Splicing it Together: Using Primary Data to Explore RNA Splicing and Gene Expression in Large-Lecture Introductory Biology
Editor: Katie Burnette
Published online:
Abstract
At the heart of scientific ways of knowing is the systematic collection and analysis of data, which is then used to propose an explanation of how the world works. In this two-day module, students in a large-lecture course are immersed in a biological problem related to the Central Dogma and gene expression. Specifically, students interpret experimental data in small groups, and then use those data to craft a scientific argument to explain how alternative splicing of a transcription factor gene may contribute to human cancer. Prior to the module, students are assigned a reading and provided PowerPoint slides outlining the basics of alternative splicing and refreshing their understanding of gene regulation. Students complete a pre-class assignment designed to reinforce basic terminology and prepare them for interpreting scientific models. Each day of the module, students are presented experimental data or biological models which they interpret in small groups, use to vote for viable hypotheses using clickers, and ultimately leverage in a culminating summary writing task requiring them to craft a data-driven answer to the biological problem. Despite the novelty of the argumentation module, students engage in all aspects (inside and outside of the classroom) of the activity and are connected across data, hypotheses, and course concepts to explain the role of alternative splicing in gene expression and cancer.
Primary image: Splicing it together. Students work together, interpreting primary data and models to investigate the effects alternative splicing may have on gene regulation and cancer.
Citation
Arneson JB, Woodbury J, Anderson J, Collins LB, Cavagnetto A, Davis WB, Offerdahl EG. 2022. Splicing it together: Using primary data to explore RNA splicing and gene expression in large-lecture introductory biology. CourseSource. https://doi.org/10.24918/cs.2022.11Society Learning Goals
Biochemistry and Molecular Biology
- Information storage and flow are dynamic and interactive
- How does the nucleotide sequence of the gene lead to biological function?
Cell Biology
- Cellular Specialization
- How can and why do cells with the same genomes have different structures and functions?
Genetics
- Gene Expression and Regulation
- How can gene activity be altered in the absence of DNA changes?
Lesson Learning Goals
Students will:- understand how multiple proteins can be produced from a single mRNA.
- connect changes in transcription factor expression to changes in gene regulation and cellular activity.
- gain experience with interpreting data and hypothesis-testing.
- understand the importance of alternative splicing in the gene regulation.
Lesson Learning Objectives
Students will be able to:- extract information from scientific models and graphical data.
- use scientific data to propose and evaluate hypotheses.
- predict the functions of proteins from different mRNA splice variants.
- identify transcription factors as products of translation and as regulators of transcription.
- describe the role of alternative splicing in gene expression within the context of human cancer.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
Introduction
There is widespread agreement that undergraduate life sciences education should develop students’ understanding of core disciplinary ideas as well their competencies with disciplinary practices (1-3). Accordingly, we designed this lesson to develop students’ competency in ideas related to information flow, exchange and storage by engaging in the disciplinary practice of scientific argumentation.
Biologists engage in argumentation every day. Every lab group meeting, research presentation, and peer-reviewed article is a form of argumentation whereby a biologist uses data to test hypotheses and make claims about the natural world. For the purposes of this lesson, we are referring to argumentation as the process of negotiating meaning as opposed to the product of the process. When applied in the classroom, scientific argumentation is a practice through which students gain deeper understanding of core disciplinary ideas (4). Indeed, developing skills necessary to “interpret, evaluate, and draw conclusions from data in order to make evidence-based arguments about the natural world” has recently been identified as a desirable learning outcome for undergraduate biology (3).
Previous efforts to engage biology students in scientific argumentation most often targeted K-12 students (e.g., 5, 6) or smaller class sizes (e.g., 7). At our university, introductory biology is a two-semester series with large enrollments (~500 students) taught in a fixed-seating auditorium. These environments have traditionally been viewed as a challenge for implementing active, student-centered pedagogies like argumentation (8). Therefore, our goal was to design an activity suitable for use in large-lecture classrooms that aligns with current initiatives in undergraduate STEM education by providing students with opportunities to develop core competencies through disciplinary practice (i.e., argumentation). Using an argumentation-to-learn framework (4), we created an argumentation module that asks students to interpret real data and craft an evidence-based argument in response to a big biological question (i.e., How does alternative splicing affect gene expression and regulation in cancer?).
The central dogma of molecular biology is a fundamental principle for biology courses (1) yet is notoriously difficult for students to learn (e.g., 9, 10). Understanding the central dogma requires knowing how interactions at the molecular level (i.e., protein binding to DNA) within a cellular context (i.e., nucleus, cell) translate to visible effects at the “macro” or organismal level (11). Research suggests that students' difficulties with central dogma are related to challenges in reasoning across levels of biological organization (12, 13). Moreover, reasoning about central dogma is difficult because it requires making sense of entities and processes that are small, hidden, or otherwise unperceivable through direct experience (14, 15).
