Teaching the Central Dogma Using a Case Study of Genetic Variation in Cystic Fibrosis
Editor: Lisa McDonnell
Published online:
Abstract
The central dogma of biology is a foundational concept that is traditionally included in genetics curricula at all academic levels. Despite its ubiquitous presence throughout genetics education, students persistently struggle with both the fundamental and advanced topics of the central dogma. In particular, students conflate the role of genomic variations in DNA replication, transcription, and translation. As research and healthcare increasingly utilizes genomic medicine to link genetic variants to clinical phenotypes, it is critically important for biology and health science students to understand the role of genetic variation in molecular genetics. This lesson focuses on the role of missense mutations in the central dogma using a case study of cystic fibrosis. The case study is paired with a creative activity in which students draw the molecular parts of the central dogma. This helps students to connect the abstract concepts of the central dogma to a real-world clinical example. The effectiveness of this lesson was evaluated for two semesters of a Human Genetics course using end-of-unit exam questions. The active-learning lesson is an engaging activity that reinforces the role of genetic variation in the central dogma and the effects on clinical phenotypes. This lesson is highly customizable and adaptable to courses of various sizes, levels, course lengths, and teaching modalities.
Primary image: Molecular View of the Central Dogma. This drawing was produced by a student at Bloomsburg University’s Human Genetics course for Part 1 of this lesson.
Citation
Hare-Harris AE. 2021. Teaching the central dogma using a case study of genetic variation in cystic fibrosis. CourseSource. https://doi.org/10.24918/cs.2021.45
Society Learning Goals
Genetics
- Nature of Genetic Material
- What are the molecular components and mechanisms necessary to preserve and duplicate an organism’s genome?
- Molecular biology of gene function
- How is genetic information expressed so it affects an organism's structure and function?
- Genetic variation
- How do different types of mutations affect genes and the corresponding mRNAs and proteins?
Lesson Learning Goals
Students will
- describe the molecular steps of the central dogma in eukaryotes including DNA replication, transcription, and translation.
- demonstrate how errors in nucleotide base substitutions that are introduced during DNA replication persist throughout transcription and translation to affect protein sequences.
- appreciate the genetic heterogeneity associated with cystic fibrosis.
Lesson Learning Objectives
Students will be able to
- label the molecular parts of DNA replication.
- label the molecular parts of transcription.
- label the molecular parts of translation.
- transcribe and translate DNA to mRNA to protein sequence.
- evaluate consequences of point mutations.
- determine an amino acid change from a point mutation in an open reading frame.
- dissect genomic variant nomenclature.
- describe the five classes of genomic variants that cause cystic fibrosis.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
Introduction
Understanding how genetic information is stored, transferred, and utilized by the cell is a critical component for understanding the basis of human genetics as well as the molecular mechanisms of genetic disorders. The molecular mechanisms of the central dogma (DNA replication, transcription, and translation) are a universal topic among undergraduate genetics courses. These processes, which describe the transfer of genetic information from DNA to mRNA to proteins, are a prime example of the “information flow, exchange and storage” category of the five core concepts for cultivating biological literacy described by the American Association for the Advancement of Science’s (AAAS) report, Vision and Change: A Call to Action (1). With the increased use of genomic technologies, it is more crucial than ever for biology students to understand the information flow of the central dogma. Furthermore, as healthcare increasingly utilizes genomic medicine to link genetic variants to clinical phenotypes, it is critically important for health science students to understand the role of point mutations in molecular genetics.
While the central dogma is a core concept of genetics curricula, the linear model of gene expression that is typically used (DNA→RNA→protein) is difficult for students to conceptualize (2). Studies using concept mapping have shown that students often make inappropriate connections between DNA and RNA and fail to conceptualize the molecular meaning of the “arrows” in this model (2). For example, students may attribute the ribosome as the molecular complex that is responsible for transcription instead of the RNA polymerase. Furthermore, students also struggle when asked to explain the molecular consequences of DNA mutations in terms of the central dogma (2, 3). A number of studies have developed concept inventories to address the origin of these misconceptions about the effects of genetic mutations on information flow in the cell (4, 5). These inventories have revealed that students often conflate misconceptions on both the fundamental and more advanced concepts of the central dogma. For example, students often incorrectly contribute a nonsense mutation to the stoppage of transcription instead of translation (4). At the same time, these students will correctly describe the effects of a nonsense mutation on translation. Lessons that address the persistence of DNA variation throughout the central dogma (from DNA to protein) are necessary to aid in the correction of these misconceptions.
To address mixed models of student thinking, Pelletreau et al, 2016 developed an interactive in-class lesson to explore the effects of premature stop codons in the central dogma (6). This clicker-based activity utilized a case study of muscular dystrophy to provide clinical context for students to describe the molecular consequences of nonsense mutations (6). Students showed significant improvement in their understanding of the role of nonsense mutations in the stoppage of translation. Additionally, by employing a case study approach, students were more engaged in the lesson since it provided a real-world example of the central dogma in action. The use of case studies in genetics curricula helps students apply their knowledge of this somewhat abstract concept to a more tangible example of genomic medicine.
