Single Cell Insights Into Cancer Transcriptomes: A Five-Part Single-Cell RNAseq Case Study Lesson
Editor: William Morgan
Published online:
Abstract
There is a growing need for integration of “Big Data” into undergraduate biology curricula. Transcriptomics is one venue to examine biology from an informatics perspective. RNA sequencing has largely replaced the use of microarrays for whole genome gene expression studies. Recently, single cell RNA sequencing (scRNAseq) has unmasked population heterogeneity, offering unprecedented views into the inner workings of individual cells. scRNAseq is transforming our understanding of development, cellular identity, cell function, and disease. As a ‘Big Data,’ scRNAseq can be intimidating for students to conceptualize and analyze, yet it plays an increasingly important role in modern biology. To address these challenges, we created an engaging case study that guides students through an exploration of scRNAseq technologies. Students work in groups to explore external resources, manipulate authentic data and experience how single cell RNA transcriptomics can be used for personalized cancer treatment. This five-part case study is intended for upper-level life science majors and graduate students in genetics, bioinformatics, molecular biology, cell biology, biochemistry, biology, and medical genomics courses. The case modules can be completed sequentially, or individual parts can be separately adapted. The first module can also be used as a stand-alone exercise in an introductory biology course. Students need an intermediate mastery of Microsoft Excel but do not need programming skills. Assessment includes both students’ self-assessment of their learning as answers to previous questions are used to progress through the case study and instructor assessment of final answers. This case provides a practical exercise in the use of high-throughput data analysis to explore the molecular basis of cancer at the level of single cells.
Citation
Samsa LA, Eslinger M, Kleinschmit A, Solem A, Goller CC. 2021. Single cell insights into cancer transcriptomes: A five-part single-cell RNAseq case study lesson. CourseSource. https://doi.org/10.24918/cs.2021.26
Society Learning Goals
Biochemistry and Molecular Biology
- Macromolecular Structure Determines Function and Regulation
- How are a variety of experimental and computational approaches used to observe and quantitatively measure the structure, dynamics and function of biological macromolecules?
Bioinformatics
- DNA - Information Storage [GENOMICS]
- Where are data about the genome found (e.g., nucleotide sequence, epigenomics) and how are they stored and accessed?
- How can bioinformatics tools be employed to analyze genetic information?
- RNA - Information Transfer [TRANSCRIPTOMICS]
- Where are data about the transcriptome found (e.g., expression, epigenomics and structure) and how are they stored and accessed?
- How can bioinformatics tools be employed to examine transfer of genetic information?
- Computational Skills
- What higher-level computational skills can be used in bioinformatics research?
Cell Biology
- Cell Communication
- How do cells send, receive, and respond to signals from their environment, including other cells?
- Multicellularity & Cell Connections
- How do cells connect to each other and organize to function as a collective entity?
- Methods & Tools of Cell Biology
- How do the methods and tools of cell biology enable and limit our understanding of the cell?
Genetics
- Molecular biology of gene function
- How is genetic information expressed so it affects an organism's structure and function?
- Methods & Tools in Genetics
- What experimental methods are commonly used to analyze gene structure and gene expression?
Lesson Learning Goals
The overarching learning goal for this case is for students to explore the interdisciplinary nature of science by analyzing a high throughput approach to identify a personalized treatment for a cancer patient. By the end of this case, students should be able to describe in detail the goals, methodology, results and analysis of a high-throughput single-cell transcriptomics study. Society learning goals addressed in this case study are listed along with corresponding parts of the lesson: From Genetics Framework
- How is genetic information expressed so it affects an organism’s structure and function? (Part 1)
- How do the methods and tools of cell biology enable and limit our understanding of the cell? (Part 1)
- How do cells connect to each other and organize to function as a collective entity? (Part 1)
- How do cells send, receive, and respond to signals from their environment, including other cells? (Part 1, Part 5)
- What experimental methods are commonly used to analyze gene structure, gene expression, gene function, and genetic variants? (Part 2)
From Genetics Core Competencies
- Students should be able to gather and evaluate experimental evidence, including qualitative and quantitative data. (Part 2)
- Students should be able to generate and interpret graphs displaying experimental results. (Part 4)
- Students should be able to critique large data sets and use bioinformatics to assess genetics data. (Part 4)
From Bioinformatics Framework
- Where are data about the genome found (e.g., nucleotide sequence, epigenomics) and how are they stored and accessed? (Part 2)
- How can bioinformatics tools be employed to analyze genetic information? (Part 3)
- Where are data about the transcriptome found (e.g., expression, epigenomics and structure) and how are they stored and accessed? (Part 3)
- How can bioinformatics tools be employed to examine transfer of genetic information? (Part 4)
- What higher-level computational skills can be used in bioinformatics research?
From Biochemistry and Molecular Biology Framework
- How are a variety of experimental and computational approaches used to observe and quantitatively measure the structure, dynamics and function of biological macromolecules? (Part 3)
Learning goals not currently included in society frameworks that are also addressed in this lesson include the following:
- Describe the cellular nature of cancer (how cancerous tumors are composed of a heterogeneous group of cells that continue to evolve over time as mutations accumulate to effect a variety of regulative physiological processes). (Part 1)
- How is individual cell behavior altered in disease and manipulated by drugs? (Part 5)
Lesson Learning Objectives
By the end of this section, students will be able to: Part 1: The patient and diagnosis
- Explain how traditional (bulk or population) RNA-seq masks intratumoral heterogeneity.
- Explain how use of techniques like single cell RNA-seq with patient tumors is an essential step toward personalized medicine.
Part 2: The technician and the samples
- Describe how single cell RNA-seq works.
Part 3: Data processing
- Summarize and interpret evidence of the quality of a single cell RNA-seq assay.
Part 4: Data visualization
- Use Granatum to create and interpret graphs displaying single cell RNA-seq experimental results.
- Interpret graphs displaying single cell RNA-seq experimental results.