Our own teaching experiences reflected the research on student learning of the central dogma. Students readily displayed basic knowledge of individual processes (e.g., transcription, translation, alternative splicing) but struggled to demonstrate more complex understanding of the relationships between processes and the link to phenotypic variation. For example, we noticed that students really grappled with the idea of transcription factors as both a product and regulator of gene expression. We thought this example was a prime opportunity to engage students in reasoning across levels of biological organization and that an argumentation-based module could help them connect what is too small to be seen with something that can be directly measured. Since readily available lessons on gene expression did not fit our needs – activities either focused solely on the consequences of changes made within the gene sequence itself (e.g., 16, 17), do not promote student consensus-seeking, or require computer access for all students (18) – we turned to primary literature to develop a novel argumentation module. Studies investigating how mRNA splice variants of a transcription factor gene led to variations in cancer tumor growth and proliferation were ideal for this activity because they require students to connect seemingly independent processes of transcription, translation, and alternative splicing to an observable phenomenon.
Intended Audience
We designed this lesson for the molecular biology and genetics semester of an undergraduate large-enrollment (~500 students) introductory biology course at a research-intensive, land-grant university. This course serves a variety of majors and pre-professional programs and consists primarily of science undergraduates ranging from freshmen to seniors.
This lesson was designed to reinforce and develop students’ understanding of introductory biology concepts by engaging them in the disciplinary practices of data interpretation and scientific argumentation. The lesson is intended to (a) reinforce prior instruction on central dogma and regulation of gene expression, (b) introduce the concept of alternative splicing, and (c) further develop student understanding of the relationship between genotype and phenotype. We designed the lesson for introductory biology students but believe the lesson could be adapted for use in sophomore or junior-level molecular and cellular biology courses. We iteratively implemented this lesson in a fixed-seating lecture hall with success but believe this lesson is particularly well suited for flexible classroom environments.
Required Learning Time
We designed this module to span two 50-minute class periods. Students completed online homework assignments before the first day to prepare for the module and after each class period to summarize their understanding of the data. Homework assignments were designed to take 15-30 minutes each.
Prerequisite Student Knowledge
This lesson took place after students had already been tested on the components and processes of transcription and translation, so students should be familiar with how information is transferred from DNA to mRNA to proteins and understand that changes in gene expression may lead to changes in protein structure and function. Further, students should be familiar with the function of transcription factors as regulators of transcription. Prior to the lesson, students are expected to review a slide packet that (a) includes information on mRNA processing, regulation of gene expression, and the hallmarks of cancer, and (b) introduces the overarching question students will use the data to answer (Supporting File S1. Splicing It Together - Pre-Class Background Slides). Students also complete a pre-class quiz (Supporting File S2. Splicing It Together - Pre-Quiz) which assesses basic understanding of the background information and provides an opportunity to interpret a simplified version of a schematic they will encounter during the lesson. Students should have some prior experience in extracting information from bar graphs and box-and-whisker plots.
Prerequisite Teacher Knowledge
Instructors should have a basic understanding of the transfer of information from DNA to proteins, with a particular emphasis on regulation of gene expression and alternative splicing. Background information for this section can be found in any introductory biology textbook (see for example, Chapter 9 of the Open Source textbook Concepts of Biology).
The lesson was based upon published research investigating the role of tumor suppressor genes on human disease. Specifically, we drew on research on Wilms’ tumor suppressor gene because it has several splice variants that exhibit different effects depending on the tissue type. Instructors should have a general understanding of the role of splice variants in human disease as well as the original studies from which the primary data for this lesson were extracted. For this activity, we named the gene WC1 to avoid confusion with a previous activity in which WT referred to the wild-type gene.
Data Set #1 was based on this paper:
Yamanouchi, K., Ohta, T., Liu, Z., Oji, Y., Sugiyama, H., Shridhar, V., ... & Kurachi, H. (2014). The Wilms' Tumor Gene WT1− 17AA/− KTS Splice Variant Increases Tumorigenic Activity Through Up-Regulation of Vascular Endothelial Growth Factor in an In Vivo Ovarian Cancer Model. Translational oncology, 7(5), 580-589.
Data Sets #2 and #3 were based on this paper:
Kramarzova, K., Stuchly, J., Willasch, A., Gruhn, B., Schwarz, J., Cermak, J., ... & Boublikova, L. (2012). Real-time PCR quantification of major Wilms’ tumor gene 1 (WT1) isoforms in acute myeloid leukemia, their characteristic expression patterns and possible functional consequences. Leukemia, 26(9), 2086-2095.
Scientific Teaching Themes
Active Learning
High-structure course designs coupled with student-centered, active learning pedagogies have been demonstrated to lower failure rates in introductory biology (19). This argumentation module reflects a high-structure design in that students are provided pre-class materials and complete a pre-class quiz. In class, students work collaboratively in small groups to (a) complete data interpretation tasks and (b) formulate a written response to make preliminary claims about what the data mean. After each data interpretation task in class, students individually answer a clicker question about the data. Students then discuss the distribution of clicker responses in their small groups and revise their arguments accordingly. Finally, the instructor calls on groups to share their ideas during whole-class discussion of the big question that frames the argumentation module.
