test protocols - oxidase, catalase, KOH). If availability of time or funds are limited, lab 9 may be removed from the module. After lab 10 (S9. CURE for Salmonella – invA PCR and gel visualization protocol), all confirmed Salmonella isolates were shipped to the Virginia Department of Consolidated Laboratory Services (DCLS) for WGS (S10. CURE for Salmonella – Preparing isolates for shipping and freezing). An overview of the Salmonella isolation protocol is shown in Figure 1.
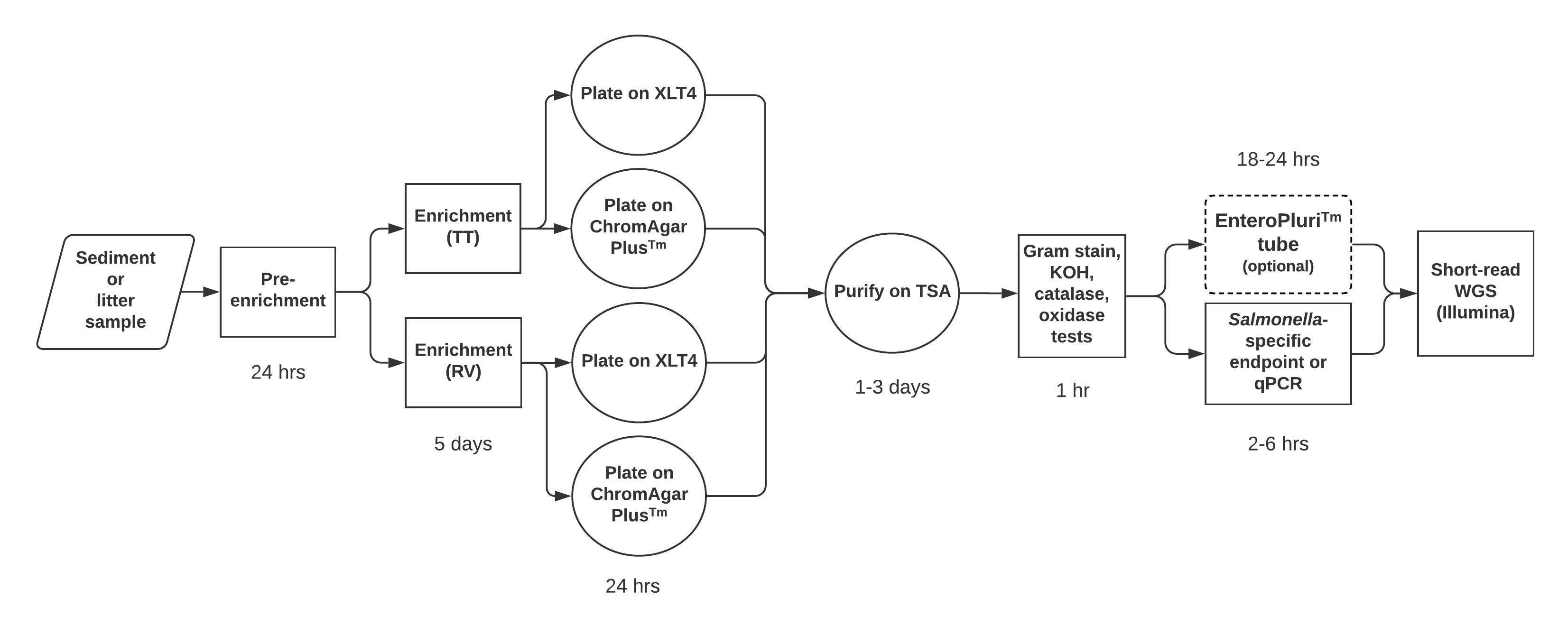
Figure 1. Overview of methodological workflow for the enrichment, isolation & purification, and identification of Salmonella used in Module 1. The estimated time expected for each step is indicated.
Module 2
If instructors wish to use module 2 as a follow up to module 1, instructors should contact their local state or other public health laboratory or the FDA Whole Genome Sequencing (WGS) Program to determine whether they will accept samples and expedite sequencing to ensure a turn-around time short enough for a semester-long lab. Some public health laboratories are particularly interested in receiving such samples because they must meet established quotas for WGS of common pathogens as a means of contributing to governmental databases. If the delay proves to be too long for a single semester course, genomes of strains isolated in a previous semester could be used for analysis.
Alternatively, module 2 can be taught as a standalone set of lessons using as raw material, the vast number of recently-sequenced Salmonella genomes available from the NCBI Sequence Read Archive (SRA). These can be downloaded within GalaxyTrakr using the accession (SRR) number as explained in S11. CURE for Salmonella – Bioinformatics Lab Guide - Navigating and using GalaxyTrakr and Galaxy. Accession numbers can be selected from the NCBI site in a number of ways, for example via the Taxonomy Browser; choose strain and then click on “SRA Experiments”) or via GenomeTrakr BioProjects.
