The Case of the Missing Strawberries: RFLP analysis
Published online:
Abstract
While solving the fictional mystery of the missing strawberries, students are engaged in a guided-inquiry lesson featuring small-group and class discussions, hands-on activities, and laboratory exercises related to molecular genotyping by restriction fragment length polymorphism (RFLP) analysis. During the lesson, students make connections between concepts to better understand abstract ideas, practice the process of science, gain hands-on experience with molecular biology techniques, collaborate as a part of a team, and elaborate their learning to other “real-life” scenarios. While intended for undergraduate students enrolled in an introductory biology or genetics course with a co-requisite lab, suggestions are presented for adapting the lesson for younger audiences and classes without a laboratory component.
Citation
Sleister, H.M. 2014. The Case of the Missing Strawberries: RFLP analysis. CourseSource. https://doi.org/10.24918/cs.2014.10Society Learning Goals
Genetics
- Nature of Genetic Material
- How is DNA organized?
- Methods & Tools in Genetics
- What experimental methods are commonly used to analyze gene structure and gene expression?
Science Process Skills
- Process of Science
- Pose testable questions and hypotheses to address gaps in knowledge
- Plan, evaluate, and implement scientific investigations
- Interpret, evaluate, and draw conclusions from data
- Construct explanations and make evidence-based arguments about the natural world
- Address novel questions through authentic research experiences
- Modeling/ Developing and Using Models
- Recognize the important roles that scientific models, of many different types (conceptual, mathematical, physical, etc.), play in predicting and communicating biological phenomena
- Make inferences and solve problems using models and simulations
- Build and evaluate models of biological systems
- Quantitative Reasoning/ Using Mathematics and Computational Thinking
- Apply the tools of graphing, statistics, and data science to analyze biological data
Lesson Learning Goals
- Students will understand the relationship between a cell, chromosomes, and DNA.
- Students will understand the structure of DNA.
- Students will demonstrate how to isolate DNA from strawberries.
- Students will demonstrate gel electrophoresis.
- Students will appreciate applications of molecular genotyping.
Lesson Learning Objectives
Students will be able to:- Describe the relationship of cells, chromosomes, and DNA.
- Isolate DNA from strawberries.
- Digest DNA with restriction enzymes.
- Perform gel electrophoresis.
- Design an experiment to compare DNAs by RFLP analysis.
- Predict results of RFLP analysis.
- Interpret results of RFLP analysis.
- Use appropriate safety procedures in the lab.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
National calls have emphasized the need to engage students in active, inquiry-driven courses that include the scientific process and opportunities to relate abstract biological concepts to real-world examples (1). Traditional laboratories can play a critical role in illustrating and extending concepts learned in the classroom. Investigative laboratories ask students to do more; they provide opportunities for students to ask scientific questions, design experiments, collect data, interpret data, and contribute to an interdisciplinary body of scientific knowledge (2).
In an attempt to connect concepts taught in lecture and experiments done in lab, Phillips et al. (3) developed exercises to connect DNA structure and replication with the techniques of PCR and gel electrophoresis. Likewise, Halme et al. (4) found that combined “hands-on and minds-on components” of a lab help to increase student learning of biological concepts. Other educators have described series of laboratory exercises including restriction fragment length polymorphism (RFLP) analysis (5-8). For example, DiBartolomeis described a semester-long Drosophila gene mapping laboratory project which included RFLP analysis for upper-level undergraduates and first-year graduate students (5). This project resulted in increased student knowledge of relevant topics and skills. Similarly, Zhang and colleagues (6) taught a molecular biology laboratory course in which students used RFLP analysis to study a SNP in the peroxiredoxin-6 gene. Students were involved in multiple steps of the project from accessing sequences from databases through analyzing DNA restriction fragments on an agarose gel. Reinking and colleagues (7) developed a shorter, two-lab period laboratory exercise in which students used RFLP analysis to determine their own genotype with respect to a single nucleotide polymorphism in the TAS2R38 gene.
The “Case of the Missing Strawberries” lesson described here extends the theme of using experimental techniques to drive learning of basic concepts related to DNA structure and function. By introducing a fictional mystery case study, students are “hooked” into wanting to learn about the biological concepts and hands-on techniques involved in molecular genotyping. In the process of solving the mystery, students make connections between concepts to better understand abstract ideas, practice the process of science, gain hands-on experience with molecular biology techniques, collaborate as a part of a team, and elaborate their learning to other “real-life” scenarios. These outcomes are aligned with several Vision and Change core concepts and competencies (1).
This guided-inquiry lesson includes a mix of small-group and class discussions, hands-on activities, and laboratory exercises. It is intended for students enrolled in an introductory biology or genetics course for science majors with a co-requisite lab. However, as described in the “Teaching Discussion” section below, the lesson can be adapted for college-level non-majors and middle/high school students. The lesson can also be modified to exclude the lab activity portions (DNA isolation, restriction digests, and gel electrophoresis) and focus on class discussion and data analysis. Since the lesson begins with a review of DNA structure and function and the relationship of DNA to chromosomes and cells, students only need basic knowledge of DNA structure prior to this lesson.
SCIENTIFIC TEACHING THEMES
Active Learning
- Students participate in think-pair-share, small group discussion, and classroom-wide discussion. They also simulate processes as a small group and complete hands-on lab activities in small groups.
Assessment
- Pre-assessment: None
- Post-assessments: Students complete short answer questions about DNA, cells, and chromosomes, and predict the outcome of RFLP analysis.