Part 5: Treatment - Back to the patient
- Describe how changes in gene expression detected by single cell RNA-seq could guide treatment selection for personalized medicine.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
Introduction
There is a growing need for integration of ‘Big Data’ concepts into the undergraduate biology curriculum (1). The slow integration of ‘Big Data’ concepts into curricula is bound by faculty perceived barriers, which include lack of access to developed curriculum resources, minimal faculty experience/expertise, and inferred absence of student interest (2). To bridge this gap, widely adoptable, engaging learning resources complete with instructor notes are integral to facilitate continued integration into life science curricula. The life sciences are also experiencing a paradigm shift from an emphasis on data acquisition to data analysis, and in many cases researchers facilitate the development of hypotheses for physical experiments with ‘big data’ analysis first (3). Thus, it is critical for student training curricula to provide students with experiences working with ‘Big Data’ and associated tools.
Transcriptomics has largely replaced microarray approaches to understanding whole genome gene expression by sequencing RNA extracted from cells (4). Recently, single cell RNA sequencing (scRNAseq) has unmasked population heterogeneity in a way that offers an unprecedented view into the inner workings of individual cells that is transforming our understanding of development, cellular identity, cell function, and disease (5). In a typical scRNAseq experiment, single cells are isolated into individual micro-wells or droplets, RNA is extracted and converted to cDNA. The cDNA is amplified and sequenced using next generation sequencing technologies. A unique barcode added to each cell’s cDNA allows for computational de-multiplexing. The next generation sequence data is mapped to the genome to yield massive quantities of information describing the expression level of each gene in the genome for each cell.
scRNAseq and other single cell technologies (e.g., single cell genomics, proteomics) are allowing the modern biologist to interrogate biological functions at a highly granular level. These technologies have led to new insights into development, disease, and cell function, and are poised to yield even more!
Cancer biology in particular has benefited from single cell technologies. Tumors are now understood to be composed of both normal and cancerous cells, both of which are highly heterogeneous. The tumor microenvironment consists of immune cells, fibroblasts, endothelial cells, and mesenchymal cells, among others, which uniquely contribute to the complex matrix influencing oncogenic potential (6). It has long been understood that cancer cells acquire mutations which afford cells hyperproliferative properties (7). Within a tumor, cancer cell clones compete as they activate oncogenes, lose tumor suppression signals, and acquire new mutations that influence survival and/or proliferation of their progeny. Thus, at any point in time, a tumor is its own experiment in natural selection as the wildly heterogenous mixture of cell types - cells with different genetic compositions and phenotypes exist and compete within the same tumor microenvironment. It is this diversity in molecular origins within the same tumor that makes prescribing the optimal treatment plan problematic. Due to unique environmental cues and genetic or epigenetic changes, therapy resistance emerges, and what works for one patient may not be effective for the same tumor type in another patient. That is, just because the tumor has the same tissue of origin, does not mean that the underlying molecular genetics are comparable, and this can impact the effect of a therapy (8).
Historically, tumors were sampled and cells analyzed on the whole, without considering the molecular difference within each cell within the tumor. This complex tumor environment must be understood for each patient to develop tailored treatment approaches. A thorough understanding on how individual cells contribute to the tumor microenvironment must guide the development of personalized cancer therapy (9). It is the fine-tuning of this understanding that will lead to effective treatments and ultimately the prognosis for primary and metastatic disease (10). Standard treatment protocols may be effective for the bulk of the cell population, but they may miss a subsection of clones with different features which survive treatment and contribute to recurrence. Thus the treatments themselves can select for resistant cell types which must be anticipated to prevent a recurrent tumor failing to respond to conventional therapy (11). To address this feature, researchers are taking a single cell approach to analyze the tumor microenvironment in the hope that that drug combinations can target the heterogeneous nature of the tumor and minimize the unintentional selection of clones which lead to relapse (12). The rationale behind this approach is that a drug (or drug combination) can be tailored to an individual patient’s tumor microenvironment and predict or adapt by predicting cell-specific responses (13). Single cell RNAseq is one such tool that can address these issues, predicting response to treatment as well as tailoring to the specific cellular features based on the transcriptional output of cells directly within the biopsied tissue (14). This is an exciting development as researchers can assist physicians by addressing questions such as, “what drug or combination of drugs is best for specific mutations?” and, from the perspective of personalized medicine, “can we predict what will work for the individual patient rather than a generalized treatment protocol?” Single-cell RNAseq is touted as one of the tools to help answer these questions. The tumor landscape can be dissociated into their single cell components and their molecular basis analyzed to make informed predictions about treatment options (15).
With the advancement of knowledge at the single cell level, it can be challenging for novice and experienced scientists to navigate the big datasets that accompany high throughput screening technology. For example, a single one centimeter tumor contains 10^9 cells (16). With each cell contributing specific genetic information that contributes functionally to the basal or tumorigenic properties, the exponential amounts of data can be intimidating for students to conceptualize, much less analyze. However, such approaches play an increasingly important role in modern biology. Additionally, it can be challenging for faculty to develop educational resources that address multiple core competencies and associated learning outcomes outlined by the 2011 AAAS Vision and Change Report and BioSkills Guide, respectively (17, 18). Williams et al. conducted a national-wide survey and found that barriers to integrating bioinformatics into undergraduate life sciences curriculum are disproportionately greater at minority-serving institutions (MSIs) compared to non-MSIs (2). To address these challenges, we created an engaging case study that guides students through an exploration of authentic scRNAseq technologies and associated datasets.
In this five-part interrupted case study (Figure 1), students work in small groups to explore a single-cell transcriptomics case study to build a personalized treatment plan for a cancer patient. This case highlights the interdisciplinary nature of science and helps students build proficiencies in both biology and bioinformatics. Working in small groups, students experience the “real” flow of pre-clinical research from multiple perspectives which is tailorable for the intended audience.