This module requires preparation to ensure that student-generated files are well organized and analyses are easy to find and use. We recommend the use of Open Science Framework (OSF), an open source data management platform, to access and store genomic data and analysis files. We also use Google Docs student electronic lab notebooks (template; S12. CURE for Salmonella – Bioinformatics lab notebook grading rubric). We have created a public OSF page to function as a living repository of protocols, templates, and instructions to be used for both modules of this course. If your institution does not already use OSF, you can work with the Center for Open Science to create a dedicated institutional OSF landing page so that students can use their university sign-in credentials to connect to the OSF, although this isn’t a requirement for its use.
For the majority of analyses in this lesson we use GalaxyTrakr (S11. CURE for Salmonella – Bioinformatics Lab Guide - Navigating and using GalaxyTrakr and Galaxy, S27. CURE for Salmonella – Intro to GalaxyTrakr mini-lecture), which is a specific implementation of Galaxy, an open, web-based platform for computational tools used to analyze genomic data. GalaxyTrakr (24) includes a relatively limited number of tools specific to microbial (and especially foodborne pathogen) genomics, including most of those used in this module, which may make it easier for students to use. GalaxyTrakr tools used in this module include SeqSero, SISTR, FastQC, Trimmomatic, SPAdes, Shovill, QUAST, PROKKA, and Abricate (Supporting Files S13, S14, S16, S17, S19, S20, and S29). Other web-based multi-tool platforms we use include PATRIC for gene annotation using RAST (S19. CURE for Salmonella – Bioinformatics Lab Guide - Gene annotation) and the website of the Center for Genomic Epidemiology for a number of tools (S21. CURE for Salmonella – Bioinformatics Lab Guide - Miscellaneous gene and genetic feature detection). Although not described in this lesson, Enterobase may also be a useful resource, particularly for multilocus sequence typing of strains. Individual web-based programs for gene feature identification include INTEGRAL and PHASTER (S21. CURE for Salmonella – Bioinformatics Lab Guide - Miscellaneous gene and genetic feature detection). Standalone programs that require installation on the computers used for computational analysis are Mauve (Supporting File S18. CURE for Salmonella – Bioinformatics Lab Guide - Selecting reference genomes/Ordering and viewing assembled contigs using Mauve), Bandage (S17. CURE for Salmonella – Bioinformatics Lab Guide - Assessing assembly quality Using QUAST and Bandage), and Artemis (S19. CURE for Salmonella – Bioinformatics Lab Guide - Gene annotation). A general overview of the computational tools used in this lesson can be seen in Figure 2.
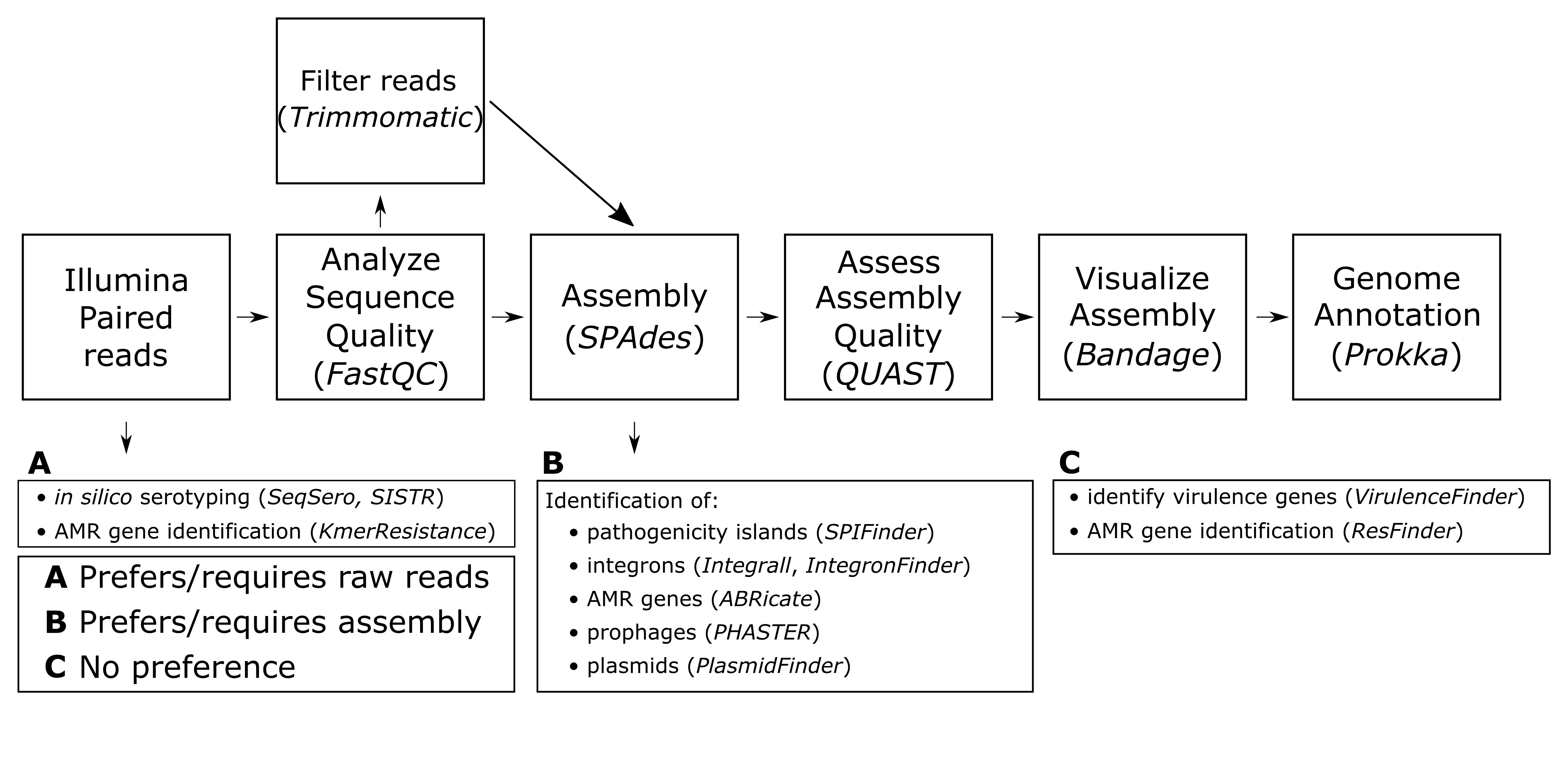