Assessment
We used a variety of assessments (see below) to diagnose student learning throughout the lesson. Most of the assessments were formative assessments, namely to reveal evidence of students’ in-progress learning for the purposes of revising instruction and providing feedback to students (20). For example, the instructor would review the pre-class quiz to determine whether or not additional class time would be needed to teach students how to interpret the alternative splicing schematics or to review content from a prior class about how transcription factors work. Clicker questions served a formative role for both the instructor and the students; they were designed to facilitate further inspection of the data or to redirect attention to the Big Question. Our design choice of including a summary writing task was informed by the science writing heuristic research (21) which encourages students’ individual consolidation of knowledge. So, the summary writing task was used both as a learning task for the individual student as much as a formative assessment for the instructor to determine if more time was needed. Only the exam data were used as a summative assessment of student learning.
-
Multiple-choice quiz (pre-class homework; Supporting File S2. Splicing It Together - Pre-Quiz) - assesses students’ proficiency with the basic biology content underlying the argumentation module (e.g., alternative splicing, transcription factors, activators, repressors) as well as their basic ability to interpret the results of biological assays (e.g., Western blots).
-
Data interpretation questions (in class; Supporting File S3. Splicing It Together - Day 1 Handouts & Supporting File S4. Splicing It Together - Day 2 Handout) - figures from the primary literature are presented to students along with guiding questions to support data interpretation.
-
Clicker questions (in-class; Supporting File S5. Splicing It Together - Day 1 Slides and Clickers & Supporting File S6. Splicing It Together - Day 2 Slides and Clickers) - students use the data to individually evaluate hypotheses presented as clicker choices.
-
Data synthesis question (homework after Day 1; Supporting File S5. Splicing It Together - Day 1 Slides and Clickers) - students individually review the figures presented in class and submit a written synthesis.
-
Summary writing task (homework after Day 2; Supporting File S6. Splicing It Together - Day 2 Slides and Clickers) - students use data from both days of the argumentation module to individually complete a summary writing task asking them to use data to propose an answer to the “Big Question”.
-
Exam questions (Supporting File S7. Splicing It Together - End of Unit Exam Questions) – multiple-choice exam questions assessing (1) conceptual understanding of alternative splicing and gene expression and (2) simple data interpretation.
Inclusive Teaching
By having students work together in small groups, we are encouraging more students to participate in the material than in a traditional lecture environment. We embedded the argumentation module within the context of a big question with real-life relevance to increase student motivation and participation. There is no single right answer that emerges from the data which invites groups to privilege sharing of different ideas. We also used multiple and varied modes of low-stakes assessments which allows learners to display diverse ideas and ways of knowing. By inviting small groups to share out to the whole class, the instructor is creating opportunities for students’ cultural funds of knowledge to gain visibility. This is particularly important for underrepresented students as without such opportunities there could be culturally driven interpretations of the biology information that may be invisible to the instructor.
Lesson Plan
Course Context
This lesson (Table 1) was integrated into the molecular and cellular biology and genetics semester of a two-semester introductory biology course for majors. The course format is generally lecture punctuated with clicker questions and whole-class discussion. The instructor delivers the course in an amphitheater-style classroom with typical enrollment of ~450 students. We implemented this lesson in spring and fall of 2019 and spring of 2020 and used a design-based research approach (22) to iteratively refine the materials each semester.
Table 1. Argumentation module implementation outline. This activity spans two 50-minute class periods with pre- and post-activity assignments but could be adapted for longer class periods or longer than two days.