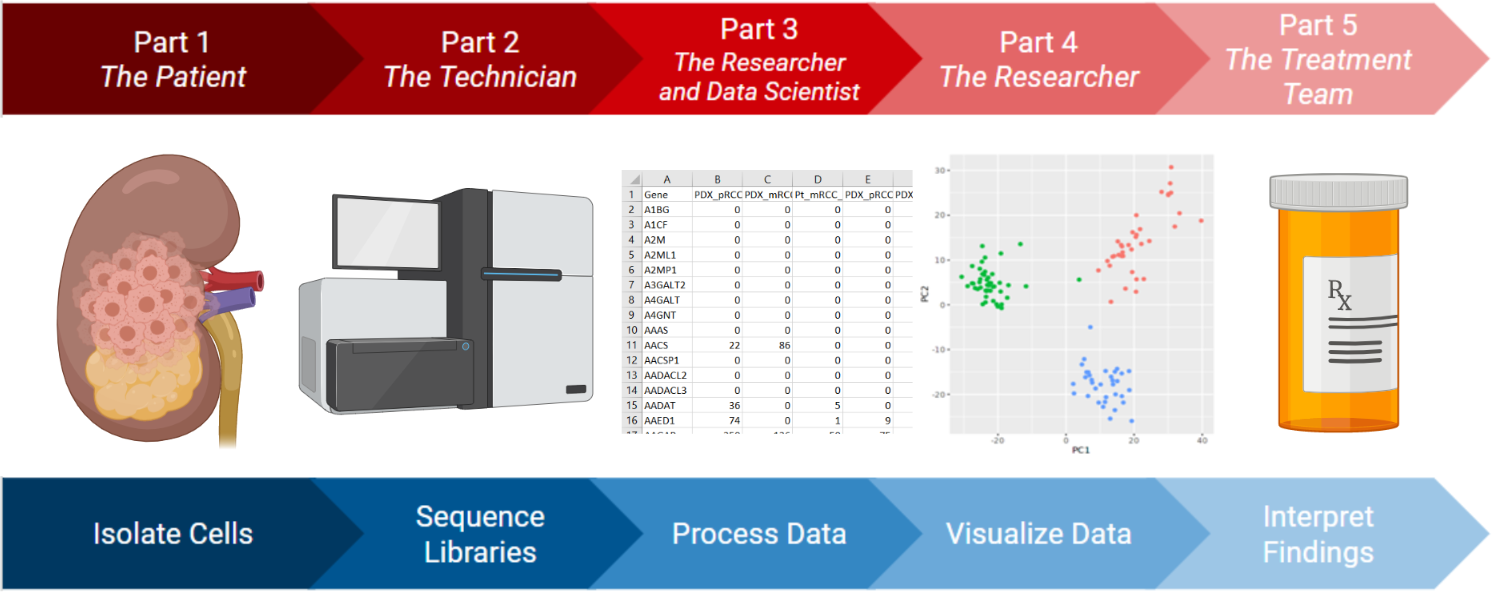
Figure 1. Schematic overview of the scRNAseq case study. Description: Schematic overview of the scRNAseq case study. Image made in ©BioRender - biorender.com.
In Part 1, they meet Gary, a 44-year old male with clear cell renal cell carcinoma who is advised by his physician, Dr. Ortiz, to allow the Genomics Resources Consortium to study a biopsy of his tumor. Students answer questions to learn about the cancer and the promise of “omics” data for personalized medicine. In Part 2, students follow the tumor into the lab where they put themselves in the shoes of a visiting student helping the technician processing the sample. They answer questions to study the technical details of how scRNAseq works and the practical details of how samples are exchanged in a research collaboration. In Parts 3-5, students assume the role of the research scientist working under Dr. Ortiz. They summarize and assess the quality of the scRNAseq data (Part 3), use Granatum, a browser-based scRNAseq data analysis platform, to analyze and graph the data (Part 4), and analyze their results to determine patterns between gene expression and appropriate therapeutic treatment (Part 5).
Students work together to solve a problem that would be too difficult for any one student to tackle alone. Student group performance and learning is assessed by evaluating responses to integrated case study questions. Students gain an appreciation for the complex, interdisciplinary nature of high-throughput research, essential bioinformatics skills (e.g., data transformation, batch variation correction), and practice locating and using relevant online resources.
To our knowledge, this is the first case study that specifically explores single cell RNAseq and one of few initial Big Data learning resources that does not require programming skills. This differs from other published RNAseq teaching resources in that it is composed of short modular units, does not require prerequisite programming skills, and focuses on a biomedical problem (19–23). Together, this modular case study can bring big data into perspective to expose students to the biology of cancer and emerging mechanisms and informatics used to reconcile the molecular differences within the tumor microenvironment.
Intended Audience
This five-part case study is intended for upper-level life science majors and graduate students in genetics, bioinformatics, molecular biology, cell biology, biochemistry, biology, and medical genomics courses. The first module (Part 1) can also be used as a standalone exercise to introduce students to concepts associated with cancer and contemporary research methodology and approaches. The lesson was originally designed for upper-level students and graduate students at a large doctoral degree-granting institution during an 8-week “High-throughput Discovery” course. In addition to this course, the case has also been taught in part or in its entirety in biology courses at a small private liberal arts school, an undergraduate military academy, and a public primarily Master’s degree granting institution.
Required Learning Time
This lesson is designed for upper-level students to complete it in five to six 60-90 minute class periods. Individual parts of the case take between 60-120 minutes to complete (Table 1). The entire case can alternatively be assigned as a one-week group challenge project culminating in a class-level discussion and individual reflections.