This lesson presented here uses a similar case study approach with a focus on the molecular consequences of missense mutations in cystic fibrosis. This lesson aims to teach the mechanics the central dogma by engaging all levels of Bloom’s Taxonomy (7) through a case study of cystic fibrosis. This lesson is composed of four parts. First, students draw and label the molecular mechanisms of DNA replication, transcription, and translation, engaging the create aspect of Bloom’s Taxonomy. Students are then given an exon sequence in the CFTR gene and the coding location of a point mutation that is known to cause cystic fibrosis. As part of the analyze and apply levels of Bloom’s Taxonomy, students transcribe and translate their assigned exon sequence with and without the genomic variant in order to identify the molecular consequence of the DNA variant on the final protein product. There are over 1,700 genetic variants in the CFTR gene that are known to cause cystic fibrosis (Cystic Fibrosis Foundation). Many of these variants are missense mutations that fall into one of five categories that relate the molecular consequence for the resulting CFTR protein (i.e., the protein is not made, the protein is misfolded, the channel gate is nonfunctional, etc.). Each category is targeted by different pharmacological treatment methods. Students research their genetic variant to determine to which class of CFTR variants it belongs and write a report describing the pharmacogenomic implications of this variant. Lastly, each student presents their drawings/reports to the class and peer reviews others’ work during an in-class gallery walk, engaging the evaluate, understand, and remember levels of Bloom’s Taxonomy.
Intended Audience
This lesson is intended as a capstone project to unify the concepts of a basic molecular genetics unit geared toward health science students. The lesson is primarily intended for and has been utilized in an introductory genetics course for health science students, but could be adapted to a general biology or more advanced genetics course at the instructor’s discretion.
Required Learning Time
The time that should be allotted for this lesson should include a 30 minute introductory lesson, a 45 minute in class working session, and a 50 minute in class gallery walk. Additional working sessions can be included at the instructor’s discretion. Students should supplement the classroom time with time spent drawing the central dogma parts. Depending on the level of the class, students may be asked to carry out literature searches and write the reflection paragraph outside of class. This assignment can be used to have students explore the role of pharmacogenomics in the treatment of cystic fibrosis.
Prerequisite Student Knowledge
Students will need a basic understanding of DNA structure, DNA replication, transcription, and translation to complete this task. This material should be covered in previous lectures and can be found in any standard genetics textbook. Students should also be familiar with gene structure, DNA coding sequences, differences between mRNA and DNA, and how to use a codon table. An example practice problem set is provided for students who need more experience with sequence analysis.
Prerequisite Teacher Knowledge
The instructor should be familiar with the molecular components of the central dogma including the initiation, elongation, and termination stages of DNA replication, as well as transcription, and translation. This information and general knowledge on the consequences of point mutations on amino acid sequences can also be found in standard genetics and molecular biology textbooks. Additionally, animated resources provided by the Cold Spring Harbor DNA Learning Center are excellent resources for students and instructors. A basic knowledge of sequence analysis, including transcription and translation processes, is needed for analysis of the CFTR exon sequence. The instructor should be able to interpret the nomenclature of a DNA sequence in a GenBank/FASTA format and basic genomic variant/amino acid change location nomenclature (i.e., c.G100A or p.R33X).
Scientific Teaching Themes
Active Learning
Students are actively engaged throughout the course of this lesson
- Out of class activities: central dogma drawing and labeling, pharmacogenomics literature search (for higher level course)
- In class activities: transcription/translation of exon, peer-reviewed gallery walk, Think-Pair-Share
Assessment
Instructors measured learning through a standard rubric to assess the accuracy of the assignment submission. Student learning was also assessed through a series of related examination questions. Prior to submission, students were able to assess their work via a peer-reviewed gallery walk, in which students presented their work and completed peer-review rubrics for others in the classroom.
Inclusive Teaching
This lesson supports the inclusion of all student participants by utilizing a multimodal approach to the learning activities. This lesson engages multiple senses and levels of Blooms Taxonomy via learning activities that involve drawing, writing, presentations, and analyzing data. This lesson has been adapted to accommodate students who cannot draw but are able to type or use talk-to-text communication devices.
In addition to engaging multiple types of students, the cystic fibrosis case study emphasizes diversity of genetic variation in genomic medicine. The prevalence of cystic fibrosis varies among different populations (1 in 3,500 in Caucasian Americans, 1 in 17,000 in African Americans, and 1 in 31,000 in Asian Americans) (8). Studies have shown that there are 1,700+ genetic variations that have been associated with cystic fibrosis, all with varying allele frequencies. Historically, the studies that classified these variant types were heavily skewed towards studies that focused on Caucasian populations. However, recent studies have shown that the frequency of these variants in non-Caucasian populations is variable (9). Furthermore, novel variants that are not present in Caucasian populations have been identified in non-Caucasian cystic fibrosis patients. Despite these recent findings, screening panels designed to test for cystic fibrosis variants are largely based upon the variants identified from the original studies of Caucasian populations (10). This lesson engages students in a discussion on the implications of study bias in medical research through an examination of the frequency of these variants in non-Caucasian populations.
Lesson Plan
The lesson is divided into four parts: Part 1) draw the molecular components of the central dogma, Part 2) transcribe and translate a CFTR exon with/without a genetic variant, Part 3) research and write a brief report of the assigned CFTR genetic variant, and Part 4) participate in a peer-reviewed gallery walk of the final project. For in person classes, students are provided an A3 size (11”x17” inches) paper on which students draw Part 1 on the front and Parts 2 and 3 on the back of the paper. For online classes, the project format can be more flexible, including both handwritten and computer- or tablet-generated projects as the instructor and students prefer. This project has been successfully completed in person and online. Table 1 provides the teaching suggested timeline.