Figure 2. Overview of computational workflow used in Module 2, as well as potential tools for further identification of genes and genome regions. A, tools that require or function best with raw reads (fastq format) as input; B, tools that require or function best with an assembly file (fasta format) as input; C, tools that can take either raw reads or assemblies equally well as input. AMR = antimicrobial resistance.
Teaching Discussion
Challenges in Implementation
A crucial step in developing Module 1 was choosing appropriate sample sources and sites. We used sediment from agriculturally-impacted streams in the Shenandoah Valley that have been regularly sampled in one of the authors’ (JBH) research lab at James Madison University. These sites were therefore relatively well characterized with respect to their potential as reservoirs of Salmonella. We also obtained poultry litter samples from local large-scale industrial and small-scale poultry farms and were successful in isolating Salmonella from some but not all of these. Since these sample sources are not available to all institutions, other sample sources (e.g., amphibians, reptiles, eggs, and raw poultry meats) could potentially be utilized.
Our isolation protocol is based on the FDA Bacteriological analytical manual (BAM) protocol for Salmonella enrichment and isolation (25). We have modified the protocol, however, to include a 5-day rather than a 24-hour enrichment period, which we have found to substantially improve recovery from sediments. We have also replaced Bismuth Sulfide agar with ChromAgar Plus plate medium, as we have found that less-experienced students can more easily distinguish putative Salmonella on this medium. We continue to use XLT-4 agar medium however, its use is optional. Included in Table 1 (Labs 6-8) are links to manufacturer’s guides to the expected appearance of Salmonella on CHROMagarPlus and XLT4 plate media. Other selective and differential media such as Brilliant Green Agar, Hektoen Enteric Agar, or Xylose Lysine Deoxycholate Agar have been successfully used by other labs for isolating Salmonella strains from various sources (26).
This CURE can be readily adapted to meet the needs of the student population and institutional resources at hand. In particular, Module 2 could be modified and implemented as a standalone online research experience. Students could rely on sequences from instructor-isolated strains or existing genomes readily available through the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) database. Here, freely accessible web-based bioinformatics tools (described in Table 1) could permit non-traditional or remote students as well as those at institutions without BSL-2 labs to engage in whole-genome research. Furthermore, while the lesson focuses entirely on Salmonella, nearly all aspects of Module 2 could be applied to other organisms including more virulent pathogens that instructors or students may be interested in studying but do not have the appropriate facilities to work with at the bench. Large numbers of genomes are being sequenced from, for example, Escherichia/Shigella, Clostridiodes, Vibrio, Yersinia, Helicobacter, and Moraxella, with sequence data publicly available.