Inclusive Teaching
- All students in the class participate in all aspects of the lesson including lab activities. The diversity and multimodal nature of learning activities (discussion, data interpretation, lab techniques) will engage a variety of learners.
LESSON PLAN
This lesson is written as a series of guided inquiry, small-group activities and class discussions and includes complementary lab exercises. While I have only taught the lesson with lab activities according to the Timeline in Table 1, I offer suggestions for teaching the lesson without the lab exercises in the “Teaching Discussion and Table 1.” The 4-hour lesson can be completed during a single 4-hour lab period or two separate 2-hour lab periods. If splitting the lesson into two days with lab activities, students should complete steps 1-6 in the first lab period. As long as they set up the DNA digests in step 6, the 45-minute digestion time can continue after they leave the lab. Without the lab activities, the total time required is approximately 1.5 hours, and this can also be split into two separate class periods if needed.
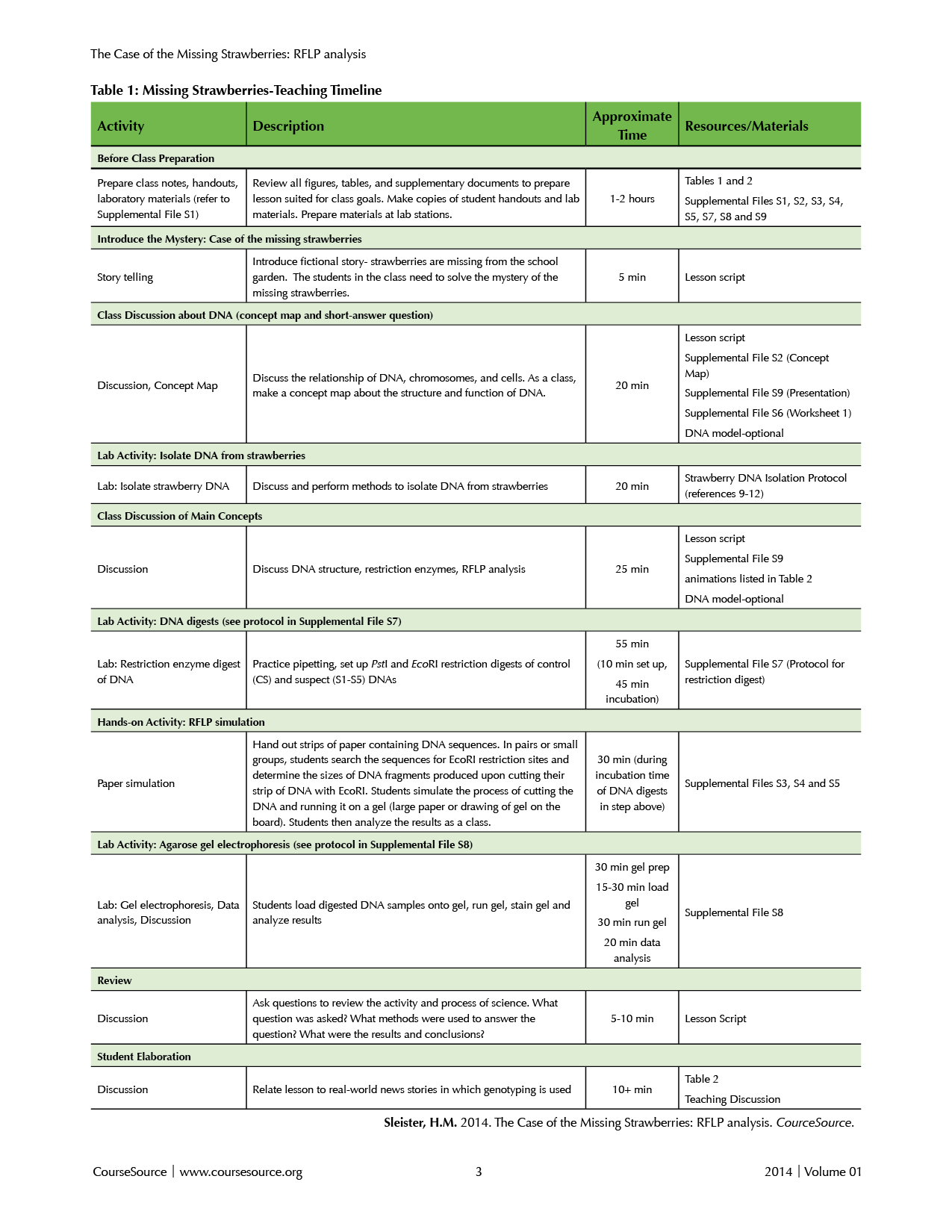
Table 1. Missing Strawberries-Teaching Timeline
Before class
Teacher preparation: Materials needed for each part of the lesson are detailed in Supplemental File S1. The teacher will need to prepare copies of Worksheet 1 (Supplemental File S6) and lab protocols (Supplemental Files S7 and S8). The teacher will also need to set up lab stations for each pair or small group of students. If the teacher chooses to prepare the agarose gel for the class, it should be ready before the “agarose gel” lab exercise in step 8 below. Depending on his/her background with the molecular methods included in this lesson, the teacher may wish to view the animations listed in Table 2 (Web Resources). Supplemental File S9 is a presentation provided as a template to which the instructor can add figures and adapt to fit the needs of his/her particular course.