Table 1. Cystic Fibrosis Case Study – Teaching Timeline
| Activity | Task | Description | Estimated Time | Notes/Suggestions |
|---|---|---|---|---|
| Pre-class Preparation | Preparation | Review materials | 15-20 minutes | If you are using this lesson for consecutive semesters, it is advised to shuffle the order of the sequence labels in the instructions to prevent academic dishonesty |
| Shuffle sequence labels | ||||
| Pre-assign Groups (if using) | ||||
| Class Day 1 | Case Study Introduction | In Class Lecture | 25 minutes | See Table 2 for lecture content |
| Assignment Instructions | Review assignment instructions and assign sequences/groups | 5 minutes | ||
| Work Session for Part 1 | Students outline the steps of the Central Dogma | 5-10 minutes | Drawings to be completed as an out of class assignment | |
| Work Session for Part 2 | Divide students into small groups | 35-40 minutes | Can have students work independently or in small groups (2-5 students) | |
| Transcription/Translation and Variant Identification | ||||
| Out of Class Assignment | Complete Assignment | Part 1 - Draw the Central Dogma | - | Students are given 2-3 weeks to complete the assignment |
| Part 3 - Research Variant Classification | ||||
| Class Day 2 | Gallery Walk | Students present their variant and peer review students with 4 other variant classifications | 50 minutes | Provide 3-4 days for students to incorporate peer feedback prior to final submission |
Pre-Class Preparation
Student preparation
Before this lesson, students should already have a working knowledge of the initiation, elongation, and termination steps of DNA replication, as well as transcription and translation. For this lesson, students are assigned pre-class lecture material and readings describing these steps. This material should include drawings/diagrams of each molecular step. The following textbooks have chapters devoted to this topic at an appropriate level for this activity: Genetics: Analysis & Principles (McGraw Hill; 7th Edition) and Human Molecular Genetics (CRC Press; 5th Edition). Students should also have a basic understanding of how to read and use a codon table. The content of the lecture and reading material is at the discretion of the instructor and should be tailored to the background level of the students.
Instructor preparation
The instructor should be familiar with the lesson material and be prepared to field questions regarding the transcription and translation of the exon sequence. Common student questions include: 1) whether the given exon sequence is the template or coding strand, 2) if the exon sequence is in frame to start, and 3) how to determine the amino acid change nomenclature. Since the classification of cystic fibrosis genomic variants is not commonly covered in traditional genetics textbooks, instructors may also wish to read the article “Types of CFTR Mutations” and the accompanying CFTR Mutation Classes Infographic (Supporting File S1. Cystic Fibrosis Case Study – CFTR Mutation Infographic; used with permission from the Cystic Fibrosis Foundation) to become familiar with the five classes of CFTR variants. Note that if this assignment is being used in consecutive semesters or among different course sections, it is advised to shuffle the order of the sequence labels in the student instructions to prevent academic dishonesty (Supporting File S2. Cystic Fibrosis Case Study – Student Instructions).
Day 1
In Class Lecture
The lesson begins with a brief overview of the steps of the central dogma as a review of previous lecture material. It is useful to emphasize that the mRNA and protein are the products and that the DNA coding sequence remains constant during transcription and translation. Next review the concept of a point mutation and the molecular consequences of a nonsense, missense, silent, and frameshift mutation on an amino acid sequence. Describe the basic clinical phenotype of cystic fibrosis and explain that there are 1700+ known genetic point mutations in the CFTR gene that cause cystic fibrosis. Depending on the level and set up of the class, you can either describe the function of the CFTR gene or have the class research the gene function in class. Have the students read the CFTR Mutation Classes Infographic describing the five classes of CFTR variants (Supporting File S1. Cystic Fibrosis Case Study – CFTR Mutation Infographic; used with permission from the Cystic Fibrosis Foundation). Discuss the molecular effects of each variant class on the CFTR protein. Table 2 lists the proposed in class lecture topics.
Table 2. Proposed In Class Lecture Topics
| Lecture Topic | Description | Time |
|---|---|---|
| Central Dogma Overview | Give an overview of the initiation, elongation, termination steps for DNA replication, transcription, and translation. | 5 minutes |
| Students should identify the template and product for DNA replication (DNA to DNA), transcription (DNA to mRNA), and translation (mRNA to protein). | ||
| Emphasize that the DNA sequence remains constant throughout the central dogma. | ||
| Emphasize the difference between the coding vs. the template strand during transcription. | ||
| Point mutations | Review the difference between a missense, nonsense, silent, and frameshift mutation. | 5 minutes |
| Cystic Fibrosis Introduction | Describe the symptoms of cystic fibrosis. | 5 minutes |
| Discuss the prevalence of cystic fibrosis among different populations. | ||
| Introduce the CFTR gene and its function. | ||
| Infographic | Discuss the five main classes of CFTR mutations and their effect on the CFTR protein. | 5 minutes |
| Review the CFTR mutation infographic. | ||
| Frequency Table | Discuss the historical bias in cystic fibrosis studies toward Caucasian populations. | 5 minutes |
| Using Table 3, demonstrate that the variants in the infographic have variable allele frequencies among different populations. | ||
| Use a Think-Pair-Share where students discuss the limitations of using a genetic screening tool that is based on data from one population in a diverse patient group. |
Following the review of the CFTR Mutation Classes Infographic, discuss the research behind the identification of cystic fibrosis variants. Historically, the studies that classified these variant types were heavily skewed towards studies that focused on Caucasian populations. However, recent studies have shown that the frequency of these variants in non-Caucasian populations is variable. Furthermore, novel variants that are not present in Caucasian populations have been identified in non-Caucasian cystic fibrosis patients. Despite these recent findings, screening panels designed to test for cystic fibrosis variants are largely based upon the variants identified from the original studies of Caucasian populations. The frequency of the variants among different populations of cystic fibrosis patients in the United States shown in the CFTR Mutation Classes Infographic are listed in Table 3. Instruct the students to examine Table 3 and discuss the distribution of these variants among these populations. Using a Think-Pair-Share approach, have the students discuss the limitations of using a genetic screening tool that is based on data from one population in a diverse patient group.