In Module 2, challenges are mainly related to a lack of student exposure to bioinformatics before taking the course. We used tools with user-friendly web-based and other graphic interfaces to reduce student intimidation. Data analyses were disseminated and stored on the Open Science Framework (OSF). We created a Project for the overall course, made a Component Project for the semester, and had students “fork” this so each group had their own page to edit. OSF allows for easy “templating” of projects and instructors are welcome to use our site and materials freely as templates.
Another challenge of Module 2 may be the acquisition of sequence data and the turnaround time of isolate sequencing. We sent isolates to the Virginia DCLS for whole-genome sequencing about halfway through the semester, after Module 1 Lab 12, so that sequence data would be available for Module 2. The DCLS turnaround is typically only two or three weeks. If this rapid turnaround isn’t possible, the students could analyze sequence data from isolates from a previous semester. While we are waiting for sequence data, we typically do antibiotic resistance phenotyping (Module 1, Lab 13). Other advanced microbiological tests and protocols for verifying and characterizing Salmonella (e.g., SIM, TSI, Voges-Proskauer tests; native plasmid preps; MICs, etc.) might also be performed. We also introduce the first few labs of Module 2 using raw read (SRR) files available from the NCBI Short Read Archive (SRA). If only Module 2 is to be utilized, there are thousands of freely available short-read sequences of Salmonella and other bacterial pathogens available in the NCBI short read archive. The NCBI Pathogen Browser might be of interest to students as it displays phylogenetic relationships, sources and other metadata of not only Salmonella but a number of bacterial pathogens (and even Candida).
Lesson Effectiveness
To complement course activities, student self-report data were gathered using pre- and post-surveys to assess the effectiveness and impact of the experience (S24. CURE for Salmonella – Sources and description of preexisting survey instruments). Here, Likert-type items were adopted from existing validated instruments (27–30) as well as open response prompts designed for this study to gather data on self-perceived cognitive and non-cognitive outcomes and perceptions of course design and instruction. Data were collected and aggregately analyzed from two semesters (Fall 2018 and Spring 2019) after a pilot semester of implementation (Spring 2018). All procedures were performed in accordance with the university’s institutional review board guidelines.
Students (n=35; all biology majors and upperclassmen, 75% female, 51% White, 31% first-generation) reported notable gains in various research-related cognitive and non-cognitive outcomes. Overall, students indicated that the course increased their confidence in being able to complete tasks individually and/or teaching others how to complete tasks using microbiology (100% of students), molecular biology (100%), and bioinformatics (94.29%) research techniques. More specifically, as recorded by 5-point Likert-type items, > 80% of students self-rated higher levels of confidence after the course in research-related outcomes including designing a research study, employing basic and advanced technical skills, working collaboratively, troubleshooting problems, using scientific literature to guide research, understanding relevant concepts knowledge, and communication with research mentors and faculty. Development of these skills were also commonly cited by students in response to an open-ended question of how the CURE may contribute to their academic and career goals – as represented in the following quote: “[this class] significantly helped me learn…how to analyze DNA sequences, and learn how to use specific databases to analyze and interpret particular data to discover what the exact genes represent” (Student A). These self-perceived gains were corroborated through direct measures such as observed in situ performance of technical practices, conceptual knowledge, and the communication of findings in oral and poster presentation format as well as in-class quizzes, homework assignments, and rubric-critiqued lab notebooks.
Multiple positive indicators drawn from student survey responses collected over the two semesters also support the instructional effectiveness of the CURE design. First, 91% of students reported that the course met or exceeded their initial expectations as represented by the following quotes: “I didn't expect to learn so many new techniques that I can now add to my CV and I am happy that I was able to participate in research as a class since some semesters, I didn't have time to dedicate to a research lab” (Student B) and “My initial expectations were this class was going to another biology class that would just teach us the methods, and approach and possibly an analysis of data; however, it gave me so much more than just the standard knowledge. It gave me actually a way to feel important that my work was done and was significant, that I am a real scientist” (Student C). For those few students (n=3) that reported that their experience was less than what was initially expected, each identified anticipating more wet lab work (rather than computational). Next, all students agreed with the statement that the course was a good way to learn the process of science, reflecting the incorporation of CURE design features recommended in the literature (4). Here, responding to a series of vetted Likert-scale items (31,29), students rated high levels of project ownership of content (4.42 [SD=0.87], on a 5-point scale) as well as collaboration (22.57 [SD=3.40], on a 6-24 point scale), discovery (26.34 [SD=5.32], on a 5-30 point scale), and iteration (28.94 [SD=4.45], on a 6-36 point scale). These results were echoed and expounded on by students in response to open-ended prompts about course characteristics that they learned or that they enjoyed, such as “I also surprisingly think that failing at getting Salmonella in our first round and having to start over completely from the beginning was helpful, because it really did show me that research is sometimes starting over and figuring it out all over again” (Student D), “In comparison to my other lab classes, we had free reign in the direction of our research that really made me excited” (Student E), and “We worked together to do everything ourselves! From the field work to the genome analysis. My group was awesome!” (Student F). In addition, students regularly highlighted the value of the CURE in exposing them to authentic research practices: “The fact that we went through the entire scientific process, from collecting data to analyzing results. There are not many classes that would allow you to do that” (Student G). Importantly, several students that had been engaged in independent research found the CURE comparable to that experience: “I enjoyed the independence of this class. As a member of a different research lab, I found this class to possess a similar environment to a research lab. This class is a valuable experience to those who have not been exposed to research (Student H).” Finally, students reported discussing their research (i.e., networking) with family (78%), friends (95%), other on-campus students (97%) and faculty (62%) not affiliated with the CURE, and other off-campus students (70%).