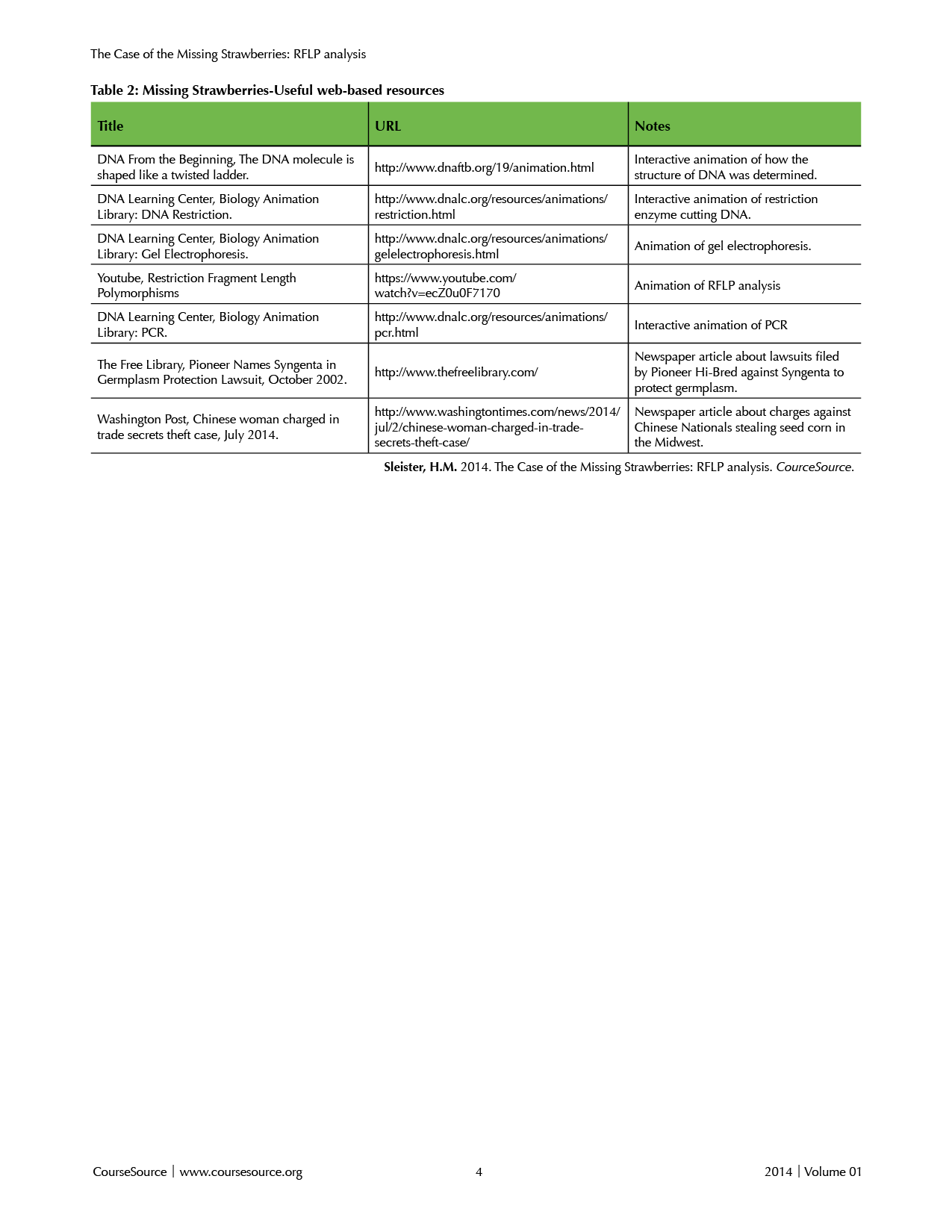
Table 2. Missing Strawberries-Useful web-based resources
During class
1. Introduce the mystery (lecture script, Supplemental File S9- slides 1-2).
This fictional story is intended to capture the interest of students and provide the motivation for learning concepts and methods related to solving the mystery. An instructor concerned with using a fictional story may wish to emphasize to students that the story is not real and adapt the scenario script to exaggerate that it is fictional. For example, the school could be referred to in the script as the “Hard Knock School.” Alternatively, an instructor could replace the mystery scenario with a discussion of how a scientist would go about determining if a plant has been genetically modified. Additional alternative scenarios are provided in the “Teaching Discussion.” (5 minutes)
“Let’s start with a fictional story. There’s a healthy food initiative service project at the [Hard Knock] school. Students taking botany this semester planted a large garden of fruits and vegetables with the intent of giving the harvested food to local people in need. The garden is located inside school grounds, so only students, faculty, and staff can access it. On July 25th someone vandalized the garden and destroyed plants, and many strawberries from the garden were missing. While looking for the missing strawberries, school officials found strawberries in the offices or backpacks of five different individuals. Let’s imagine that our biology class has been asked to figure out if any of the strawberries found were taken from the garden. How would we go about doing this?” [Give students time to think of possible answers. Example student responses: compare strawberries from the garden and the “found” strawberries for appearance (macroscopic and microscopic), taste, DNA.] “Fortunately, we have the equipment needed to compare DNA. Let’s first talk about the structure and function of DNA, and then we’ll isolate DNA from strawberries.”
2. Class discussion about DNA (Concept map, Supplemental File S6, Supplemental File S9, optional- model of DNA). (20 minutes)
As a class, make a concept map about DNA structure/function during this discussion (see Supplemental File S2 for sample concept map). While the level of detail in the concept map will vary depending on the level of the class, I suggest including at a minimum the following concepts.