Table 3. Frequency of CFTR mutations in non-Caucasian populations of cystic fibrosis patients. (This table has been adapted from Scrijver et al., 2016 (9).
| Variant | Variant Class | Variant prevalence (% of CF population) among US cystic fibrosis patients | ||||
|---|---|---|---|---|---|---|
| Caucasian | Hispanic | African/African American | Asian/Asian American | Native American | ||
| G542X | I | 2.3% | 5.2% | 1.6% | 0.5% | 2.1% |
| R553X | I | 1.0% | 0.7% | 1.2% | 0.5% | 0.6% |
| W1282X | I | 1.5% | 0.8% | 0.4% | 1.1% | - |
| F580del | II | 73.4% | 54.0% | 46.5% | 43.4% | 65.2% |
| I507del | II | 0.4% | 1.3% | 0.4% | - | 0.2% |
| N1303K | II | 1.4% | 1.5% | 0.8% | 0.3% | 0.4% |
| S549N | III | - | 1.2% | 0.8% | 4.0% | 0.2% |
| D1152H | IV | 0.3% | 0.6% | - | - | - |
| G551D | IV | 2.6% | 0.7% | 1.4% | 1.1% | 0.8% |
| R117H | IV | 1.8% | 0.6% | 0.7% | - | 1.3% |
| R347P | IV | 0.2% | - | - | 1.3% | 0.2% |
| A455E | V | 0.3% | - | - | 0.8% | 0.8% |
Assignment Instructions and Work Session for Part 1
Explain the instructions for Part 1 of the lesson: drawing the central dogma (Supporting File S2. Cystic Fibrosis Case Study – Student Instructions). In class, ask the students to list the steps of the central dogma in order to identify the nine drawings that are required. There should be a total of nine diagrams at the completion of the project: initiation, elongation, and termination steps for DNA replication, transcription, and translation. Students work independently outside of class to draw and label the steps of the central dogma. Note that this lesson has been implemented in a 75-minute class session with ample time for a brief work session for Part 1 during class (Table 1). For 50-minute classes, instructors may wish to limit outlining the Central Dogma steps to the Case Study introduction to allow for more in class time for the work session for Part 2.
Work Session for Part 2
Divide the class into five groups (one for each exon sequence/CFTR variant class) and explain the lesson instructions for Parts 2 and 3. Groups are assigned randomly, however this can be adjusted according to the instructor’s discretion. Each group is given the coding sequence for an exon in the CFTR gene (sequences are listed in the Supporting File S2. Cystic Fibrosis Case Study – Student Instructions). Students will transcribe and translate the sequence to determine the amino acid sequence of that exon. Discuss the difference between the coding and the template strand of the DNA. Using the example sequence in the instructions, explain the exon sequence format to the students as follows: “Each exon sequence is the coding sequence of an exon and is shown in the 5’ to 3’ direction. The sequence is in frame to start and is in a standard GenBank format. The numbers on the left represent the coding position of the first base in the corresponding line. For example, if the number is 360, the first base on that line is found at position 360 in the coding sequence of the gene. There are 50 bases in each sequence line, so the last base in the first line is position 409.”
Each group is also given the position of a nucleotide base substitution in their exon that is known to cause cystic fibrosis (Supporting File S2. Cystic Fibrosis Case Study – Student Instructions). Explain how a nucleotide base substitution in a coding region creates a point mutation that can alter the resulting amino acid sequence. Then explain how to read the variant nomenclature using the instruction example T361C. In the example, the original T is changed to a C. This T is located at position 361, or the second base in the example sequence. The students will identify which base is to be changed, make the change, and transcribe/translate the new sequence to observe the consequence of the variant. The variant may cause either a missense or a nonsense mutation. Have each student report the amino acid change using standard nomenclature. Explain this process using the example variant as follows: “The original sequence ATC codes the amino acid isoleucine (I). The variant changes this sequence to ACC, resulting in a threonine (T). To determine the position of the amino acid change in the protein sequence, divide the variant position by three since there are three bases for each codon. If the number is not a whole number, round up to the next whole number. In this case, the amino acid position is 121. To write this variant in terms of amino acid change, you would write the original amino acid, the position, and the new amino acid (I121T). If your variant creates a nonsense mutation, the new amino acid is denoted with an X (i.e., I121X).”
Depending on the size and level of the class, students may work in groups or individually to determine their amino acid change during class. This assignment has been implemented in class sizes ranging from 25-50 students in both an in person and online environment. Groups of 4-5 students per group is ideal for larger classes. For online environments, breakout sessions should be utilized to facilitate group discussions. Once the student/group has determined their amino acid change, have the students determine to which CFTR variant class their variant belongs. The amino acid change is listed in the examples in the CFTR variant infographic (Supporting File S1. Cystic Fibrosis Case Study – CFTR Mutation Infographic). If the student’s variant is not listed in the infographic, then they made a mistake in their analysis or nomenclature. Common mistakes include assuming that the given exon is the template strand instead of the coding strand and using the nucleotide position instead of the amino acid position in the amino acid change nomenclature. The key for Part 2 and 3 is provided (Supporting File S3. Cystic Fibrosis Case Study – Instructor Key for Parts 2 and 3) and an example of a completed student submission is provided in Supporting File S4. Cystic Fibrosis Case Study – Example Student Submission.