Alternative implementations
As stated previously, for Module 1 if environmental Salmonella sources are unavailable, sources such as the feces of captive amphibians and reptiles (especially turtles and snakes), backyard poultry, or raw eggs or poultry meat from the grocery may be utilized. Also, other members of the Enterobacteriaceae may be easier than Salmonella to isolate in some areas. E. coli in particular is often found in fecal-contaminated environmental waters and sediments. Protocols for the isolation and identification of E. coli are readily available (32). However, state and federal labs may be unwilling to provide short-read sequencing of these for free, since their main interest is in the pathogenic E. coli strains.
If only Module 2 is to be utilized, there are thousands of freely available short-read sequences of Salmonella and other bacterial pathogens available in the NCBI short read archive. The NCBI Pathogen Browser might also be of interest to students as it displays phylogenetic relationships, sources and other metadata of a number of bacterial and fungal pathogens.
Conclusion
This lesson offers students an authentic research experience, valuable skills in pathogen microbiology and genomics, and results that are potentially of use to the wider community of microbiologists, particularly those studying the distribution and genomic epidemiology of foodborne pathogens. Students ask important questions such as ‘Are there Salmonella in this stream or fecal sample?’, ‘Based on their serotypes, what is the potential for human infection from these isolates?’, ‘Are these Salmonella related to strains isolated from similar environments or regions?’, ‘Do they exhibit antibiotic resistance and, if so, what is the genetic basis of the resistance?’, and so forth. They learn methods and approaches in field sampling, enrichment and culturing, identification and characterization, and the safe handling of pathogens. Students learn how to assemble, annotate, and analyze microbial genomes. Additionally, students learn to work in teams to solve problems and to present their work orally and in poster form. The results students gather -- particularly the genome sequences of their isolates -- are of genuine value to genomic epidemiologists at the CDC, the FDA, and other public health labs who seek to track outbreaks in real time (for example, we were directly contacted by the CDC about one of our isolates that was potentially linked to a Salmonella outbreak in poultry). Some of the results of this study have been published (33).
Students are also prepared for careers in public health and infectious disease. For example, a number of our students have already gone on to jobs or to graduate programs in public health, epidemiology, and infectious disease. This lesson provides students a powerful mix of field, wet lab, genomic and bioinformatics experiences and skills not typically seen in an undergraduate microbiology curriculum. In one student’s words, this lesson “gave me so much more than just the standard knowledge. It gave me … a way to feel important, that my work was done and was significant, that I am a real scientist.”