- Each cell contains a nucleus.
- Inside the nucleus are chromosomes.
- Each chromosome is a long double-stranded DNA molecule.
- DNA is a helical double-stranded molecule made up of four different kinds of nucleotides (A,C,G,T).
- An organism’s characteristics are determined by its specific order of nucleotides.
- Using humans as an example, the DNAs from two unrelated individuals are ~99.9% identical. It’s the small number of differences in DNA sequence between two people that make us different (and also the environment). So, no two people (except identical twins) will have exactly the same DNA. The same is true for other organisms- even strawberry plants.
In addition to the concept map, the instructor can refer to Supplemental File S9 (Slides 3-4) which includes illustrations related to the conversation.
Following the concept map discussion, students should be able to answer questions in Worksheet 1 (Supplemental File S6). These questions are intended to reinforce the relationships among cells, chromosomes, and DNA as well as the structure of DNA. An instructor can modify the questions for the level of his/her students. Students can complete the worksheet in pairs or small groups. Alternatively, the questions on this worksheet can be used later to evaluate learning.
3. Process of science class discussion- introduction (script, Supplemental File S9- slide 5). (3 minutes)
“Earlier you suggested one way to figure out whether any of the strawberries were taken from the garden would be to compare the DNA from the strawberries in the backpacks or offices to the DNA in strawberry plants from the garden. Let’s call the garden strawberry DNA the “control DNA”. How would we determine if the DNA from two different strawberries is the same or different?” [Let students brainstorm ideas.] “Good, so I heard that we’d somehow need to compare the sequence of nucleotides in the DNAs from different strawberry plants. So, first we’d need to isolate DNA from strawberries, and then use a method to detect differences in the DNA sequence.”
4. Lab activity: Isolate DNA from strawberries (20 minutes)
Use a protocol for isolating DNA from strawberries. I adapted a method described by Diane Sweeney (9), and other very similar protocols are available (10-12). Depending on time available, after isolating strawberry DNA, the students can either view the DNA by eye or put a small amount of the DNA on a slide (with coverslip) and view it on a light microscope. The benefit of using a microscope is that students will learn a little about scale. A microscope, of course, can magnify objects, but light microscopy is not a tool that can be used to determine DNA sequence.
5. Class discussion of main concepts: DNA structure, restriction enzymes, gel electrophoresis, RFLP analysis (script), also refer to Supplemental File S9-Slides 5-9 (script) and optional model of DNA. (25 minutes)
“We can’t see the sequence of nucleotides when we look at strawberry DNA by eye or using a light microscope, so how do you know if the DNA sequences of two strawberries are the same or different?” [Make a list of student ideas on the board.] “There are a variety of molecular methods we can use to compare the DNA sequences from two strawberries. For example, we can determine the actual DNA sequence (i.e., ACGT) of the strawberries or we can determine a molecular genotype for the strawberries. One method of genotyping is called restriction fragment length polymorphism (RFLP) analysis.”
“There are many different applications of RFLP analysis. [Refer to Supplemental File S9 slide 6.] For example, RFLP analysis can be used to detect individuals with the genetic disorder sickle cell anemia. Let’s talk about how RFLP analysis is done.”
[Using pictures and/or models, review the overall structure of DNA.] “As we talked about in our concept map, DNA is a double-stranded, helical molecule (similar to a twisted ladder). Each side of the ladder consists of a single strand of DNA which is a polymer (chain) of nucleotides. Each nucleotide contains a deoxyribose sugar, a phosphate group, and one of four nitrogenous bases (A-adenine, C-cytosine, G-guanine, T-thymine). The two strands are held together by hydrogen bonds that form between the paired nitrogenous bases (A pairs with T, G pairs with C). DNA has an overall negative charge (from the phosphates).”
“Using humans as an example, the DNA nucleotide sequences from two unrelated individuals are ~99.9% identical. In addition to features of our environment, the small number of differences in DNA sequence between two people is what makes each of us unique. The same is true for strawberry plants. While two strawberries from the same plant should be genetically identical, strawberries from two different plants may differ genetically. So, let’s talk about how we can detect these differences in DNA sequence using RFLP analysis. We need to first understand a little about restriction endonucleases and gel electrophoresis.” [See Supplemental File S9 slides 7-8 and animations listed in Table 2]. “A restriction enzyme is a protein that acts like molecular scissors. There are hundreds of different restriction enzymes. Each enzyme recognizes a very specific sequence of base pairs in double-stranded DNA. For example, the restriction enzyme EcoRI recognizes the sequence GAATTC, but note that only the sequence of one strand is written here. The enzyme binds this region of the DNA, called the restriction site, and cuts both strands of the DNA molecule. If a specific restriction site occurs in more than one location in a DNA molecule, a restriction enzyme will cut each of those sites, resulting in multiple fragments of DNA. If a linear piece of DNA is cut with a restriction enzyme whose specific recognition site is found at one location in the DNA molecule, the result will be two DNA fragments [Supplemental File S9, slide 7]. The length of each fragment will depend on the location of the restriction site in the DNA molecule. DNA fragments that result from cutting with restriction enzymes can be observed using a technique called agarose gel electrophoresis.”