Out-of-Class Assignment
Students will complete the remainder of this assignment outside of class. Students are given 2-3 weeks (depending on the length of the course) to complete the assignment outside of class. Part 1 of the assignment is to illustrate and label the central dogma by drawing the initiation, elongation, and termination steps of DNA replication, transcription, and translation. Each step and molecular component should be labeled clearly. A color coded key is the most commonly used method of labeling. Each step should be accompanied by a brief explanation of the step.
Part 3 of the assignment is to research the CFTR genetic variant class identified during the in-class portion of the lesson. The student should prepare a ½ page to 1 page report describing the effect that the genetic variant has on the CFTR protein and describe what medications have been developed to target this class of variants. Emphasis should be placed on the role of pharmacogenomics in the report. This exercise can be tailored to the level of the class to increase or decrease the difficulty as the instructor sees fit. This lesson can also be extended to include a comparison of allele frequencies among populations listed in the gnomAD databases to the allele frequencies listed in Table 3 for advanced classes. Likewise, advanced students may also wish to use the ClinVar database to assess the clinical evidence for pathogenicity for their variant. The gnomAD database is a resource of aggregated sequencing data from control samples from large genomic sequencing studies (11). Conversely, ClinVar is a repository of genomic variation identified in clinical patient samples (12). ClinVar and gnomAD accession numbers for each variant are listed in Supporting File S3. Cystic Fibrosis Case Study – Instructor Key for Parts 2 and 3.
Day 2
Gallery Walk
In a second class session, prior to turning the assignment in, hold a gallery walk for students to present and peer review each other’s final project. Assign students with different variant classes to peer review each other. Each student should present their project and peer review up to four different projects, one of each variant that is not their assigned variant/class. The number of peer reviews is determined by the number of students in the course and length of the class time. Allow 8 minutes per peer review and 1-2 minutes for transitions for a total of 50 minutes for all five variants (four peer reviews and their own presentation). Students are provided with the peer review rubric to complete for each student that they assess (Supporting File S5. Cystic Fibrosis Case Study – Peer Review Rubric). All completed rubrics are collected at the conclusion of the gallery walk and returned to the presenting students. This allows students to make revisions to their work based on their peer’s feedback prior to submission.
Student Assessment
An example of a correct student submission is provided in Supporting File S4. Cystic Fibrosis Case Study – Example Student Submission. Students provide feedback to each other using the provided peer review rubric in Supporting File S5. Cystic Fibrosis Case Study – Peer Review Rubric. The final assignment submission is graded using the rubric in Supporting File S6. Cystic Fibrosis Case Study – Grading Rubric, which can be altered as the instructor sees fit.
Teaching Discussion
Implementation of Lesson
This lesson has been implemented in the Human Genetics course at Bloomsburg University for five semesters. Assessment of the effectiveness of this lesson for two of these semesters is discussed below. On average, this course included 48 students (range 24-51 students) and has been taught in both an in person and online format. The lesson was originally designed for implementation in a 15-week in person course that met for 1 hour and 15 minutes twice a week. In this format, the lesson consisted of two dedicated class periods (one for group work/discussion of Part 2 and one for the gallery walk) and an out of classroom assignment (Parts 1 and 3). Prior to completing this assignment, students attended/viewed three lectures on DNA replication, transcription, and translation as part of the course sequence. In each cohort, students completed the assignment independently, but were encouraged to discuss their results with students who were assigned the same exon sequence for Part 2. The lesson has been adapted to an online format using Zoom breakout rooms to facilitate group discussions. This lesson has also been implemented in a 7-week accelerated format. Due to time constraints in the accelerated format, the gallery walk was omitted.
Each student cohort consisted primarily of second- and third- year health science students. A small portion of each cohort consists of health science students who are enrolled in the pre-Genetic Counseling/Medical Genomics advising track. Other student majors/advising tracks include pre-physician assistant, pre-physical therapy, biology, exercise science, and natural history. The overall goal of this lesson was to use a relatable case study of genomic variation to emphasize the steps of the central dogma for health science students. This lesson emphasizes the role of genomic medicine in diagnosing and treating cystic fibrosis. The medical case study helped to keep students engaged in the learning process and facilitated critical thinking. Students were able to relate the abstract concept of molecular genetics to a tangible outcome in pharmacogenomics.
