Teaching Genetic Linkage and Recombination through Mapping with Molecular Markers
Published online:
Abstract
Most introductory genetics courses cover genetic linkage, a core concept in the CourseSource genetics learning outcome framework. Although it is a classical genetics topic, genetic linkage remains an important concept to understand in order to grasp modern genetics research approaches including Single Nucleotide Polymorphism (SNP) mapping, Genome Wide Association Studies (GWAS), and gene discovery. Typically, genetic linkage is taught in a very traditional way within our introductory genetics classes. Invariably, we see students struggling with the same aspects of linkage: how to distinguish between parental and recombinant combinations of alleles and how to relate phenotype proportions to meiotic processes and outcomes. We designed a lesson that provides a practical and experimental context to target these common student difficulties in learning about linkage and recombination. This student-centered interactive lesson and associated post-class problem set teaches genetic linkage through mapping a gene by determining co-segregation of a phenotype with microsatellite sequences revealed by gel electrophoresis banding patterns. This lesson includes very interactive class sessions and a follow-up problem set and post-test that allows students to develop a deeper understanding of genetic linkage, and provides instructors with insights about student thinking. When we implemented this lesson, we observed a dramatic increase in student understanding of genetic linkage and how to use molecular markers to map the location of genes.
Citation
McDonnell, L. and Klenz, J. 2015. Teaching Genetic Linkage and Recombination through Mapping with Molecular Markers. CourseSource. https://doi.org/10.24918/cs.2015.13Society Learning Goals
Genetics
- Transmission - Patterns of Inheritance
- How can one deduce information about genes, alleles, and gene functions from analysis of genetic crosses and patterns of inheritance?
- How does the phenomenon of linkage affect the assortment of alleles during meiosis?
- Methods & Tools in Genetics
- What experimental methods are commonly used to analyze gene structure and gene expression?
Science Process Skills
- Process of Science
- Interpret, evaluate, and draw conclusions from data
- Construct explanations and make evidence-based arguments about the natural world
- Modeling/ Developing and Using Models
- Recognize the important roles that scientific models, of many different types (conceptual, mathematical, physical, etc.), play in predicting and communicating biological phenomena
- Make inferences and solve problems using models and simulations
- Build and evaluate models of biological systems
- Quantitative Reasoning/ Using Mathematics and Computational Thinking
- Apply the tools of graphing, statistics, and data science to analyze biological data
Lesson Learning Goals
Students will understand how the genetic linkage of loci on chromosomes, such as genes and microsatellite molecular markers, affects the assortment of alleles during meiosis and phenotypes observed in a population.Lesson Learning Objectives
Students will be able to:- Explain how recombination can lead to new combinations of linked alleles.
- Explain how molecular markers (such as microsatellites) can be used to map the location of genes/loci, including what crosses would be informative and why.
- Explain how banding patterns on an electrophoresis gel represent the segregation of alleles during meiosis.
- Predict how recombination frequency between two linked loci affects the genotype frequencies of the products of meiosis compared to loci that are unlinked (or very tightly linked).
- Analyze data from a cross (phenotypes and/or genotypes) to determine if the cross involves linked genes.
- Calculate the map distance between linked genes using data from genetic crosses, such as gel electrophoresis banding patterns.
- Justify conclusions about genetic linkage by describing the information in the data that allows you to determine genes are linked.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Genetic linkage and recombination are core concepts in an undergraduate genetics curriculum(1), but many educators find the traditional method of teaching this topic is problematic. Despite the common challenges encountered when teaching genetic linkage, we were surprisingly unable to find published information providing teaching strategies or tips to improve student understanding of genetic linkage and recombination. Typically, students analyze crosses involving somewhat random phenotypes, such as 'Waltzing' or 'bent-tail' mice(2), and even after instruction and practice, they are rarely able to articulate the utility of understanding or using genetic linkage. Through the observation of students solving genetic linkage problems, we noted that most simply memorize a strategy or formula to solve genetics linkage problems. As a result, they are unable to connect the segregation of alleles at the chromosome level to the final genotypes and phenotypes observed, and struggle to understand how phenotype proportions in a cross relate to meiosis and chromosomes. To tackle some of these challenges we developed a more modern, context rich, and active learning-focused lesson to teach genetic linkage.