“Agarose gel electrophoresis separates DNA fragments by size. DNA fragments are loaded into an agarose gel slab, which is placed into a chamber filled with a buffer solution. A direct current is passed between wire electrodes at each end of the chamber. Since DNA fragments are negatively charged, they will be drawn toward the positive end when placed in an electric field. The DNA fragments have to weave their way through the agarose gel matrix, and smaller DNA fragments can move more easily than larger ones. Therefore, DNA fragments are separated on an agarose gel based on size, with the smaller fragments moving faster than larger ones. Thus, when you look at a gel of DNA fragments after exposing it to current for 45 minutes to an hour, the smaller fragments will be closer to the positive electrode than larger fragments. The DNA fragments can be seen in the gel after the DNA in the gel is stained.”
“If the human genome which is about 3 billion basepairs in length contained equal amounts of each of the nucleotides, then digesting human DNA with the restriction enzyme EcoRI would be expected to produce over 700,000 different fragments of DNA! When run on an agarose gel, this very large number of fragments would appear as a smear rather than distinct bands. Fortunately, a technique called polymerase chain reaction (PCR) can be used to amplify (or make lots of copies of) a particular region of the genome of interest so that it can be analyzed apart from the rest of the genome.”
“Let’s focus on a single gene which would occupy a relatively small region of a chromosome. If you were comparing the DNA sequences of a single gene from two different people, you would likely see several differences in the gene’s DNA sequence. These differences are called polymorphisms. While the frequency of polymorphisms varies greatly across the human genome, on average, a 2,000-3,000 basepair human gene contains 10 different polymorphic sites (13). Different forms (sequences) of a gene are called alleles.” [Refer to Supplemental File S9, slide 9. Remind students that this figure shows only one small part of the genome.] “In the figure, notice there are two different alleles (alleles 1 and 2), and they are identical in sequence except for one basepair. Notice that this difference is located within the EcoRI restriction site in allele 1. The enzyme EcoRI can recognize and cut Allele 1 DNA. In contrast, because of a single base pair change in sequence, Allele 2 DNA is not cut by EcoRI. Remember, digesting human genomic DNA with EcoRI would result in hundreds of thousands of DNA fragments, but for now let’s focus on the gene illustrated here. [Refer to Supplemental File S9, slide 9.] After digesting Allele 1 and Allele 2 DNAs with restriction enzyme EcoRI, different numbers and lengths of DNA fragments are produced. Allele 1 is cut into two fragments (fragment A1-1, fragment A1-2), whereas Allele 2 which is uncut is a larger DNA fragment (fragment A2-1). Notice that the difference in number and lengths of fragments produced after cutting Alleles 1 and 2 DNAs with EcoRI is detected using gel electrophoresis. However, to visualize the DNA fragments on the gel corresponding to alleles 1 and 2 (and to avoid seeing DNA fragments produced by digesting DNA from the rest of the genome), one would need to do PCR to amplify a small region of the genome where this gene is located before digesting the DNA with EcoRI and running the gel. In the figure, you can differentiate Allele 1 and Allele 2 on the gel.”
“To have enough DNA to analyze, a technique called polymerase chain reaction (PCR) is often used to amplify (or make more copies of) a specific region of DNA from an organism. The region that’s selected is one that tends to vary in DNA sequence among individuals (so that differences in DNA sequence can be detected among individuals). Today let’s imagine that we’re analyzing a small region of the strawberry genome that has already been amplified by PCR, so we can focus on differences in this small region.”
As a class, discuss how you would design an experiment to compare “suspect” strawberries to the missing strawberries from the garden using RFLP analysis. Based on previous discussions, students should realize that they will need to analyze a small region of the strawberry genome. Help them understand that the region that’s selected would be one that’s previously been reported to vary in DNA sequence among individuals (so that differences in DNA sequence can be detected among individuals). Emphasize that you would need to amplify this small region of the strawberry genome by PCR to focus on the polymorphic region of interest. Students should also realize that they will need to digest the PCR-amplified strawberries with a restriction enzyme, run the digested DNAs on an agarose gel, visualize the DNA fragments in the gel, and compare the band pattern of the garden strawberry plant DNA (you can call this the control strawberry DNA) with the patterns of DNA isolated from strawberries found at the school (“suspect” strawberry DNAs). The progression of these steps will be reinforced in the “RFLP Simulation” activity described in Step 7 below.
After discussing the experimental design, tell students that the class will carry out the progression of steps discussed for the experimental design, but you will be using already-prepared plasmid DNAs instead of testing the strawberry DNAs they isolated so that the results of RFLP analysis will be clear. For RFLP analysis, I use plasmid DNAs from the BioRad Forensic DNA Fingerprinting Kit (BioRad #166-0007EDU) (14). This kit contains five different plasmids (named S1-S5 for “suspect” DNAs), and plasmid S3 is also used as the control (described in the BioRad kit as “CS” for crime-scene DNA). A different piece of lambda DNA has been cloned into each plasmid S1-S5 such that a different pattern of DNA fragments is produced upon double-digestion of the plasmids with restriction enzymes PstI and EcoRI. In the next lab activities, students will digest these plasmid DNAs (intended to represent strawberry DNAs) with restriction enzymes, run the digested DNAs on a gel to separate DNA fragments by size, and compare the restriction fragment patterns produced for plasmids S1-S5 to the control (CS) (See Supplemental File S7: Protocol: Restriction digests and Supplemental File S8: Protocol: Agarose gel electrophoresis).