Learning Outcomes from the Lesson
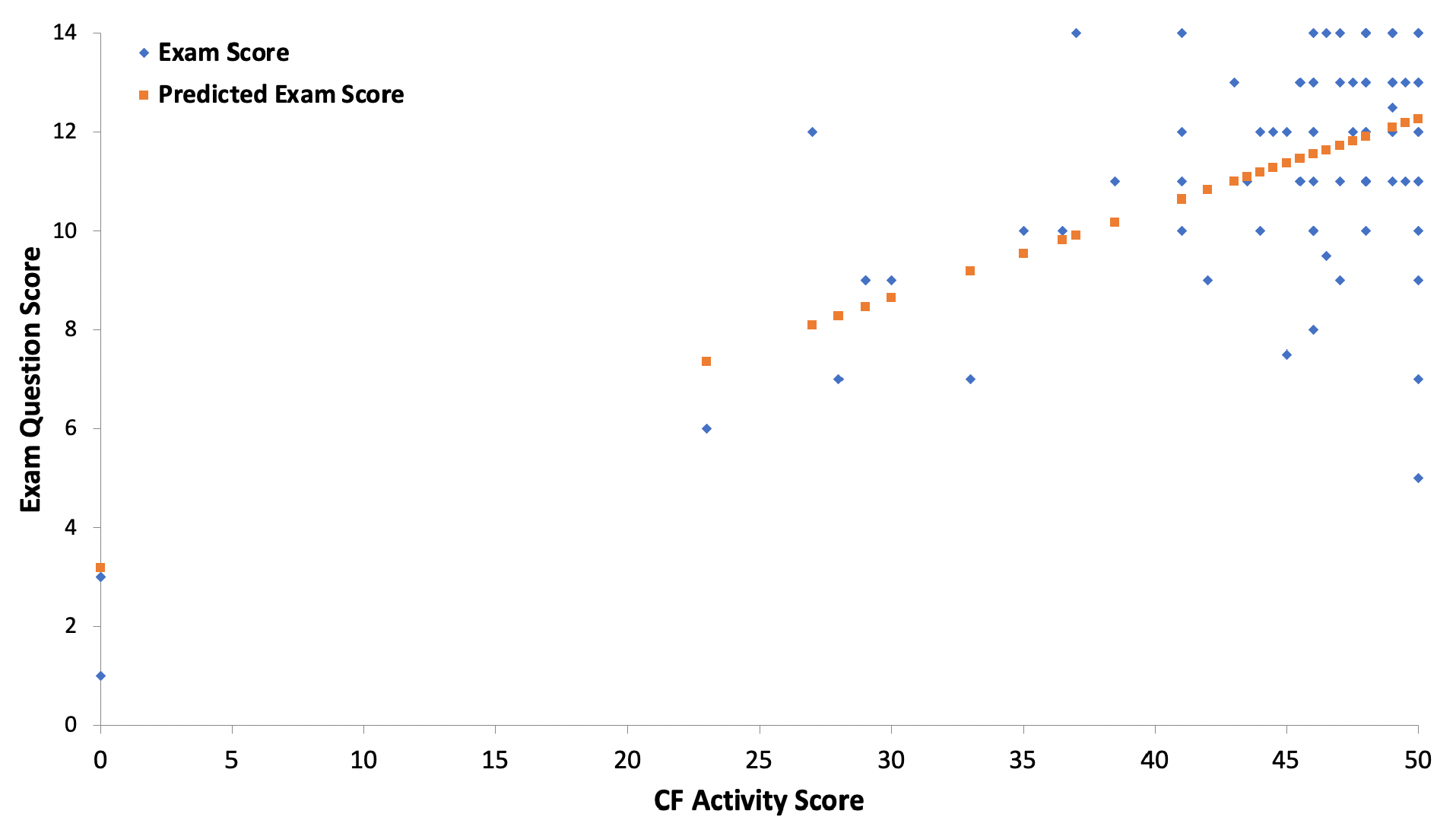
This lesson has been developed and implemented in the Human Genetics course at Bloomsburg University for five semesters. The lesson was primarily developed over the course of three semesters (Fall 2018, Spring 2019, and Fall 2019) and the effectiveness of this lesson was assessed using two sets of questions from the Unit 3 examination for the Spring 2020 and Fall 2020 semesters (Supporting File S7. Cystic Fibrosis Case Study – Examination Questions for Assessment). In these semesters, the course was taught online in a flipped classroom environment. As such, the students did not complete the gallery walk during these semesters. All students who completed the examination were included in this analysis (n=86 students). This examination occurred within one week of the final submission of the lesson. The first set of questions addressed the learning outcomes for labeling the molecular parts of the central dogma. The second set of questions addressed the learning outcomes regarding transcription, translation, and genomic variant consequences. Overall, performance on the lesson was highly correlated with student exam performance (R = 0.70; p<0.001; Figure 1). Within the exam, 72% of students correctly labeled the molecular parts of the central dogma. Students were proficient in identifying the steps of the central dogma, but struggled with the identification of specific molecular components, such as DNA polymerase δ vs. DNA polymerase ε. Overall, 86% of students correctly identified the molecular consequence of a genomic variant in a coding sequence. In this section, students excelled at identification of the type of point mutation (missense, nonsense, silent), with 96% of students correctly identifying the variant type in this question set. The most common mistakes in both question sets stemmed from not distinguishing between the coding and the template strand during transcription. Among the students who struggled with this concept, it was clear that students were able to translate a sequence correctly, but were unable to distinguish between the coding and template strand both visually and analytically, making their mRNA sequence incorrect.
Student Reactions to Lesson
Bloomsburg University students responded positively to the lesson in each implementation of this course. Student reflections were gathered as part of the overall course evaluation conducted at the end of each semester and through personal correspondence. As this class was geared toward a health science audience, students were most excited to use a “real world” application to see how genetics can be applied to real life scenarios in genomic medicine. Students also indicated that this lesson was effective in helping them review the material for the examination and many used the completion of this assignment as their primary study method. Students who were interested in pursuing careers in genomic medicine and/or genetic counseling were most interested in learning how to decipher the genetic code and to learn about the clinical consequences of genomic variations. The most common critique of the lesson was the time consuming nature of completing Part 1 outside of class. If this is a concern, suggestions for adaptations are discussed below.