SUPPORTING MATERIALS
Module 1
S1. CURE for Salmonella – Laboratory safety contract
S2. CURE for Salmonella – Pre-enrichment and enrichment media preparation
S3. CURE for Salmonella – Field sampling protocol
S4. CURE for Salmonella – Lab notebook grading rubric
S5. CURE for Salmonella – Pre-enrichment and enrichment inoculation protocol
S6. CURE for Salmonella – Plate media preparation
S7. CURE for Salmonella – Plating and purification protocol
S8. CURE for Salmonella – Miscellaneous test protocols - oxidase, catalase, KOH
S9. CURE for Salmonella – invA PCR and gel visualization protocol
S10. CURE for Salmonella – Preparing isolates for shipping and freezing
Module 2
S11. CURE for Salmonella – Bioinformatics Lab Guide - Navigating and using GalaxyTrakr and Galaxy
S12. CURE for Salmonella – Bioinformatics lab notebook grading rubric
S13. CURE for Salmonella – Bioinformatics Lab Guide - Salmonella serotyping
S14. CURE for Salmonella – Bioinformatics Lab Guide - Assessing & filtering illumina data using FastQC and Trimmomatic
S15. CURE for Salmonella – Bioinformatics Lab Guide - File naming conventions
S16. CURE for Salmonella – Bioinformatics Lab Guide - Genome assembly using SPAdes and Shovill
S17. CURE for Salmonella – Bioinformatics Lab Guide - Assessing assembly quality Using QUAST and Bandage
S18. CURE for Salmonella – Bioinformatics Lab Guide - Selecting reference genomes/Ordering and viewing assembled contigs using Mauve
S19. CURE for Salmonella – Bioinformatics Lab Guide - Gene annotation
S20. CURE for Salmonella – Bioinformatics Lab Guide - Antibiotic resistance gene detection
S21. CURE for Salmonella – Bioinformatics Lab Guide - Miscellaneous gene and genetic feature detection
S22. CURE for Salmonella – Presentation evaluation rubric
S23. CURE for Salmonella – Poster evaluation rubric
Miscellaneous
S24. CURE for Salmonella – Sources and description of preexisting survey instruments
S25. CURE for Salmonella – Course Overview
S26. CURE for Salmonella – Media, Isolation, Sampling
S27. CURE for Salmonella – Intro to GalaxyTrakr mini-lecture
S28. CURE for Salmonella – Overview of Next-Gen Sequencing and Assembly
S29. CURE for Salmonella – FastQC & Trimmomatic
Acknowledgments
This development and implementation of this course would not have been possible without the assistance of Dr. Rebecca Bell and Dr. Marc Allard of the U.S. FDA and especially Dr. Lauren Turner and her team at the Virginia Department of Consolidated Laboratory Services. We also acknowledge former members of the Herrick laboratory, especially Charles Holmes II and Curtis Kapsak, for their assistance in the development and optimization of many of the methods outlined here. The authors wish to acknowledge the financial support of the Madison Trust and the JMU Department of Biology in the development and implementation of this course. Lastly, we express our appreciation for the students of Bio 346 Bacterial Discovery who helped with not only the scientific aspects but also the assessment of the course.
References
- President’s Council of Advisors on Science and Technology (U.S.). 2012. Report to the President, Engage to Excel: Producing One Million Additional College Graduates with Degrees in Science, Technology, Engineering, and Mathematics. Executive Office of the President, President’s Council of Advisors on Science and Technology.
- Brewer CA, Smith D. 2011. Vision and change in undergraduate biology education: a call to action. American Association for the Advancement of Science, Washington, DC.
- Bangera G, Brownell SE. 2014. Course-based undergraduate research experiences can make scientific research more inclusive. CBE Life Sci Educ 13:602–606. https://doi.org/10.1187/cbe.14-06-0099
- Auchincloss LC, Laursen SL, Branchaw JL, Eagan K, Graham M, Hanauer DI, Lawrie G, McLinn CM, Pelaez N, Rowland S, Towns M, Trautmann NM, Varma-Nelson P, Weston TJ, Dolan EL. 2014. Assessment of course-based undergraduate research experiences: a meeting report. CBE Life Sci Educ 13:29–40. https://doi.org/10.1187/cbe.14-01-0004
- Hyman OJ, Doyle EA, Harsh J, Mott J, Pesce A, Rasoul B, Seifert K, Enke RA. 2019. CURE-all: large scale implementation of authentic DNA barcoding research into first-year biology curriculum. CourseSource https://doi.org/10.24918/cs.2019.10.
- Fisher GR, Olimpo JT, McCabe TM, Pevey RS. 2018. The Tigriopus CURE - A course-based undergraduate research experience with concomitant supplemental instruction. J Microbiol Biol Educ 19. https://doi.org/10.1128/jmbe.v19i1.1503
- Hanauer DI, Graham MJ, SEA-PHAGES, Betancur L, Bobrownicki A, Cresawn SG, Garlena RA, Jacobs-Sera D, Kaufmann N, Pope WH, Russell DA, Jacobs WR Jr, Sivanathan V, Asai DJ, Hatfull GF. 2017. An inclusive Research Education Community (iREC): Impact of the SEA-PHAGES program on research outcomes and student learning. Proc Natl Acad Sci U S A 114:13531–13536. https://doi.org/10.1073/pnas.1718188115
- Varner J, Connors PK, Brown JS, Dizney L, Duggan JM, Flaherty EA, Hanson J, Lanier HC, Yahnke CJ. 2019. Squirreling around for science: Incorporating sciurid behavioral research into undergraduate curriculum, p. E427–E427. In Integrative and Comparitive Biology. Oxford Univ Press Inc Journals Dept, Cary, NC.