Table 1. Lesson timeline with a summary of each part of the case study. It is recommended that the parts be completed in sequence over five class periods or assigned as a week-long activity. Individual parts may be extracted for adaptation and use as a single lesson in suggested courses.
|
Activity |
Description |
Estimated Time |
Notes |
|
Part 1. The patient and diagnosis |
Story: Meet the patient, physician, and research consortium. Activity: Students research and answer questions about cancer, tumor heterogeneity, and “omics” data for personalized medicine. |
60-90 min |
|
|
Part 2. The technician and the samples |
Story: Follow the tumor into the lab for scRNAseq. Activity: Students research and answer questions to study how scRNAseq works. |
60-90 min |
|
|
Part 3. Data processing |
Story: Determine if the scRNAseq experiment worked. Activity: Students conduct summary analyses to determine the overall quality of the data. |
60-90 min |
|
|
Part 4. Data visualization |
Story: Process and visualize the scRNAseq experiment data Activity: Students use Granatum, a browser-based scRNAseq data analysis platform, to process and graph the data. |
90-120 min |
|
|
Part 5. Treatment of the patient |
Story: Analyze the scRNAseq to predict which drug will work for the patient Activity: Students analyze their results from Part 4 to make the case for an appropriate therapeutic treatment. |
90-120 min |
|
Prerequisite Student Knowledge
Students who have taken advanced courses in cell biology, molecular biology, genetics, and biostatistics should have adequate background knowledge to complete this lesson as an asynchronous, online activity. Students who do not have this background may benefit from synchronous distance learning sessions or a face-to-face classroom setting for successful completion. Parts 3-5 involve data analysis activities that require students to have intermediate skills in using spreadsheet software (e.g., Microsoft Excel) and in the analysis and interpretation of data. Students with limited computer competency will struggle with these parts of the case but can be assigned the case in an adapted format (see Teaching Discussion). Many questions throughout the case require students to have intermediate to advanced ability to search the internet and identify relevant information from FASTQ sequence quality sources. Students who lack these skills would greatly benefit from Part 1, which provides for highly structured inquiry.
Prerequisite Teacher Knowledge
In addition to the student knowledge, the instructor should have an intermediate understanding of scRNAseq and RNAseq technologies to be able to evaluate student answers. A good starting point to address knowledge gaps is Haque et al. “A practical guide to single-cell RNA sequencing for biomedical research and clinical applications.” (24) The case is loosely based on a published research study by Kim et al. “Application of single-cell RNA sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma” and uses the tutorial dataset provided by Granatum (25, 26). We highly recommend that the instructor review this manuscript in preparation for guiding students through the case study (25). We recommend that instructors complete the case independently to identify conceptual gaps and practice using the Granatum analysis software (26). While instructions are current as of publication, bioinformatics software is not static; it is possible that updates and upgrades to the software will require ad hoc modifications to the analysis instructions provided in Part 4. Alternatively, instructors could provide students with previously generated output data visualizations from Granatum (see Instructor Versions of supplemental documents) to achieve the same learning gains with analyzing large data sets. Instructors who require software version control or who prefer to use the software locally may follow instructions provided at Granatum’s website to download and host the freely-available software.
Scientific Teaching Themes
Active Learning
Regardless of delivery format, this case study actively engages student learning by prompting students to explore outside resources, involving real-world situations, and providing scaffolded questions. Engaging visual content or interaction with realistic data promotes deeper understanding of material and appeals to a variety of learning preferences (27). Students work in groups to complete the case, which provides social stimulation and helps students solve problems that might otherwise be too difficult for any one student to solve alone. This collaboration across disciplines is an important skill and highlights the multidisciplinary nature of science and medicine (17). If taught in a synchronous classroom setting, case questions provide many opportunities for students to express their opinions and explain their answers to the class.
Assessment
The Next Generation Science Standards (NGSS) publishes guidelines on the engagement of students in modeling science curriculum (28). Pedagogical approaches focused on engagement of students by answering realistic questions, using guided instructions to analyze and synthesize data and generate models has been particularly effective in teaching skills to emerging scientists (29, 30). When students connect ideas during multiple levels of analysis and link concepts to specific ideas, these learning exercises are effective at promoting student learning (31). Several intrinsic assessment mechanisms are embedded within the case modules to provide instructor opportunities for formative learning assessments. Instructors evaluate achievement of learning objectives by evaluating the case analysis (correct/incorrect answers to case questions and strength of argument for open-ended questions). Instructors also have the opportunity to assess student generated spreadsheets, charts, graphs, and sketches throughout the case for both completion and accuracy. In doing so, the instructor measures knowledge and understanding, critical-thinking skills, problem-solving skills, and (for student groups) collaborative learning skills. Quantitative scoring of the case may vary for different courses, leaving the instructor latitude to adapt the case for their student population and course structure. The use of de-identified student quotes was covered by: NCSU IRB protocol #11758.
Inclusive Teaching
The case study is designed as a multipart highly structured series of tasks that require positive collaboration between group participants. Fostering such experiences promotes inclusion of all team members while using a highly adaptable active learning technique (32). Inclusion in science underscores the collaborative nature of the discipline. The essential points of evidence-based teaching seek to provide a resource for designing an inclusive teaching experience for the classroom (33). Importantly, the social relationships developed during the dialogue discussing this case among groups promotes an environment where the knowledge of each participant is heard (32). In particular, students can empathize with the patient, Gary, the researcher or the students engaging in the data analysis portions of the case. This crucial component promotes diversity through the active learning experience (34). The inclusive dialogues and relationship between characters within the case has students navigating the informatic activities as they explicitly address emerging developments within the field of informatics and molecular biology. Moreover, this dialogue between both characters and students promotes engagement and trains self-efficacy when navigating large data sets associated with biological enquiry. In return, this promotes an experience that reveals aspects of clinical and translations research processes: it shows some of the professional obligations clinical providers and researchers have to patients during the cancer treatment and provides students a taste of what it might be like to perform the underlying research and analysis necessary to advance personalized patient care.
Lesson Plan
Summary
In this five-part case study, students work in small groups to explore the interdisciplinary nature of science and build bioinformatics proficiencies. Students experience the “real” flow of pre-clinical research. The case study is intended to be completed in sequence across five to six 60–90-minute class periods in asynchronous online or synchronous face-to-face formats. Part I has been used as a stand-alone lesson for introductory level students. Student learning and progress may be assessed after each part or upon completion of the whole case study.
Preparation
Each case section and associated data or metadata files should be posted on a course learning management system prior to implementation. See Prerequisite Teacher Knowledge section for instructor preparation recommendations and resources. We recommend that the instructor run through the entire activity in advance of beginning the lesson in class or online and make any ad hoc adjustments necessary to account for changes in website content for materials linked within the case. Though all links and internet resources are up to date at the time of submission, we are unable to provide continuous curation of content beyond the time of submission.