Suggestions for Adaptations
The flexibility of this lesson allows for adaptation to courses of various sizes, levels, course lengths, and teaching modalities. At Bloomsburg University, the lesson has been implemented in its entirety for a sophomore-level Human Genetics course of 25-50 students. This lesson assumes basic knowledge of the central dogma and each part of this assignment may be adjusted to meet the level of the intended audience. For example, in larger or lower level classes, the instructor may wish to emphasize the molecular mechanisms in Part 1 and assign Parts 2 and 3 as a group project instead of an individual assignment. Alternatively, in upper level courses where students have a more advanced working knowledge of the central dogma, the instructor may wish to emphasize Part 2 in class and adjust Part 3 to be a more comprehensive, independent term paper. In this instance, instead of providing the Cystic Fibrosis Infographic (Supporting File S1. Cystic Fibrosis Case Study – CFTR Mutation Infographic), the students may be instructed to research the different classes of cystic fibrosis variants independently.
Instructors may also wish to instruct advanced students (seniors and/or graduate students) to compare the information in large genomic databases such as gnomAD and ClinVar to assess the pathogenicity of their given variant as part of a comprehensive term paper. The gnomAD database is a resource of aggregated sequencing data from control samples from large genomic sequencing studies (11), while, ClinVar is a repository of genomic variation identified in clinical patient samples (12). Instructors may wish to guide their students to focus on the ACMG guidelines for variant interpretation. Additionally, students can expand the discussion of the role of ascertainment bias in genetic studies of cystic fibrosis in Caucasian populations by incorporating the allele frequencies of their variant among different ancestral groups (9). The variant IDs for gnomAD and ClinVar are provided in Supporting File S3. Cystic Fibrosis Case Study – Instructor Key for Parts 2 and 3.
The lesson also has the flexibility to be implemented for courses of various teaching modalities. For in person iterations at Bloomsburg University, each student was given an A3 sized (11”x17”) paper for completion of the assignment (Part 1 is drawn on one side and Parts 2/3 are included on the other side of the paper). In online adaptations of this course, students were given more freedom in their medium choices. Many students opted to draw Part 1 using a tablet, such as an iPad, or a computer program such as Paint. Other students submitted photographs or scanned images of their hand-drawn mechanisms. In addition to adaptations to an online format, this lesson has also been implemented in an accelerated 7-week course format. In full 15-week semester iterations of the course, two full lectures were dedicated for working on Part 1 and 2 of the lesson, one 50-minute lecture was dedicated to the gallery walk, and Part 3 was completed outside of the classroom. If time constraints are a concern, each part of this lesson can be conducted as standalone assignments and the in-class portions can be adjusted as the instructor sees fit. For example, in the accelerated format at Bloomsburg University, Parts 1 and 3 were completed as an out of class assignment, Part 2 was completed in class in small groups, and the gallery walk was not implemented.
This assignment can also be adapted to accommodate students with a variety of disabilities that prohibit the student from completing the assignment as written. For example, students who are unable to draw may be provided a set of unlabeled Central Dogma steps (Supporting File S8. Cystic Fibrosis Case Study – Alternative Student Instructions and Supporting File S9. Cystic Fibrosis Case Study – Alternative Student Assignment Slides). The student places the slides in order and labels each molecular component electronically.
Conclusions
This lesson is an engaging, adaptable classroom activity that reinforces the role of genetic variation in the central dogma and the effects on clinical phenotypes. This activity helps to connect the abstract concepts of the central dogma to a real-world clinical example using a case study of cystic fibrosis. Using drawings, sequence analysis, group discussions, essay writing, and a gallery walk, students are engaged in active learning throughout the process while engaging each level of Bloom’s Taxonomy. This lesson is highly customizable and adaptable to courses of various sizes, levels, course lengths, and teaching modalities.
SUPPORTING MATERIALS
- S1. Cystic Fibrosis Case Study – CFTR Mutation Infographic
- S2. Cystic Fibrosis Case Study – Student Instructions
- S3. Cystic Fibrosis Case Study – Instructor Key for Parts 2 and 3
- S4. Cystic Fibrosis Case Study – Example Student Submission
- S5. Cystic Fibrosis Case Study – Peer Review Rubric
- S6. Cystic Fibrosis Case Study – Grading Rubric
- S7. Cystic Fibrosis Case Study – Examination Questions for Assessment
- S8. Cystic Fibrosis Case Study – Alternative Student Instructions
- S9. Cystic Fibrosis Case Study – Alternative Student Assignment Slides
Acknowledgments
The author would like to acknowledge Ms. Katelyn Kelchner (Bloomsburg University) for contributing the primary image and example student work for this lesson. The assessment protocol of this lesson was approved by the Bloomsburg University IRB (BU-IRB #2021-1).
References
- AAAS. 2009. Vision & change in undergraduate biology education. AAAS, Washington, DC.
- Kate Wright L, Nick Fisk J, Newman DL. 2014. DNA→RNA: What do students think the arrow means? CBE Life Sci Educ 13:338–348.
- Briggs AG, Morgan SK, Sanderson SK, Schulting MC, Wieseman LJ. 2016. Tracking the resolution of student misconceptions about the central dogma of molecular biology. J Microbiol Biol Educ 17:339–350.
- Smith M, Knight J. 2012. Using the Genetics Concepts Assessment to document persistent conceptual difficulties in undergraduate genetics courses. Genetics 191:21–32.
- Prevost LB, Smith MK, Knight JK. 2016. Using student writing and lexical analysis to reveal student thinking about the role of stop codons in the central dogma. CBE Life Sci Educ 15:1–13.
- Pelletreau KN, Andrews T, Armstrong N, Bedell MA, Dastoor F, Dean N, Erster S, Fata-Hartley C, Guild N, Greig H, Hall D, Knight JK, Koslowsky D, Lemons P, Martin J, McCourt J, Merrill J, Moscarella R, Nehm R, Northington R, Olsen B, Prevost L, Stolzfus J, Urban-Lurain M, Smith MK. 2016. A clicker-based case study that untangles student thinking about the processes in the central dogma. CourseSource 3.