| Activity | Description | Estimated Time | Notes |
|---|---|---|---|
| Preparation for Class Day 1 | |||
| Instructor Preparation |
1. Post slide packet and pre-class quiz (S1. Splicing it together - Pre-Class Background Slides and S2. Splicing it together - Pre-Quiz) 2. Print Data Sets 1 and 2 (S3. Splicing it together - Day 1 Handouts) 3. Review papers on Wilms’ Tumor gene expression |
2-4 hours, depending on size of class | Count the number of seats in each row and collate print outs for quick distribution. |
| Student Pre-Class Preparation |
1. Review slide packet with background information 2. Complete pre-class quiz |
1-2 hours | |
| In-Class Day 1 | |||
| Group Formation | Instruct students to self-assemble into small groups | <2 minutes | Project instructions before class to reduce time needed |
| Introduce the Argumentation Module | Discuss background information from the slides. Contextualize and introduce the “Big Question” | 5 minutes | |
| Data Interpretation 1 | Display and distribute Data Set 1. Students work in groups to answer Data Set 1 questions. | 10 minutes | Data Set 1 on worksheet found in S3. Splicing it together - Day 1 Handouts. |
| Clicker Question 1 & Follow-Up |
1. Present Clicker Question 1. 2. Poll class and ask a few students to share reasoning. |
2-3 minutes for CQ + 7-8 minutes to discuss | CQ 1 found in S5. Splicing it together - Day 1 Slides & Clickers |
| Data Interpretation 2 | Display and distribute Data Set 2. Students work in groups to answer the questions about Data Set 2. | 10 minutes | |
| Clicker Question 2 & Follow-Up |
1. Present Clicker Question 2. 2. Poll class and elicit student reasoning in whole class discussion. |
2-3 minutes for CQ + 7-8 minutes to discuss |
CQ 2 found in S5. Splicing it together - Day 1 Slides & Clickers. Students should discuss how they could rule out the other hypotheses. |
| Day 1 Wrap-Up |
1. Summarize students’ data interpretation and hypothesis-testing 2. Introduce the homework for Day 2 |
3 minutes | |
| Preparation for Class Day 2 | |||
| Instructor Preparation |
1. Post Pre-Class Homework 2. Print out and prepare Data Set handouts (S4. Splicing it together - Day 2 Handouts) 3. Review student pre-class answers to gather themes for in-class discussion. |
2-4 hours | Homework prompt found in S5. Splicing it together - Day 1 Slides & Clickers. |
| Student Pre-Class Preparation |
1. Review figures from Data Activities 1 and 2. 2. Respond to essay prompt. |
15-30 minutes | Essay prompt is found on final slide in S5. Splicing it together - Day 1 Slides & Clickers. |
| In-Class Day 2 | |||
| Introduction |
1. Instruct students to re-form their groups from Day 1 2. Review Day 1 activities and class conclusions. 3. Highlight important themes observed in the pre-class homework. |
10 minutes | |
| Data Set 3 | Display and distribute Data Set 3. Students work in groups to answer the questions about Data Set 3. | 10+ minutes | |
| Clicker Question 3 & follow-up |
1. Present Clicker Question 3. 2. Poll class and ask students to share their reasoning. |
2-3 minutes for CQ + 7-8 minutes for discussion | CQ3 is found in S6. Splicing it together – Day 2 Slides & Clickers. |
| Answering the Big Question | Display the Big Question and lead class discussion | 15 minutes | |
| Wrap-up | Inform students of summary writing activity and wrap-up the activity. | 5 minutes | |
| Post-Argumentation Activity | |||
| Summary Student Activity | Students use Data Sets 1-3 to respond to the Big Question | 15-30 minutes | Essay prompt can be found in S6. Splicing it together – Day 2 Slides & Clickers. |
Pre-Class Day 1
Pre-class materials and quiz were designed to introduce students to the biology topic of the day. The materials summarize key ideas such as transcription factors, gene expression, and cancer that students need for interpreting the data in class (Supporting File S1. Splicing It Together - Pre-Class Background Slides). Prior to class, students should take a quiz on the pre-class materials (Supporting File S2. Splicing It Together - Pre-Quiz). The pre-class quiz reinforces students’ understanding of the experimental techniques and graphs that they will be interpreting in-class.
In Class Day 1
Introduction
As students enter the class, instructors should tell them to form small groups of 2-4 students. A PowerPoint slide can be used to broadcast instructions to the whole class (Supporting File S5. Splicing It Together - Day 1 Slides and Clickers). Once students are organized into groups, the instructor should introduce the Big Question (“How does alternative splicing of WC1 pre-mRNA affect gene expression and regulation in cancer?”) and explain that students will act as members of the scientific community to investigate that question. Specifically, they will be expected to interpret data, make predictions, evaluate hypotheses, and seek alternative explanations.
Data Set 1
As Data Set 1 is handed out, the instructor should emphasize the importance of coming to a group consensus for each prompt and recording those ideas on their handouts or notes. The figures should also be projected on the screen so students can start thinking about the task without waiting to receive their physical copy. Students work as a group to unpack the information in the first figure and use it to address the prompt:
How would you explain the effect of WC1 transcription factor A on ovarian tumor growth?
During this time, the instructor and learning assistants, if available, should move through the classroom to answer questions, clarify potential confusion, and develop an idea of what the students are paying attention to in the data or background materials. Groups should be able to work through Data Set 1 in about 10 minutes, but additional time can be granted if many groups are still discussing the data.
Next, the instructor should present Clicker Question 1 to the class (Supporting File S5. Splicing It Together - Day 1 Slides and Clickers) to be answered individually by students.
Which do you think best explains how WC1 Transcription Factor A affects ovarian tumor growth?
-
Transcription Factor A activates expression of the VEGF gene.
-
Transcription Factor A represses expression of the VEGF gene.
-
Transcription Factor A activates expression of another protein, which is an activator of VEGF expression.
-
Transcription Factor A represses expression of another protein, which is a repressor of VEGF expression.
At this point, one explanation (Answer B) can be ruled out, so responses will be split between the remaining options. After revealing the distribution, ask for volunteers or call on students to share their reasoning with the class. The instructor could seek input on each clicker option or use ideas collected when engaging with student groups during Data Set 1 to spark additional discussion. In our last implementation, the instructor also diagrammed a hypothesis on the overhead (see Slide 8 in Supporting File S5. Splicing It Together - Day 1 Slides and Clickers) and suggested students could sketch their own models to help visualize the gene regulation events described. We did not require students to do so, nor did we collect any student sketches.