6. Lab activity: DNA digests (see Supplemental File S7: Protocol: Restriction Digests). (10 minutes to set up digests, 45 minutes digestion time)
Provide time for students to practice using a micropipettor if they are not already familiar with it. For example, after demonstrating how to use a micropipettor, you could instruct students to work in pairs and ask each student to practice pipetting 10 µl water colored with food coloring onto a piece of plastic wrap or parafilm. Paired students can compare the sizes of dispensed drops.
Students digest plasmid S3 (labeled CS to represent control strawberry plant DNA) and plasmids S1, S2, S3, S4, S5 (representing “suspect” strawberry DNAs from lockers) with restriction enzymes EcoRI and PstI. (See Supplemental File S7 Protocol: Restriction Digests.) I give each student group a single tube containing 10 µl of plasmid DNA (tubes labeled CS, S1, S2, S3, S4, S5 to represent strawberry DNAs). Students add 10 µl pre-aliquoted restriction enzymes to the tube containing the DNA, mix by gently tapping the tube, and incubate the restriction digest reaction at 37oC for 45 min. If you routinely do molecular work in your lab, I recommend digesting the plasmids from the kit with EcoRI and PstI restriction enzymes you likely already have on hand (see Materials List in Supplemental File S1).
While the DNAs are digesting, the class should prepare a gel (see step 8 below) and/or do the hands-on RFLP analysis simulation activity (step 7 below).
7. Hands-on activity: RFLP Simulation. (See Supplemental Files S3,S4, S5) (30 minutes)
To help students make the connection between differences in DNA and seeing differences in band patterns on a gel, I created a hands-on activity for students to simulate RFLP analysis. This activity can be completed either while DNAs are being digested or while the gel is running. I give students the background information that they will use for the RFLP simulation as a class. Each group (pair or small group) is given a strip of paper which has one of six 100 basepair double-stranded DNA sequences (Supplemental File S3). One of the sequences is named “Evidence,” and the others are named “Sample 1” through “Sample 5”. Samples 1 through 5 represent different alleles. Although each strip of paper looks like a single piece of DNA, when we actually do RFLP analysis in the lab, we digest billions of copies of the same DNA, so that the band we see on the gel contains billions of DNA fragments of the same length. In our paper simulation, we are trying to determine if one (or more) of the suspect “sample” DNAs match the evidence DNA. The five suspect sample DNAs are nearly identical, but there are a few differences (i.e., polymorphisms) among the five sequences, and some of these differences fall within the EcoRI restriction enzyme site GAATTC. The students are asked to search their assigned 100 basepair sequence for the sequence GAATTC and to draw a box around each site they find (similar to what they saw in Supplemental File S9, Slide 7]. Next, they cut their paper strip vertically at each GAA/TTC (at the location of the slash) to produce restriction fragments. Note that EcoRI creates sticky ends, so the slash does not accurately indicate the exact location where the enzyme cuts. While this works fine for the purpose of this activity, students could also be directed to cut the DNA sequence to more accurately reflect how EcoRI would cut, creating sticky ends. Next, students count and write down the number of basepairs in each fragment. I draw a large picture of an agarose gel with molecular weight marker bands on the whiteboard and ask students to simulate the process of running their DNA fragments on the gel. So that their DNA bands are visible and similar in size, I have students tape each fragment onto a rectangular piece of colored paper the shape of a gel band. Each group “loads” the digested paper DNA fragments into the gel well (depicted as light gray band in Supplemental File S4) and moves their negatively-charged DNA fragments in their lane of the gel toward the positive electrode until they reach the appropriate location relative to the molecular weight marker in the first lane of the gel. Students tape their paper DNA fragments at the appropriate location on the “gel” as it would appear at the conclusion of running the gel as illustrated in Supplemental Files S4 and S5. After each group has simulated gel electrophoresis, we review the process. Each original sequence of DNA on the strip of paper was nearly identical to the other sequences, but DNA polymorphisms (i.e., differences) were detected by digesting the DNAs with EcoRI and running them on a gel. Next, we ask if any of the “suspect” samples match the evidence (the paper “gel” should look like the one in Supplemental Files S4 and S5). This is the same kind of thinking the students will need to do after seeing their actual gel in step 8 below.
8. Lab activity: agarose gel electrophoresis (see protocol in Supplemental File S8: Protocol: Agarose gel electrophoresis). (30 minutes gel preparation which overlaps step 6, 60 minutes gel loading/running, 20 minutes data analysis)
While the students’ DNAs are being digested with restriction enzymes, demonstrate for the class how to prepare a 1% agarose gel using a comb with at least 7 wells (refer to Supplemental File S8: Protocol: Agarose Gel Electrophoresis). The gel should be ready by the time the digests are finished. Alternatively, the gel can be prepared in advance by the instructor. I prepare two gels for the class- one to practice loading dye alone into and one for the digested DNA samples.
After 45 minutes of DNA digestion (set up in step 6 above), students should add 5 µl loading dye (pre-aliquoted for students) into each sample of digested DNA. The students can quickly spin the tube in a microcentrifuge or flick the tube by hand to move contents to the bottom of the tube. After they have practiced loading dye into the extra “practice” gel, each group loads their single digested DNA sample into the gel using a P20 micropipettor. The class can decide the order for loading samples on the gel, but remind them to load a molecular weight marker (such as Lambda-HindIII) and the EcoRI-PstI-digested control (CS) DNA. Next, run the gel about 30 minutes at 100 Volts. While the gel is running, review important concepts, the question being asked, experimental methods, and expected gel results.