We wanted to provide students with a compelling practical and experimental context that targeted these common difficulties in learning about linkage and recombination. The solution we developed is a student-centered, interactive lesson with additional tutorial (i.e. recitation or discussion section) activities to teach genetic linkage using data from molecular marker mapping experiments. Specifically, we used microsatellites so students would be able to analyze images of gel electrophoresis banding patterns, a technique that our students often encounter in high school or as undergraduates. We also found that working with molecular markers helps students develop a better understanding of the types of DNA sequences found in the genome, particularly that the genome consists of more than protein-coding gene sequences. Using microsatellite markers to learn linkage also provided a nice bridge to discuss the use of single nucleotide polymorphisms (SNPs) in genetic mapping and identifying loci associated with particular traits.
Microsatellites are tandem repeats of two to six base pairs. They are also called short tandem repeats (STRs), simple sequence repeats (SSRs), simple sequence length polymorphisms (SSLPs), and variable number tandem repeats (VNTRs). Various microsatellites occur in species from bacteria(3) to plants(4, 5) and humans(6), scattered throughout the genomes. The number of nucleotide repeats in a single microsatellite locus is highly polymorphic, often producing different alleles (distinguished by different repeat number) of a single locus in a population (Figure 1A). Microsatellite alleles can be detected via PCR, using primers designed to sequences flanking the microsatellite locus. Separating the PCR products using agarose gel electrophoresis produces a banding pattern that represents the microsatellite genotype of each DNA sample (Figure 1B).
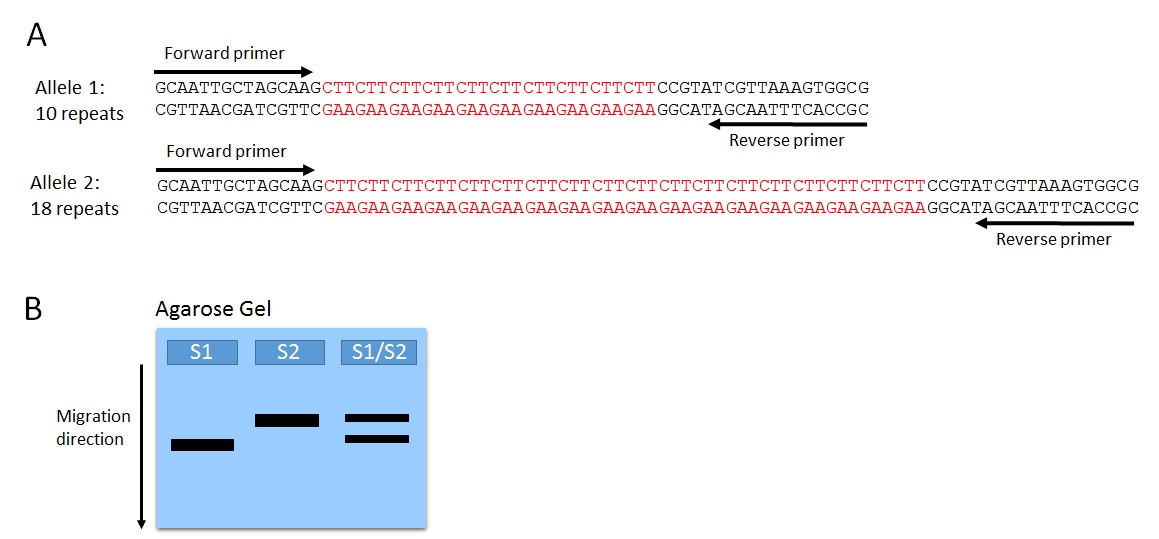
Figure 1. Variation in microsatellite alleles can be detected by analyzing polymerase chain reaction (PCR) products using agarose gel electrophoresis. (A) For example, two different alleles of the CTT microsatellite, composed of multiple CTT trinucleotide repeats. Alleles differ by the number of CTT repeats. Amplification of the microsatellite locus can be accomplished using DNA as a template for PCR and primers designed to bind to the region flanking the microsatellite. (B) PCR products can be separated using agarose gel electrophoresis. The allele with more repeats produces a larger (more nucleotides) PCR product, which remains closer to the top of the gel compared to a PCR product composed of fewer nucleotides. The banding pattern represents the microsatellite genotype of the sample: Strain 1 (S1), homozygous for allele 1; Strain 2 (S2), homozygous for allele 2; Strain 1/2 heterozygote (S1/S2), carrying both allele 1 and 2.