Data Set 2
After evaluating the first round of hypotheses, distribute and display Data Set 2. Again, instructors and learning assistants should move through the classroom while students spend approximately 10 minutes working through the new data to answer the following question (Supporting File S3. Splicing It Together - Day 1 Handouts):
Though only one protein product is shown in the schematic, each of the four mature mRNAs are translated into protein. Based on Figure 2, how do you think each of the four proteins function in the cell?
Next, the instructor should present Clicker Question 2 (Supporting File S5. Splicing It Together - Day 1 Slides and Clickers):
Given information in Data Sets 1 and 2, which hypothesis do you think best explains how WC1 Transcription Factor A affects ovarian tumor growth?
-
Transcription Factor A activates expression of the VEGF gene.
-
Transcription Factor A represses expression of the VEGF gene.
-
Transcription Factor A activates expression of another protein, which is an activator of VEGF expression.
-
Transcription Factor A represses expression of another protein, which is a repressor of VEGF expression.
This clicker question is designed so students should choose one explanation and rule out the others (Supporting File S5. Splicing It Together - Day 1 Slides and Clickers). When polling has ended, ask a few students to explain or justify their choice. After a few students have shared their reasoning, ask the same clicker question again, allowing students to change their answers if they would like. This allows students to be persuaded and incorporate new interpretations that were brought up by their classmates in the discussion.
Day 1 Wrap-up
Walk students through the conclusions they may have come to so far, what they have learned about transcription factor A, and their ideas about the functions of the other 3 proteins. Introduce the pre-class activity for the next day, which students should complete between Day 1 and Day 2 of the argumentation module (Supporting File S5. Splicing It Together - Day 1 Slides and Clickers).
Pre-Class Day 2
Students should complete the Day 2 pre-class activity, which is a single open-ended question (Supporting File S5. Splicing It Together - Day 1 Slides and Clickers):
Using the information provided in Data Sets 1 & 2: How does alternative splicing of WC1 pre-mRNA affect gene expression and regulation in cancer?
This activity requires students to articulate their current understanding of the data, without being restricted by clicker options. Review a handful of responses prior to class to help frame class discussions on Day 2.
In Class Day 2
Day 2 Introduction
The instructor should remind the class about the context of the argumentation module and the Big Question. Next, the instructor should review the figures from Day 1 and discuss confusing aspects or prevailing ideas observed in pre-class activity responses. Before introducing new data, remind students to form groups if they have not already done so.
Data Set 3
As Data Set 3 handouts (Supporting File S4. Splicing It Together - Day 2 Handout) are distributed, project the data on a slide (Supporting File S6. Splicing It Together - Day 2 Slides and Clickers). Again, students should be given 10 minutes to discuss and record their group answers to the following prompts:
How does the amount of each type of WC1 mRNA expressed in the leukemia patients’ bone marrow compare to the expression of WC1 in normal bone marrow?
Many of you just determined that WC1 Transcription Factor A most likely represses the expression of proteins that repress genes (i.e., VEGF) that promote tumor growth. Given the data in Figure 3, does this hypothesis explain the function of WC1 in both leukemia and ovarian cancer cells? Why or why not?
Monitor student groups to evaluate how students are working with the data, what explanations they are reaching, and if they are confused about anything they are encountering in the data. Through discussion, students should be able to conclude that the hypothesis that best fits the ovarian cancer data does not explain what they are observing in the data regarding WC1 expression in leukemia.
Following discussion, begin Clicker Question 3, which presents new explanations for the effect of WC1 transcription factors on gene expression (Supporting File S6. Splicing It Together - Day 2 Slides and Clickers):
Based on Figures 2 & 3, which hypothesis about the role of WC1 Transcription Factors in leukemia can be ruled out?
-
Transcription Factors B and C activate the expression of genes that lead to cancer cell growth.
-
Transcription Factor B activates the expression of genes that lead to cancer cell growth.
-
Transcription Factor C represses the expression of genes that lead to cancer cell growth.
This clicker question is meant to inspire discussion as students must use structural/functional information from Data Set 2 and expression levels from Data Set 3 to evaluate the hypotheses. We also used common interpretation errors (e.g., a significant difference must mean an increase) from students in previous semesters to help craft possible explanations. Using the data provided, students should be able to rule out only one of the hypotheses (Answer A).
During clicker question follow-up, have students share out why they selected their answer or why they could not rule out the other hypotheses. During discussion, ask students about what they noticed in Data Set 3 to ensure that class members share an understanding of the figure.
Answering the Big Question & Activity Wrap-Up
Display the Big Question and instruct groups to come to consensus on an answer using the information they have acquired over the past two days. Students should have several minutes to assemble their responses, but the instructor should reserve time at the end of class to wrap-up discussion and introduce the final homework assignment (Supporting File S6. Splicing It Together - Day 2 Slides and Clickers). While groups are discussing this question, move through the room again and ask for groups’ explanations. Again, gather themes to inform the subsequent whole-class discussion.