Assign the case
Divide the class into groups of 3-4. Communicate your course-specific expectations for the assignment in writing and face-to-face (if possible). When you assign the lesson, make sure to explain (in writing and in person) how the case aligns with course goals and provides experience with analysis of genomics data; this is important for student engagement. Also, make sure to communicate expectations for time commitment, group work, data analysis, use of external resources as reference, and deliverables. Students should enter the assignment knowing that it will be challenging and require the collective effort of all members and time, and in return they will solve an exciting real-world application of this technology that will provide data analysis skills that are in high-demand.
Regardless of whether you are teaching this case in an online format or in the classroom, students will need access to electronic documents. Use your online learning management system (LMS, e.g., Canvas, Moodle, Blackboard, etc...) or GoogleDrive (in combination with Doctopus) to deliver electronic copies of the case and associated data files in advance. See Supporting Files S1-S12 for student and instructor versions of the case, provided both as an all-in-one file (S1. scRNAseq – scRNAseq Case Study Parts 1-5 - Student version, S2. scRNAseq – scRNAseq Case Study Parts 1-5 - Answer key) for the five-part case and individual files for each part (Supporting Files S3-S12). Additional files necessary for each Part are provided in Supporting Files S13-26. We recommend using GoogleDocs for online collaboration in the distance education environment – this format facilitates easy sharing between group members and allows you to easily track student progress in real time.
Lesson
Your main role as instructor is to guide students through the case, answering questions and helping troubleshoot any problems they may have. Many of the questions are open-ended and prompt student exploration of cancer biology, pre-clinical research, bioethics, scRNAseq technologies, and data science. If you have expertise in any of these areas, student questions are a great launching point for special topics teaching moments. If you do not have expertise in these areas, help students find reliable sources where they (and you) can learn more. Answer keys to each Part are provided in Supporting File S2. scRNAseq – scRNAseq Case Study Parts 1-5 - Answer key and Supporting Files S8-12). You may choose to provide students with answers from that key if they get stuck or as part of a classroom discussion/wrap up for each part. Parts 4 and 5 build on files that students create in Parts 3 and 4, respectively. You may choose to provide students with answer-key files between parts so that errors they may make in the previous Part are not carried over. Halfway through the case assignment, review student progress and provide encouraging comments. This can help encourage groups to complete the case and identify any potential technical or conceptual issues that can be addressed at the class level.
After students submit their work for assessment, a class-level discussion of the overall findings and reflections on the process is important to emphasize key concepts and procedures. In an online environment, a short video or audio recording along with a class announcement can help start discussions in a forum or stimulate discussion in the classroom. Individuals should be provided the opportunity to reflect on their experience and how it ties into the course goals and their career paths. This can be a short reflection or one-minute paper. After that, the instructor should post a key and encourage students to review it, posting additional questions for discussion.
Assessment
Evaluate student group answers against the provided answer key. Feedback should be provided to each group and emphasized through class-level discussions
Teaching Discussion
Observations
The case study was effective in achieving the course goals and lesson objectives of an upper-level undergraduate and graduate High-throughput Discovery course. Students realized the application of this technology and how it is high-throughput after having to analyze the data. In other courses that the case was taught, students pursuing professional health science and biomedical careers were interested in the real-world applications of single cell transcriptomics. This case is challenging even for teams with several graduate students, as the datasets are large and the analyses complex; one out of three groups struggled with the data analysis part of the case study. With instructor encouragement, this case is effective in achieving the stated objectives, and most (9/11) students found the challenge worthwhile. Students commented on the connection of the case to cancer and precision medicine (5/11), data analysis and spreadsheets (4/11), and the comparison of single-cell and bulk RNA transcriptomics (3/11). These comments and the evidence provided by accurate completion of the case study worksheet, support achievement of key learning outcomes 1-4.
Representative student feedback is provided below:
I learned about what sc transcriptomics is and how it differs from the bulk version. It was interesting to see how the technologies we are learning about in class can be used in the real world, [and] I wonder how accurate some of the case study scenarios were to real life. The case study also helped me to learn about how isolation of single cells occurs on a high-throughput basis.
I learned more about cancer cells and how they differ from normal cells and how they work in the body. I also learned more about how sequencing works and how people use the data it provides for application, in this case for determining the best course of treatment for a specific person (precision medicine).
(…) I really liked the fact that it could have felt like it was a real case study. By doing so it allows us to step into our future roles/ careers and that truly changes the way that we view the question/ scenario. I did really enjoy that.
I thought it was cool that we had to talk about what Gary's diagnosis was, and that we got to look at carcinoma cells under a microscope. I thought this was a very unique and fun lab. I thought that the picture under the microscope could have had some more labels on it to show everything that was present. Yes, I thought this case was very effective in learning about single cell transcriptomics.
The case was very interesting with a lot of information. I liked how there was just so much background information, and provided details in the case study, but it was very confusing… I believe it was an effective way to learn about single cell transcriptomics, but personally I am a hands on learner so I would have loved to hear about it or see it in person. I did find the topic interesting in the end though, and it had me on the edge of my seat. I was intrigued by the such difficult concept of cancer information. It was crazy to see how it is all analyzed after all.
This case study was very interesting to me and it helped me better understand single cell transcriptomics. The topic is a little bit confusing so opening many websites and videos was helpful to not only find the answers to the questions but it taught me more than just what the questions were asking. The videos were very helpful and I liked how they were informational with providing general definitions but also giving examples. I found the information on stages of cancer to be very interesting to me and I spent some time exploring that website because it taught me a lot!
Suggestions for possible improvements or adaptations
Pre-lesson activity
In line with student feedback when field-testing the case, for some courses and learners, it may be appropriate to assign a low-stakes pre-lesson to familiarize participants with the type of data resulting from scRNAseq and how it is analyzed. For example, we suggest assigning some of the videos or readings a week before administering the case in class. This assignment can be un-graded or a short formative assessment can be included. Use of the software and familiarity with its key features are important for the successful completion of the case. Instructors may wish to assign as a pre-lesson activity the use of Granatum and require a deliverable to help troubleshoot any technical issues that may arise.