- Gin LE, Rowland AA, Steinwand B, Bruno J, Corwin LA. 2018. Students who fail to achieve predefined research goals may still experience many positive outcomes as a result of CURE participation. CBE Life Sci Educ 17:ar57. https://doi.org/10.1187/cbe.18-03-0036
- Pogoda CS, Keepers KG, Stanley JT, Kane NC. 2019. A CURE-based approach to teaching genomics using mitochondrial genomes. CourseSource. https://doi.org/10.24918/cs.2019.33
- Gastreich KR. 2020. Assessing urban biodiversity with the eBird citizen science project: A course-based undergraduate research experience (CURE) module. CourseSource https://doi.org/10.24918/cs.2020.18
- Dolan EL. 2016. Course-based undergraduate research experiences: Current knowledge and future directions. Natl Res Counc Comm Pap 1–34.
- Armstrong GL, MacCannell DR, Taylor J, Carleton HA, Neuhaus EB, Bradbury RS, Posey JE, Gwinn M. 2019. Pathogen genomics in public health. N Engl J Med 381:2569–2580. https://doi.org/10.1056/NEJMsr1813907
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States--major pathogens. Emerg Infect Dis 17:7–15. https://doi.org/10.3201/eid1701.P11101
- Bell RL, Zheng J, Burrows E, Allard S, Wang CY, Keys CE, Melka DC, Strain E, Luo Y, Allard MW, Rideout S, Brown EW. 2015. Ecological prevalence, genetic diversity, and epidemiological aspects of Salmonella isolated from tomato agricultural regions of the Virginia Eastern Shore. Front Microbiol 6:415. https://doi.org/10.3389/fmicb.2015.00415
- Li B, Vellidis G, Liu H, Jay-Russell M, Zhao S, Hu Z, Wright A, Elkins CA. 2014. Diversity and antimicrobial resistance of Salmonella enterica isolates from surface water in Southeastern United States. Appl Environ Microbiol 80:6355–6365. https://doi.org/10.1128/AEM.02063-14
- Winfield MD, Groisman EA. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl Environ Microbiol 69:3687–3694. https://doi.org/10.1128/AEM.69.7.3687-3694.2003
- Burgess BA, Noyes NR, Bolte DS, Hyatt DR, van Metre DC, Morley PS. 2015. Rapid Salmonella detection in experimentally inoculated equine faecal and veterinary hospital environmental samples using commercially available lateral flow immunoassays. Equine Veterinary Journal 47:119–122. https://doi.org/10.1111/evj.12234
- Goodwin S, McPherson JD, McCombie WR. 2016. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 17:333–351. https://doi.org/10.1038/nrg.2016.49
- Edwards DJ, Holt KE. 2013. Beginner’s guide to comparative bacterial genome analysis using next-generation sequence data. Microb Inform Exp 3:2. https://doi.org/10.1186/2042-5783-3-2
- Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Grüning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D. 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Research. https://doi.org/10.1093/nar/gky379
- Brownell SE, Hekmat-Scafe DS, Singla V, Chandler Seawell P, Conklin Imam JF, Eddy SL, Stearns T, Cyert MS. 2015. A high-enrollment course-based undergraduate research experience improves student conceptions of scientific thinking and ability to interpret data. CBE Life Sci Educ 14:14:ar21. https://doi.org/10.1187/cbe.14-05-0092
- Wiggins G, McTighe J. 1998. What is backward design? Understanding by design 1:7–19.
- Gangiredla J, Rand H, Benisatto D, Payne J, Strittmatter C, Sanders J, Wolfgang WJ, Libuit K, Herrick JB, Prarat M, Toro M, Farrell T, Strain E. 2021. GalaxyTrakr: a distributed analysis tool for public health whole genome sequence data accessible to non-bioinformaticians. BMC Genomics 22, 114. https://doi.org/10.1186/s12864-021-07405-8
- Andrews WH, Jacobson A, Hammack T. 2014. Bacteriological analytical manual: Salmonella. FDA Med Bull.