Elicit student reasoning by asking how they answered the Big Question:
How does alternative splicing of WC1 pre-mRNA affect gene expression in cancer?
During wrap-up, remind them of the data they interpreted and which hypotheses they were able to support or exclude based on that data. If time allows, ask them what additional data they might pursue to rule out any remaining explanations or solidify their current explanation.
After Day 2
Students will individually answer the Big Question outside of class (Supporting File S6. Splicing It Together - Day 2 Slides and Clickers):
Using the information provided in Data Sets 1, 2 and 3, how does alternative splicing of WC1 pre-mRNA affect gene expression and regulation in cancer?
Teaching Discussion
Iterative Design Process
Our goal was to generate an activity suitable for large lecture that would promote understanding of information flow by engaging students in data interpretation and collaborative argumentation. Crafting this argumentation module was a highly iterative process. Our team scoured primary literature to create an initial draft of the activity, which we implemented, observed, and then used instructor feedback and student responses to revise over the course of several semesters. For instance, after our first implementation we improved the pre-class quiz to better support student thinking about transcription factors, gene expression, and cancer. Before deploying the module in the third semester, we changed the order in which we presented the figures to students and reworked the clicker questions to provide the more cohesive, hypothesis-driven story outlined in this lesson plan. To aid in the revision process, we required groups to enter their consensus answers for each round of data interpretation into an online Qualtrics form. We did not use this information to alter instruction in real-time, so we did not include it as a supplementary document. Using an online form in a large-lecture hall is not always feasible, as it would require every group to have stable internet access to complete the assignment.
Student Engagement in the Argumentation Module
Argumentation is not the standard pedagogical approach in this intro biology course. Students are regularly assigned multiple-choice pre-class quizzes and receive participation points for responding to clicker questions embedded in the lecture. In contrast, extensive group work only happens during the argumentation modules. Given the novelty of the argumentation module, which followed the first exam, we were concerned that students would not be motivated to exert the “extra” effort to discuss data, evaluate hypotheses, and write essay responses given the small incentive (the three homework assignments and two days of class participation together comprised ~1.5% of the overall course grade). However, we were pleased to see that students in all three semesters did engage in all aspects of the argumentation session.
When the final version of the activity was implemented in the Spring 2020 semester, clicker question participation for both days of the argumentation module was consistent with other lecture days in that exam unit (Table 2). To guide our observation of student behavior, we used a modified form of the Classroom Observation Protocol for Undergraduate STEM (23) to characterize and compare argumentation days to other lecture periods. In typical class sessions, students spent most of their time listening and taking notes to the lecture and follow-up discussions on instructor- or student-posed questions (Figure 1). We observed little discussion between students during clicker questions, and only a few students tended to answer other questions posed by the instructor.
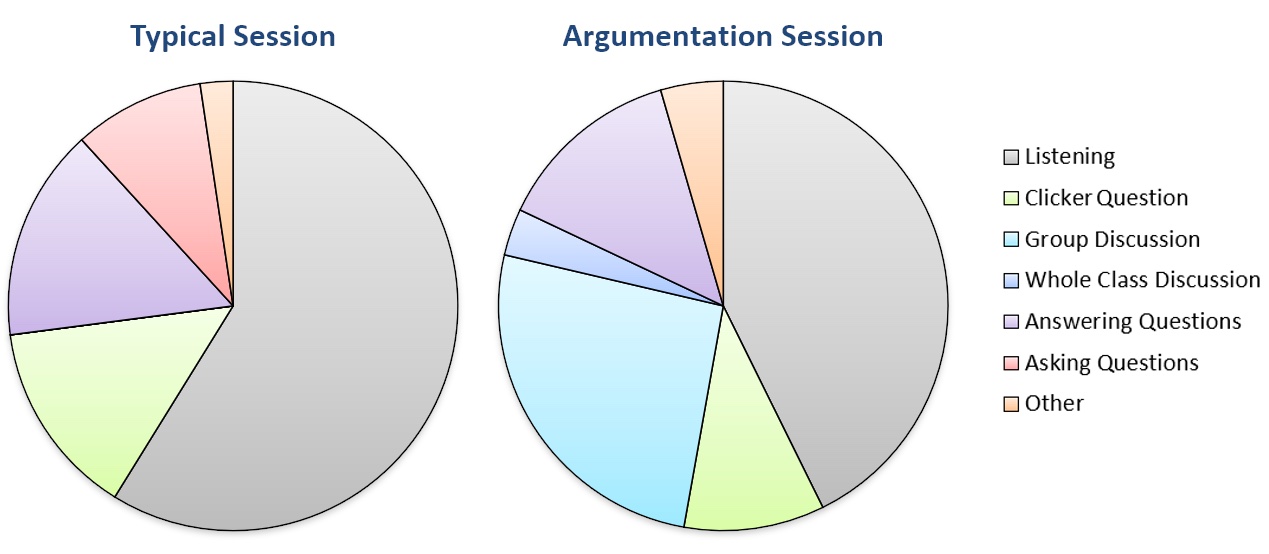
Table 2. Daily clicker participation and homework completion rates for this activity were consistent with typical interactive lectures within the same exam unit. Numbers reflect the percentage of all students enrolled in the course (N=504). Homework completion rates were higher for students who attended class sessions.