Modifications
If course learning goals do not include data analysis, we suggest focusing primarily on Part 1. Part 1 is an appropriate launching point for exposing undergraduate biology students to concepts in cancer biology and the underlying, interrelated concepts in cell biology, genetics, and physiology. To retain learning outcomes about how cancer is treated, omit Parts 2-4 and complete Part 5 using graphs and data extracted from the Part 4 answer key. In this case, students analyze pre-processed data.
For example, Part 1 of the case study can be used within an introductory cellular/molecular biology course to introduce contemporary biological tools used to characterize a diverse group of cells (e.g., cancer). Indeed, the case was used as a capstone exploration of a modern technique used by researchers and clinicians to understand the dynamics of cancer biology and to assist in the formulation of an individual treatment plan respectively. Throughout the course students explored the essential tenets of cell biology (e.g., signaling, regulation of the cell cycle, apoptosis) with a focus on applications associated with cancer. Additionally, students engaged in societal and ethical issues associated with cancer research in humans through reading The Immortal Life of Henrietta Lacks by Rebecca Skloot (35). Toward the end of the term, the scRNA-seq case study provided a means to learn about tools and techniques for characterizing a tumor, while touching on informed consent.
Advanced bioinformatics courses could extend Part 3 to include data processing activities whereby students can replicate published methods to clean and process raw RNAseq data into a read count matrix. The original next generation sequencing files used to generate the gene counts in the processed data are publicly available in Gene Expression Omnibus repository under accession number GSE73122.
Part 5 requires students to analyze data to predict which compound is likely to work to treat the patient’s tumor. There are many “right” answers and multiple approaches to this task. Students are evaluated based on the strength of their rationale for their selection. This could be extended to include group presentations where students present a report and defend their rationale. Similarly, students could write a mini-manuscript to report their scRNAseq analysis, findings, and predictions.
If this activity is implemented in an online asynchronous course, we suggest creating a short instructor video describing the goals of the case study (learning objectives) and how it aligns with the course goals. In that overview, the instructor should highlight how this realistic scenario will expose students to single-cell RNA-seq data they will have to analyze. It is highly recommended that instructors tell students that they will use Granatum to analyze the data and will use Excel spreadsheets. We found that an online student help forum helped encourage students to post and answer technical questions, often promoting peer-to-peer communication. In the spring of 2020, we implemented this lesson by assigning a GoogleDoc editable version of the case study to groups of three or four undergraduate and graduate students. Using the Learning Management System (LMS) we sent information about the goals of the case and use of Granatum via class announcements. Students were told this would be a challenging case and required their work as a team. Mid-week, a second announcement and instructor comments on the Google Docs encouraged students to continue making progress on the case. After submission of the cases by the groups, we shared a summary of the major findings and goals. We also summarized the individual reflections students submitted in response to a prompt about what they learned from this experience. While students found this experience to be challenging, all responses were positive and highlighted how single-cell technologies could be used in clinical settings – a fact many of them had not considered.
SUPPORTING MATERIALS
- S1. scRNAseq – scRNAseq Case Study Parts 1-5 - Student version
- S2. scRNAseq – scRNAseq Case Study Parts 1-5 - Answer key
- S3. scRNAseq – Part 1 The patient and diagnosis - Student version
- S4. scRNAseq – Part 2 The technician and the samples - Student version
- S5. scRNAseq – Part 3 Data processing - Student version
- S6. scRNAseq – Part 4 Data visualization - Student version
- S7. scRNAseq – Part 5 Treatment - Student version
- S8. scRNAseq – Part 1 The patient and diagnosis - Answer key
- S9. scRNAseq – Part 2 The technician and the samples - Answer key
- S10. scRNAseq – Part 3 Data processing - Answer key
- S11. scRNAseq – Part 4 Data visualization - Answer key
- S12. scRNAseq – Part 5 Treatment - Answer key
- S13. scRNAseq – File for Part 2 - Sequencing Metadata - Student version
- S14. scRNAseq – File for Part 2 - Sequencing Metadata - Instructor version
- S15. scRNAseq – File for Part 2 - Processing Datasheet - Student version
- S16. scRNAseq – File for Part 2 - Processing Datasheet - Instructor version
- S17. scRNAseq – File for Part 3 - Expression - Student version
- S18. scRNAseq – File for Part 3 - Expression - Instructor version
- S19. scRNAseq – File for Part 3 - Metadata - Student version
- S20. scRNAseq – File for Part 3 - Metadata - Instructor version
- S21. scRNAseq – File for Part 3 - Processing Notes - Student version
- S22. scRNAseq – File for Part 3 - Processing Notes - Instructor version
- S23. scRNAseq – File from Part 4 - Normalized Expression - Instructor version
- S24. scRNAseq – File from Part 4 - Metadata with Clusters - Instructor version
- S25. scRNAseq – File from Part 4 - DE PDX meta vs PDX primary- Instructor Version
- S26. scRNAseq – File for Part 5 - Normalized Expression annotated for instructor
Acknowledgments
We are grateful to students in the BIT 479/579 Spring 2020 High-throughput Discovery course, CH388 Spring 2020 Genetics, BIO 235 Spring 2020 Cell Biology, who participated in this case study. Additionally, we are thankful to Dr. Kathleen McAdams for her input during case study development.
References
- Wilson Sayres MA, Hauser C, Sierk M, Robic S, Rosenwald AG, Smith TM, Triplett EW, Williams JJ, Dinsdale E, Morgan WR, Burnette JM, Donovan SS, Drew JC, Elgin SCR, Fowlks ER, Galindo-Gonzalez S, Goodman AL, Grandgenett NF, Goller CC, Jungck JR, Newman JD, Pearson W, Ryder EF, Tosado-Acevedo R, Tapprich W, Tobin TC, Toro-Martínez A, Welch LR, Wright R, Barone L, Ebenbach D, McWilliams M, Olney KC, Pauley MA. 2018. Bioinformatics core competencies for undergraduate life sciences education. PLOS ONE 13:e0196878. https://doi.org/10.1371/journal.pone.0196878.