- Bloom B. 1956. Taxonomy of educational objectives, Handbook I: The cognitive domain. David McKay Co Inc, New York.
- Genetics Home Reference. Cystic Fibrosis. MedlinePlus.
- Schrijver I, Pique L, Graham S, Pearl M, Cherry A, Kharrazi M. 2016. The spectrum of CFTR variants in nonwhite cystic fibrosis patients. J Mol Diagnostics 18:39–50.
- Hughes EE, Stevens CF, Saavedra-Matiz CA, Tavakoli NP, Krein LM, Parker A, Zhang Z, Maloney B, Vogel B, DeCelie-Germana J, Kier C, Anbar RD, Berdella MN, Comber PG, Dozor AJ, Goetz DM, Guida L, Kattan M, Ting A, Voter KZ, van Roey P, Caggana M, Kay DM, Cutting GR, Bashwinger S, DuJack H, Langfelder-Schwind E, Young KG, Bonforte RJ, Quinones Y, Reyes N, Boyer J, Welter J, Westby D, French T, Jacobs K, Fortner C, Soultan Z, Forell M, Grabowski L, Rinn A, Bonitz L, Carney T, Ren C, Platt M, Borowitz D, Harris M, Fisher L, Ozarowski AL, Schaefer AM, Gomez J, Pena A, Peters L, Giusti R, Mavaro C. 2016. Clinical sensitivity of cystic fibrosis mutation panels in a diverse population. Hum Mutat 37:201–208.
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, Walters RK, Tashman K, Farjoun Y, Banks E, Poterba T, Wang A, Seed C, Whiffin N, Chong JX, Samocha KE, Pierce-Hoffman E, Zappala Z, O’Donnell-Luria AH, Minikel EV, Weisburd B, Lek M, Ware JS, Vittal C, Armean IM, Bergelson L, Cibulskis K, Connolly KM, Covarrubias M, Donnelly S, Ferriera S, Gabriel S, Gentry J, Gupta N, Jeandet T, Kaplan D, Llanwarne C, Munshi R, Novod S, Petrillo N, Roazen D, Ruano-Rubio V, Saltzman A, Schleicher M, Soto J, Tibbetts K, Tolonen C, Wade G, Talkowski ME, Aguilar Salinas CA, Ahmad T, Albert CM, Ardissino D, Atzmon G, Barnard J, Beaugerie L, Benjamin EJ, Boehnke M, Bonnycastle LL, Bottinger EP, Bowden DW, Bown MJ, Chambers JC, Chan JC, Chasman D, Cho J, Chung MK, Cohen B, Correa A, Dabelea D, Daly MJ, Darbar D, Duggirala R, Dupuis J, Ellinor PT, Elosua R, Erdmann J, Esko T, Färkkilä M, Florez J, Franke A, Getz G, Glaser B, Glatt SJ, Goldstein D, Gonzalez C, Groop L, Haiman C, Hanis C, Harms M, Hiltunen M, Holi MM, Hultman CM, Kallela M, Kaprio J, Kathiresan S, Kim BJ, Kim YJ, Kirov G, Kooner J, Koskinen S, Krumholz HM, Kugathasan S, Kwak SH, Laakso M, Lehtimäki T, Loos RJF, Lubitz SA, Ma RCW, MacArthur DG, Marrugat J, Mattila KM, McCarroll S, McCarthy MI, McGovern D, McPherson R, Meigs JB, Melander O, Metspalu A, Neale BM, Nilsson PM, O’Donovan MC, Ongur D, Orozco L, Owen MJ, Palmer CNA, Palotie A, Park KS, Pato C, Pulver AE, Rahman N, Remes AM, Rioux JD, Ripatti S, Roden DM, Saleheen D, Salomaa V, Samani NJ, Scharf J, Schunkert H, Shoemaker MB, Sklar P, Soininen H, Sokol H, Spector T, Sullivan PF, Suvisaari J, Tai ES, Teo YY, Tiinamaija T, Tsuang M, Turner D, Tusie-Luna T, Vartiainen E, Ware JS, Watkins H, Weersma RK, Wessman M, Wilson JG, Xavier RJ, Neale BM, Daly MJ, MacArthur DG. 2020. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581:434–443.
- Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. 2014. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42:980–985.
Article Files
Login to access supporting documents
Hare-Harris-Teaching central dogma using a case study.pdf(PDF | 273 KB)
S1. Cystic Fibrosis Case Study - CFTR Mutation Infographic.pdf(PDF | 938 KB)
S2. Cystic Fibrosis Case Study - Student Instructions.docx(DOCX | 22 KB)
S3. Cystic Fibrosis Case Study - Instructor Key to Parts 2 and 3.docx(DOCX | 30 KB)
S4. Cystic Fibrosis Case Study - Example Student Submission.pdf(PDF | 4 MB)
S5. Cystic Fibrosis Case Study - Peer Review Rubric.docx(DOCX | 13 KB)
S6. Cystic Fibrosis Case Study - Grading Rubric.xlsx(XLSX | 10 KB)
S7. Cystic Fibrosis Case Study - Examination Questions for Assessment.pdf(PDF | 1 MB)
S8. Cystic Fibrosis Case Study - Alternative Student Instructions.docx(DOCX | 22 KB)
S9. Cystic Fibrosis Case Study - Alternative Student Assignment Slides.pptx(PPTX | 1 MB)
- License terms

Comments
Comments
Comment removed by original commenter @ on (Edited: @ on )