- Lee K-M, Runyon M, Herrman TJ, Phillips R, Hsieh J. 2015. Review of Salmonella detection and identification methods: aspects of rapid emergency response and food safety. Food Control 47:264–276. https://doi.org/10.1016/j.foodcont.2014.07.011
- Corwin LA, Runyon C, Robinson A, Dolan EL. 2015. The laboratory course assessment survey: A tool to measure three dimensions of research-course design. CBE Life Sci Educ 14:ar37. https://doi.org/10.1187/cbe.15-03-0073
- Hanauer DI, Dolan EL. 2014. The project ownership survey: Measuring differences in scientific inquiry experiences. CBE Life Sci Educ. 13:1, 149-158. https://doi.org/10.1187/cbe.13-06-0123
- Hanauer DI, Graham MJ, Hatfull GF. 2016. A measure of college student persistence in the sciences (PITS). CBE Life Sci Educ 15:ar54. https://doi.org/10.1187/cbe.15-09-0185
- Maltese A, Harsh J, Jung E. 2017. Evaluating undergraduate research experiences—development of a self-report tool. Education Sciences 7:87. https://doi.org/10.3390/educsci7040087
- Corwin LA, Graham MJ, Dolan EL. 2015. Modeling course-based undergraduate research experiences: an agenda for future research and evaluation. CBE Life Sci Educ 14:es1. https://doi.org/10.1187/cbe.14-10-0167
- Feng P, Weagant SD, Grant MA, Burkhardt W, Shellfish M, Water B. 2002. BAM: enumeration of Escherichia coli and the coliform bacteria. U.S. Food and Drug Administration.
- Greenman NA, Jurgensen SK, Holmes II CP, Kapsak CJ, Davis RE, Maza WM, Edemba D, Esser BA, Hise SM, Keen TN, Larson HG, Lockwood DJ, Wang B, Harsh JA, Herrick JB. Genomics of environmental Salmonella: engaging students in the microbiology and bioinformatics of foodborne pathogens. Frontiers in Microbiology. 12, 592422-592422. https://doi.org/10.3389/fmicb.2021.592422
Article Files
Login to access supporting documents
Jurgensen-A CURE for Salmonella.pdf(PDF | 260 KB)
S1. CURE for Salmonella - Laboratory safety contract.docx(DOCX | 281 KB)
S2. CURE for Salmonella - Pre-enrichment and enrichment media preparation.docx(DOCX | 21 KB)
S3. CURE for Salmonella - Field sampling protocol.docx(DOCX | 18 KB)
S4. CURE for Salmonella - Lab notebook grading rubric.docx(DOCX | 29 KB)
S5. CURE for Salmonella - Pre-enrichment and enrichment inoculation protocol.docx(DOCX | 18 KB)
S6. CURE for Salmonella - Plate media preparation.docx(DOCX | 24 KB)
S7. CURE for Salmonella - Plating and purification protocol.docx(DOCX | 19 KB)
S8. CURE for Salmonella - Miscellaneous test protocols - oxidase catalase KOH.docx(DOCX | 17 KB)
S9. CURE for Salmonella - invA PCR and gel visualization protocol.docx(DOCX | 39 KB)
S10. CURE for Salmonella - Preparing isolates for shipping and freezing.docx(DOCX | 37 KB)
S11. CURE for Salmonella - Bioinformatics Lab Guide - Navigating and using GalaxyTrakr and Galaxy.docx(DOCX | 49 KB)
S12. CURE for Salmonella - Bioinformatics lab notebook grading rubric.docx(DOCX | 31 KB)
S13. CURE for Salmonella - Bioinformatics Lab Guide - Salmonella serotyping.docx(DOCX | 21 KB)
S14. CURE for Salmonella - Bioinformatics Lab Guide - Assessing X filtering illumina data using FastQC and Trimmomatic.docx(DOCX | 4 MB)
S15. CURE for Salmonella - Bioinformatics Lab Guide - File naming conventions.docx(DOCX | 307 KB)
S16. CURE for Salmonella - Bioinformatics Lab Guide - Genome assembly using SPAdes and Shovill.docx(DOCX | 108 KB)
S17. CURE for Salmonella - Bioinformatics Lab Guide - Assessing assembly quality Using QUAST and Bandage.docx(DOCX | 340 KB)
S18. CURE for Salmonella - Bioinformatics Lab Guide - Selecting reference genomes_Ordering and viewing assembled contigs using Mauve.docx(DOCX | 647 KB)
S19. CURE for Salmonella - Bioinformatics Lab Guide - Gene annotation.docx(DOCX | 19 KB)
S20. CURE for Salmonella - Bioinformatics Lab Guide - Antibiotic resistance gene detection.docx(DOCX | 89 KB)
S21. CURE for Salmonella - Bioinformatics Lab Guide - Miscellaneous gene and genetic feature detection.docx(DOCX | 243 KB)
S22. CURE for Salmonella - Presentation evaluation rubric.docx(DOCX | 16 KB)
S23. CURE for Salmonella - Poster evaluation rubric.docx(DOCX | 13 KB)
S24. CURE for Salmonella - Sources and description of preexisting survey instruments.docx(DOCX | 13 KB)
S25. CURE for Salmonella - Course_Overview.pptx(PPTX | 6 MB)
- License terms

Comments
Comments
There are no comments on this resource.