After the gel has run 30 min, stop it by turning off the power supply. Remove the gel from the electrophoresis chamber and stain/destain the gel as described in Supplemental File S8. Once bands are visible, have students record and analyze data. What conclusions would they make? (Do any of the samples match sample CS representing the garden strawberry control?) They should observe that the sizes of DNA fragments in the gel lane containing S3 DNA match the control DNA. For a sample photo of the completed gel, see the BioRad #166-0007EDU manual, page 7 (14).
9. Review discussion. (5-10 minutes)
Following are questions to review and wrap-up. What was the question we were asking? What methods did we use to answer this question? Be sure to make connections between the properties of DNA, restriction enzymes, gel electrophoresis, and RFLP analysis. What did we find? How certain are we that sample X matches the control (CS) DNA? Thinking back to the missing strawberry scenario, could there be other explanations (e.g., the suspect by chance purchased the same type of strawberry from the store as the ones in the school garden, someone else put the strawberries in their locker as a joke)?
10. Student elaboration. (10 minutes+)
“Can you think of other real-life scenarios in which you’d use the types of techniques and thinking that you used today?” [Discuss as class.] “These same techniques are used in real life to determine whether a suspect committed a crime. Let’s imagine a murder scene in which DNA evidence (e.g., blood) is collected from a crime scene and from several suspects. Ideally, the DNA from the crime scene “matches” one of the suspects. If this person is convicted, he/she could spend the rest of his/her life in jail, or even worse! If you were a juror, what would be important to know about the DNA evidence for you to confidently conclude the person committed the crime? (e.g., the DNA match should be significant---only one person in > 6 billion would be expected to have DNA with this specific profile; the DNA evidence was collected in an appropriate manner; there are no other reasonable explanations for this person’s DNA to be at the crime scene).” Depending on the level of the class, this discussion could lead into an exploration of other types of molecular genotyping such as simple tandem repeats used in forensic analysis and the use of population genetics (allele and genotype frequencies) to calculate the likelihood of a particular genotype.
The lesson can be extended by relating it to news stories students might be familiar with such as the BTK killer and plant biotechnology theft (see Table 2. Useful Web-based Resources). Another logical extension is to discuss the ethical implications of genotyping or genetic testing with respect to forensic science, paternity issues, and disease diagnosis.
TEACHING DISCUSSION
Early on in the lesson and during the review near the end of the lesson, students demonstrated understanding of the relationship between a cell, chromosomes, and DNA through verbal and written responses (Supplemental File S6: Worksheet 1). Students shared knowledge of the structure of DNA while completing a concept map and participating in a large group discussion of the structure of DNA. Each pair of students successfully isolated DNA from strawberries, and each individual student demonstrated technical proficiency with pipetting and loading a gel. Students also readily simulated the process of RFLP analysis and correctly interpreted the data from both the simulation and the actual experiment. Importantly, students were engaged throughout the lesson, connected ideas throughout the activities, and seemed to enjoy discovering the answer to the mystery presented.
For teachers wishing to adopt this lesson, if time allows I recommend including more opportunities for students to demonstrate their understanding through writing (e.g., worksheets, written think-pair-share responses) and elaboration. In addition, I suggest using a pre and post-quiz to assess learning of concepts included in the lesson. Although we discussed concepts and our experimental approach throughout the lesson, it would be helpful for students to outline the process of science used in the lesson to further connect ideas and reinforce the purpose of each activity. Teachers who would like to focus on writing skills in their courses could have students prepare a written lab report and/or elaborate on a related topic (e.g., using molecular genotyping to diagnose disease).
This lesson can be modified for different levels of students. For example, the lesson was originally designed for a single day of a summer science camp for middle school students from groups underrepresented in science. The fictional story used for the science camp included a description of a middle school building and finding strawberries in student lockers. An instructor concerned about using a fictional story will want to remind students that the mystery is made-up and is intended to pique their interest and provide an opportunity for them to apply molecular techniques and science process thinking.
The lesson can also be adapted for a course without lab activities. This modification will significantly shorten the time required for the lesson (see Table 1. Teaching Timeline), but it will still encourage students to think about how one would design an experiment, learn the sequence of events required to solve the mystery, discuss molecular techniques, and analyze results (the teacher should show sample results such as the gel photo on page 7 of the BioRad #166-0007EDU Manual).
While I implemented learning molecular genotyping through a “case of the missing strawberries,” there are a variety of applications of RFLP analysis (Supplemental File S9, Slide 6), and alternative scenarios such as forensics, disease diagnosis, and detection of genetically modified organisms would work for the same lesson. Several good suggestions are included in the BioRad #166-0007EDU Manual.
The lesson provides opportunities to help students connect concepts described (e.g., DNA structure and RFLP analysis), as well as additional concepts. Following are questions an instructor could ask students to help them connect ideas, interpret data, and elaborate on their understanding.
- (Refer to Supplemental File S9, Slide 9) How would the DNA banding pattern on the gel appear for an individual homozygous for allele 1, homozygous for allele 2, or heterozygous for alleles 1 and 2?
- A person is diploid, so how is it that the DNA from a person homozygous for allele 2 would appear as a single band on the gel?
- Strawberries are octaploid (i.e. contain eight sets of chromosomes). In contrast, humans are diploid (contain two sets of chromosomes). In humans, two haploid gametes- egg and sperm- each of which contains a single set of chromosomes, unite to produce a diploid zygote. How do you think an organism could become octaploid?
- How could Southern Blotting technique be used to detect RFLPs?