| Typical Sessions | Argumentation Session | |
|---|---|---|
| Clicker Participation | 77-90% |
Day 1: 87% Day 2: 80% |
| Homework Completion | 81-92% |
Pre-Quiz for Day 1: 94% Pre-Class for Day 2: 87% Summary Activity: 89% |
In contrast, students spent less time passively listening and more time actively engaging in the alternative splicing argumentation session (Figure 1). Many groups used most if not all of the allotted time to discuss interpretation of the data, and more students volunteered to share their reasoning about including/excluding hypotheses than on a typical day. At times, students from three or more separate groups interacted in response to a question, which we considered a whole-class discussion.
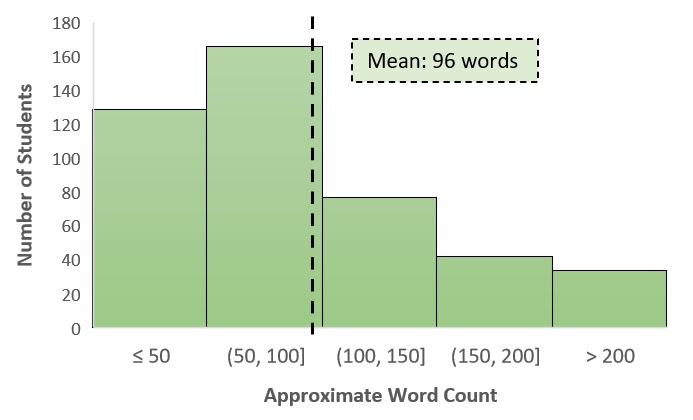
Students also spent time outside of class to prepare for both days of the argumentation module. Most students took the Day 1 pre-quiz, which had the highest completion rate for daily assignments in that unit, and responded to the Day 2 pre-class essay (Table 2). The response rate for the summary writing assignment after Day 2 was similarly high, and despite knowing they were simply scored on completion, many students were thoughtful in crafting their explanations. Students used an average of 96 words, though several were more verbose, in answering the Big Question (Figure 2). In their summaries, students clearly drew on appropriate cognitive resources (e.g., data figures, hypotheses, background information) and looked across levels of biological organization to explain the role of alternative splicing in the context of cancer. Many students summarized the in-class discussion in their own words, while others expanded the scope of the question by generating alternative hypotheses that could be tested with additional data.
Adaptations
This lesson could be of value to other mid- to upper-level courses (e.g., genetics, molecular biology) that cover gene expression and regulation. It could easily be adapted for smaller sized courses, where it might be more manageable to collect and review group consensus answers in addition to the individual responses. If clicker technology or mobile voting is not available, the clicker questions could be answered using voting cards or by raising hands.
The lesson could easily be expanded to cover a third day, allowing more time to unpack student reasoning during follow-up and discussion, especially if the instructor would like students to model the proposed hypotheses. In some of our iterations, students either requested more time for data interpretation or the follow-up discussion extended beyond the scheduled time, which meant the Wrap-Up discussion was delayed until the next lecture day. On a third day, students could also discuss their summary writing activities with their group members by collaborating and contrasting their summaries or drawing a figure that synthesizes their summaries into a single interpretation. In an upper-division course, the activity could be extended by instructing students to search for other examples of alternative splicing in a gene database or transcript database such as RefSeq. Students could compare and contrast the effects of alternative splicing between WC1 and their example gene.
We recommend that students have some opportunity to engage in data interpretation and/or collaboration in class before they begin this argumentation module. In our first semester, this activity was the only argumentation module within the two-semester intro bio sequence. In subsequent semesters, we introduced another argumentation module on mutation and the central dogma prior to the first exam. When they had the chance to practice using these skills with more familiar content prior to this activity, students seemed to engage more in discussion and spend more time thinking about alternative splicing data than the novelty of the activity.
SUPPORTING MATERIALS
- S1. Splicing it together – Pre-Class Background Slides
- S2. Splicing it together – Pre-Quiz
- S3. Splicing it together – Day 1 Handouts
- S4. Splicing it together – Day 2 Handouts
- S5. Splicing it together – Day 1 Slides & Clickers
- S6. Splicing it together – Day 2 Slides & Clickers
- S7. Splicing it together – End of Unit Exam Questions
Acknowledgments
This material is based upon work supported by the National Science Foundation under Grant No. 1822490. This work was reviewed by the WSU Office of Research Assurances and determined to satisfy the criteria for Exempt Research at 45 CFR 46.101 (b)(1) and 45 CFR 46.101(b)(2).
References
- American Association for the Advancement of Science. 2011. Vision and Change in Undergraduate Biology Education: A Call to Action. Washington, DC.