- Williams JJ, Drew JC, Galindo-Gonzalez S, Robic S, Dinsdale E, Morgan WR, Triplett EW, Burnette JM, Donovan SS, Fowlks ER, Goodman AL, Grandgenett NF, Goller CC, Hauser C, Jungck JR, Newman JD, Pearson WR, Ryder EF, Sierk M, Smith TM, Tosado-Acevedo R, Tapprich W, Tobin TC, Toro-Martínez A, Welch LR, Wilson MA, Ebenbach D, McWilliams M, Rosenwald AG, Pauley MA. 2019. Barriers to integration of bioinformatics into undergraduate life sciences education: A national study of US life sciences faculty uncover significant barriers to integrating bioinformatics into undergraduate instruction. PLOS ONE 14:e0224288. https://doi.org/10.1371/journal.pone.0224288.
- Dolinski K, Troyanskaya OG. 2015. Implications of Big Data for cell biology. Mol Biol Cell 26:2575–2578. https://doi.org/10.1091/mbc.E13-12-0756.
- Wang B, Kumar V, Olson A, Ware D. 2019. Reviving the Transcriptome Studies: An Insight Into the Emergence of Single-Molecule Transcriptome Sequencing. Front Genet 10:384. https://doi.org/10.3389/fgene.2019.00384.
- Hwang B, Lee JH, Bang D. 2018. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med 50:96. https://doi.org/10.1038/s12276-018-0071-8.
- Runa F, Hamalian S, Meade K, Shisgal P, Gray PC, Kelber JA. 2017. Tumor Microenvironment Heterogeneity: Challenges and Opportunities. Curr Mol Biol Rep 3:218–229. https://doi.org/10.1007/s40610-017-0073-7.
- Hanahan D, Weinberg RA. 2011. Hallmarks of Cancer: The Next Generation. Cell 144:646–674. https://doi.org/10.1016/j.cell.2011.02.013.
- Yuan S, Norgard RJ, Stanger BZ. 2019. Cellular Plasticity in Cancer. Cancer Discov 9:837–851. https://doi.org/10.1158/2159-8290.CD-19-0015.
- Hanahan D, Coussens LM. 2012. Accessories to the Crime: Functions of Cells Recruited to the Tumor Microenvironment. Cancer Cell 21:309–322. https://doi.org/10.1016/j.ccr.2012.02.022.
- De Palma M, Hanahan D. 2012. The biology of personalized cancer medicine: Facing individual complexities underlying hallmark capabilities. Mol Oncol 6:111–127. https://doi.org/10.1016/j.molonc.2012.01.011.
- Fillon M. 2012. Cancer and Natural Selection. JNCI J Natl Cancer Inst 104:1773–1774. https://doi.org/10.1093/jnci/djs496.
- Kim K-T, Lee HW, Lee H-O, Kim SC, Seo YJ, Chung W, Eum HH, Nam D-H, Kim J, Joo KM, Park W-Y. 2015. Single-cell mRNA sequencing identifies subclonal heterogeneity in anti-cancer drug responses of lung adenocarcinoma cells. Genome Biol 16:127. https://doi.org/10.1186/s13059-015-0692-3.
- Marquina-Sanchez B, Fortelny N, Farlik M, Vieira A, Collombat P, Bock C, Kubicek S. 2020. Single-cell RNA-seq with spike-in cells enables accurate quantification of cell-specific drug effects in pancreatic islets. Genome Biol 21:106. https://doi.org/10.1186/s13059-020-02006-2.
- Suvà ML, Tirosh I. 2019. Single-Cell RNA Sequencing in Cancer: Lessons Learned and Emerging Challenges. Mol Cell 75:7–12. https://doi.org/10.1016/j.molcel.2019.05.003.
- Lee HW, Chung W, Lee H-O, Jeong DE, Jo A, Lim JE, Hong JH, Nam D-H, Jeong BC, Park SH, Joo K-M, Park W-Y. 2020. Single-cell RNA sequencing reveals the tumor microenvironment and facilitates strategic choices to circumvent treatment failure in a chemorefractory bladder cancer patient. Genome Med 12:47. https://doi.org/10.1186/s13073-020-00741-6.
- Talmadge JE. 2007. Clonal Selection of Metastasis within the Life History of a Tumor. Cancer Res 67:11471–11475. https://doi.org/10.1158/0008-5472.CAN-07-2496.
- American Association for the Advancement of Science. 2011. Vision and change in undergraduate biology education: A call to action. http://visionandchange.org/files/2010/03/VC_ report.pdf
- Clemmons AW, Timbrook J, Herron JC, Crowe AJ. 2020. BioSkills Guide: Development and National Validation of a Tool for Interpreting the Vision and Change Core Competencies. CBE—Life Sci Educ 19:ar53. https://doi.org/10.1187/cbe.19-11-0259
- Peterson MP, Malloy JT, Marden JH, Buonaccorsi VP. 2015. Teaching RNAseq at Undergraduate Institutions: A tutorial and R package from the Genome Consortium for Active Teaching. CourseSource 2. https://doi.org/10.24918/cs.2015.14.
- Wachira JM, Hughes-Darden CA, Nkwanta A. 2019. Investigating Cell Signaling with Gene Expression Datasets. CourseSource 6. https://doi.org/10.24918/cs.2019.1.
- Makarevitch I, Frechette C, Wiatros N. 2015. Authentic Research Experience and “Big Data” Analysis in the Classroom: Maize Response to Abiotic Stress. CBE—Life Sci Educ 14:ar27. https://doi.org/10.1187/cbe.15-04-0081.
- Procko C, Morrison S, Dunar C, Mills S, Maldonado B, Cockrum C, Peters NE, Huang SC, Chory J. 2019. Big Data to the Bench: Transcriptome Analysis for Undergraduates. CBE—Life Sci Educ 18:ar19. https://doi.org/10.1187/cbe.18-08-0161.