Furthermore, the lesson plan can be used as a springboard for student elaboration as described in step 10 above. To make the lesson more interdisciplinary, it can be extended to include the calculation of probabilities of specific genotypes and the ethics of genetic testing.
SUPPLEMENTAL MATERIALS
Table 1. Missing Strawberries-Teaching Timeline
Table 2. Missing Strawberries-Useful web-based resources
Supplemental File S1. Missing Strawberries-Materials list
Supplemental File S2. Missing Strawberries-Sample concept map of DNA structure/function
Supplemental File S3. Missing Strawberries-DNA sequences for RFLP simulation
Supplemental File S4. Missing Strawberries-Agarose gel picture for RFLP simulation-Template and answers
Supplemental File S5. Missing Strawberries-Photo of completed RFLP simulation
Supplemental File S6. Missing Strawberries-Worksheet 1-Questions about DNA location, structure and function
Supplemental File S7. Missing Strawberries-Protocol-Restriction digests
Supplemental File S8. Missing Strawberries-Protocol-Agarose gel electrophoresis
Supplemental File S9. Missing Strawberries-Missing Strawberries-PowerPoint presentation
ACKNOWLEDGEMENTS
The method for isolating strawberry DNA was adapted from Diane Sweeney (9). DNA samples used in this lesson are from BioRad Forensic DNA Fingerprinting Kit (14), and the manual that accompanies this kit provides good background information and protocols for DNA digestion and gel electrophoresis (adapted for the lesson described here). Financial support for development of related curriculum for the “genetics day” of a summer science camp for middle-school students was received from NASA-ISGC.
References
- AAAS. 2010. Vision and Change: A Call to Action. Washington, DC: AAAS. Accessed July 27, 2014. http://visionandchange.org/files/2011/03/Revised-Vision-and-Change-Final-Report.pdf.
- Bell E. 2001. The future of education in the molecular life sciences. Nature Reviews Molecular Cell Biology 2:221-225.
- Phillips AR, Robertson AL, Batzli J, Harris M, Miller S. 2008. Aligning goals, assessments, and activities: an approach to teaching PCR and gel electrophoresis. CBE Life Sciences Education 7:96-106.
- Halme DG, Khodor J, Mitchell R, Walker GC. 2006. A small-scale concept-based laboratory component: the best of both worlds. CBE Life Sciences Education 5:41-51.
- DiBartolomeis SM. 2011. A semester-long project for teaching basic techniques in molecular biology such as restriction fragment length polymorphism analysis to undergraduate and graduate students. CBE Life Sciences Education 10:95-110.
- Zhang B, Wang Y, Xu X, Guan X, Bai Y. 2013. Using PCR-RFLP technology to teach single nucleotide polymorphism for undergraduates. Biochemistry and Molecular Biology Education 41:262-266.
- Reinking JL, Waldo JT, Dinsmore J. 2013. A trio of human molecular genetics PCR assays. Biochemistry and Molecular Biology Education 41:173-179.
- Tait RC. 2000. Introductory experiments in recombinant DNA. Current Issues in Molecular Biology 2:71-85.
- Sweeney D. Isolation of Strawberry DNA. Accessed October 21, 2014. http://www.gs.washington.edu/outreach/dhillon_dnaprocedure.pdf.
- Squishy Science: Extract DNA from Smashed Strawberries. Accessed October 21, 2014. http://www.scientificamerican.com/article/squishy-science-extract-dna-from-smashed-strawberries/.
- Strawberry DNA Extraction Lesson Plan. Accessed October 21, 2014. http://www.shsu.edu/~agr_www/documents/DNALAB.pdf. Steve
- Spangler Science: Strawberry DNA- Food Science. Accessed October 21, 2014. http://www.stevespanglerscience.com/lab/experiments/strawberry-dna.
- Brooker R. 2009. Genetics: Analysis and Principles. McGraw Hill, New York, New York.
- BioRad Forensic DNA Fingerprinting Kit (BioRad #166-0007EDU) Accessed July 27, 2014. http://www.bio-rad.com/webroot/web/pdf/lse/literature/1660077EDU.pdf.
Article Files
Login to access supporting documents
The Case of the Missing Strawberries: RFLP analysis(PDF | 168 KB)
Supplemental File S1. The Case of the Missing Strawberries-Materials list.docx(DOCX | 28 KB)
Supplemental File S2. The Case of the Missing Strawberries-Example concept map of DNA structure-function.jpg(JPG | 500 KB)
Supplemental File S3. The Case of the Missing Strawberries-DNA sequences for RFLP simulation.docx(DOCX | 19 KB)
Supplemental File S4. The Case of the Missing Strawberries-Agarose gel picture for RFLP simulation-template and answers.pptx(PPTX | 57 KB)
Supplemental File S5. The Case of the Missing Strawberries-Photo of completed RFLP simulation.jpg(JPG | 228 KB)
Supplemental File S6. The Case of the Missing Strawberries-Worksheet 1.docx(DOCX | 17 KB)
Supplemental File S7. The Case of the Missing Strawberries-Protocol-Restriction digests.docx(DOCX | 15 KB)
Supplemental File S8. The Case of the Missing Strawberries-Protocol-Agarose gel electrophoresis.docx(DOCX | 18 KB)
Supplemental File S9. The Case of the Missing Strawberries-PowerPoint presentation-RFLP analysis.pptx(PPTX | 131 KB)
- License terms

Comments
Comments
There are no comments on this resource.