- Brownell SE, Freeman S, Wenderoth MP, Crowe AJ. 2014. BioCore Guide: a tool for interpreting the core concepts of Vision and Change for biology majors. CBE Life Sci. Educ. 13(2): 200-211. doi: 10.1187/cbe.13-12-0233
- Clemmons AW, Timbrook J, Herron JC, Crowe AJ. 2020. BioSkills guide: Development and national validation of a tool for interpreting the vision and change core competencies. CBE Life Sci. Educ. 19(4):ar53.
- Asterhan CS, Schwarz BB. 2016. Argumentation for learning: Well-trodden paths and unexplored territories. Educ. Psychol. 51(2):164-87.
- Sampson V, Gerbino F. 2010. Two instructional models that teachers can use to promote & support scientific argumentation in the biology classroom. Am. Biol. Teach. 72(7):427-31.
- Sampson V, Schleigh S. 2013. Scientific argumentation in biology: 30 classroom activities. Arlington, Va: NSTA Press.
- Lammers A, Goedhart MJ, Avraamidou L. 2019. Reading and synthesising science texts using a scientific argumentation model by undergraduate biology students. Int. J. Sci. Educ. 41(16):2323-46.
- Allen D, Tanner K. 2005. Infusing active learning into the large-enrollment biology class: seven strategies, from the simple to complex. Cell Biol. Educ. 4(4):262-8.
- Lewis J, Kattmann U. 2004. Traits, genes, particles and information: re-visiting students’ understandings of genetics. Int. J. Sci. Educ. 26: 195–206.
- Marbach-Ad G. 2001. Attempting to break the code in student comprehension of genetic concepts. J. Biol. Educ. 35: 183-189.
- Duncan RG, Reiser BJ. 2007. Reasoning across ontologically distinct levels: students’ understandings of molecular genetics. J. Res. Sci. Teach. 44(7): 938–959.
- Marbach-Ad G, Stavy R. 2000. Students’ cellular and molecular explanations of genetic phenomena. J. Bio. Educ. 34: 200–205.
- Reinagel A, Bray Speth E. 2016. Beyond the central dogma: Model-based learning of how genes determine phenotypes. CBE – Life Sci. Educ. 15(1):ar4.
- Horwitz P, Neumann E, Schwartz J. 1996. Teaching science at multiple space time scales. Comm. ACM. 39(8):100-2.
- Kapteijn M. 1990. The functions of organizational levels in biology for describing and planning biology education. Relating macroscopic phenomena to microscopic particles. CD‐Press, Utrecht, The Netherlands. 139-50.
- Pelletreau, K. N., et al. 2016. A clicker-based case study that untangles student thinking about the processes in the central dogma. CourseSource. https://doi.org/10.24918/cs.2016.15 .
- Ross, J.A. 2016. Predicting and classifying effects of insertion and deletion mutations on protein coding regions. CourseSource. https://doi.org/10.24918/cs.2016.18
- Laakso, M.M., et al. 2017. An undergraduate bioinformatics curriculum that teaches eukaryotic gene structure. CourseSource. https://doi.org/10.24918/cs.2017.13
- Freeman S, Haak D, Wenderoth MP. 2011. Increased course structure improves performance in introductory biology. CBE—Life Sci.Educ. 10(2):175-86.
- Offerdahl EG, McConnell M, Boyer J. 2018. Can I have your recipe? Using a fidelity of implementation (FOI) framework to identify the key ingredients of formative assessment for learning. CBE—Life Sciences Education, 17(4), es16.
- Burke KA, Greenbowe TJ, Hand BM. 2006. Implementing the science writing heuristic in the chemistry laboratory. Journal of Chemical Education, 83(7), 1032.
- Barab S. 2014. Design-based research: A methodological toolkit for engineering change, pp. 151–170. In The Cambridge handbook of the learning sciences 2nd ed. Cambridge University Press. Cambridge, United Kingdom. https://doi.org/10.1017/CBO9781139519526.011
- Smith MK, Jones FH, Gilbert SL, Wieman CE. 2013. The Classroom Observation Protocol for Undergraduate STEM (COPUS): A new instrument to characterize university STEM classroom practices. CBE –Life Sci. Educ. 12(4):618-27.
Article Files
Login to access supporting documents
Arneson-Splicing it together - Using primary data to explore RNA splicing and gene expression.pdf(PDF | 339 KB)
S1. Splicing it Together - Pre-Class Background Slides.pptx(PPTX | 566 KB)
S2. Splicing it Together - Pre-Quiz.docx(DOCX | 65 KB)
S3. Splicing it Together - Day 1 Handouts.docx(DOCX | 69 KB)
S4. Splicing it Together - Day 2 Handouts.docx(DOCX | 35 KB)
S5. Splicing it Together - Day 1 Slides and Clickers.pptx(PPTX | 1 MB)
S6. Splicing it Together - Day 2 Slides and Clickers.pptx(PPTX | 216 KB)
S7. Splicing it Together - End of Unit Exam Questions.docx(DOCX | 45 KB)
- License terms

Comments
Comments
There are no comments on this resource.