- Escobar MA, Morgan W, Makarevitch I, Robertson SD. 2019. Tackling “Big Data” with Biology Undergrads: A Simple RNA-seq Data Analysis Tutorial Using Galaxy. CourseSource 6. https://doi.org/10.24918/cs.2019.13.
- Haque A, Engel J, Teichmann SA, Lönnberg T. 2017. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med 9:75. https://doi.org/10.1186/s13073-017-0467-4.
- Kim K-T, Lee HW, Lee H-O, Song HJ, Jeong DE, Shin S, Kim H, Shin Y, Nam D-H, Jeong BC, Kirsch DG, Joo KM, Park W-Y. 2016. Application of single-cell RNA sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma. Genome Biol 17:80. https://doi.org/10.1186/s13059-016-0945-9.
- Zhu X, Wolfgruber TK, Tasato A, Arisdakessian C, Garmire DG, Garmire LX. 2017. Granatum: a graphical single-cell RNA-Seq analysis pipeline for genomics scientists. Genome Med 9:108.
- Bobek E, Tversky B. 2016. Creating visual explanations improves learning. Cogn Res Princ Implic 1:27. https://doi.org/10.1186/s41235-016-0031-6.
- NGSS Lead States. 2013. Next Generation Science Standards: For States, By States. The National Academies Press. Available at: https://www.nextgenscience.org/.
- Abd-El-Khalick F, Lederman NG. 2000. Improving science teachers’ conceptions of nature of science: a critical review of the literature. Int J Sci Educ 22:665–701. https://doi.org/10.1080/09500690050044044
- Lederman NG, Abd-El-Khalick F, Bell RL. 2000. If we want to talk the talk we must also walk the walk: The nature of science, professional development, and educational reform.Issues in Science Education: Professional Development Planning and Design. National Science Teachers Association, Arlington, VA.
- Stone EM. 2014. Guiding Students to Develop an Understanding of Scientific Inquiry: A Science Skills Approach to Instruction and Assessment. CBE—Life Sci Educ 13:90–101. https://doi.org/10.1187/cbe-12-11-0198.
- Tanner KD. 2013. Structure matters: twenty-one teaching strategies to promote student engagement and cultivate classroom equity. CBE Life Sci Educ 12:322–331. https://doi.org/10.1187/cbe.13-06-0115.
- Dewsbury B, Brame CJ. 2019. Inclusive Teaching. CBE—Life Sci Educ 18:fe2. https://doi.org/10.1187/cbe.19-01-0021.
- Haak DC, HilleRisLambers J, Pitre E, Freeman S. 2011. Increased Structure and Active Learning Reduce the Achievement Gap in Introductory Biology. Science 332:1213–1216. https://doi.org/10.1126/science.1204820.
- Skloot R. 2010. The immortal life of Henrietta Lacks. Crown Publishers, New York.
Article Files
Login to access supporting documents
Samsa-Single Cell Insights Into Cancer Transcriptomes.pdf(PDF | 341 KB)
S1. scRNAseq-scRNAseq Case Study Parts 1-5 - Student version.docx(DOCX | 613 KB)
S2. scRNAseq-scRNAseq Case Study Parts 1-5 - Answer key.docx(DOCX | 4 MB)
S3. scRNAseq-Part 1 The patient and diagnosis - Student version.docx(DOCX | 569 KB)
S4. scRNAseq-Part 2 The technician and the samples - Student version.docx(DOCX | 589 KB)
S5. scRNAseq-Part 3 Data processing - Student version.docx(DOCX | 567 KB)
S6. scRNAseq-Part 4 Data visualization - Student version.docx(DOCX | 570 KB)
S7. scRNAseq-Part 5 Treatment - Student version.docx(DOCX | 571 KB)
S8. scRNAseq-Part 1 The patient and diagnosis - Answer key.docx(DOCX | 571 KB)
S9. scRNAseq-Part 2 The technician and the samples - Answer key.docx(DOCX | 1 MB)
S10. scRNAseq-Part 3 Data processing - Answer key.docx(DOCX | 646 KB)
S11. scRNAseq-Part 4 Data visualization - Answer key.docx(DOCX | 3 MB)
S12. scRNAseq-Part 5 Treatment - Answer key.docx(DOCX | 674 KB)
S13. scRNAseq-File for Part 2 - Sequencing Metadata - Student version.xlsx(XLSX | 14 KB)
S14. scRNAseq-File for Part 2 - Sequencing Metadata - Instructor version.xlsx(XLSX | 15 KB)
S15. scRNAseq-File for Part 2 - Processing Datasheet - Student version.xlsx(XLSX | 11 KB)
S16. scRNAseq-File for Part 2 - Processing Datasheet - Instructor version.xlsx(XLSX | 17 KB)
S17. scRNAseq-File for Part 3 - Expression - Student version.csv(CSV | 6 MB)
S18. scRNAseq-File for Part 3 - Expression - Instructor version.csv(CSV | 6 MB)
S19. scRNAseq-File for Part 3 - Metadata - Student version.csv(CSV | 9 KB)
S20. scRNAseq-File for Part 3 - Metadata - Instructor version.csv(CSV | 11 KB)
S21. scRNAseq-File for Part 3 - Processing Notes - Student version.txt(TXT | 4 KB)
S22. scRNAseq-File for Part 3 - Processing Notes - Instructor version.txt(TXT | 4 KB)
S23. scRNAseq-File from Part 4 - Normalized Expression - Instructor version.csv(CSV | 22 KB)
S24. scRNAseq-File from Part 4 - Metadata with Clusters - Instructor version.csv(CSV | 10 KB)
S25. scRNAseq-File from Part 4 - DE PDX meta vs PDX primary - Instructor Version.csv(CSV | 1 MB)
S26. scRNAseq-File for Part 5 - Normalized Expression annotated for instructor.xlsx(XLSX | 16 MB)
- License terms

Comments
Comments
There are no comments on this resource.