Microsatellites are a type of "molecular marker" that can be used in a variety of applications including forensics analysis, population genetics, detecting certain human diseases, and linkage mapping(7). The focus of this lesson is the use of microsatellite loci to map the location of a locus/gene/mutation involved in a phenotype of interest. For example, if an interesting new phenotype were identified, one may want to map the location of the mutation responsible for the phenotype to facilitate determining the molecular mechanisms that underlie the phenotype. When using microsatellite markers for genetic mapping, we are ultimately analyzing the segregation of a particular microsatellite allele (represented as a band on a gel) with the phenotype of interest (Figure 2).
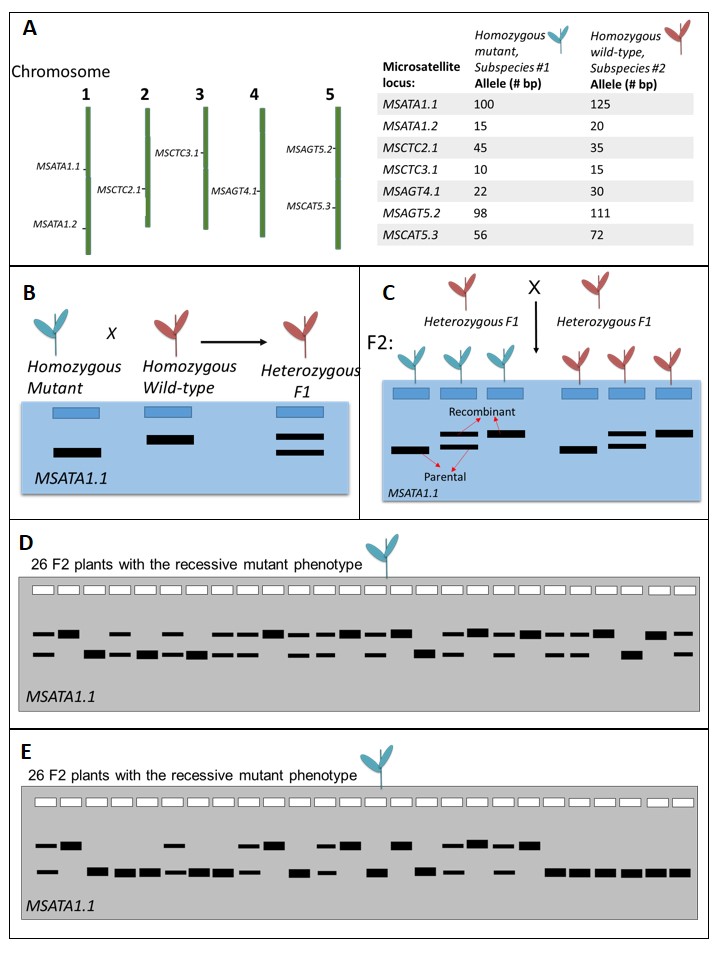
Figure 2. An example of mapping with molecular markers to determine the locus involved in a recessive phenotype of interest (blue flowers). (A) We begin with a map of known molecular markers for the species we are working with. Normally there are dozens or hundreds of known markers; here we illustrate a few for simplicity. We will focus on one microsatellite locus, MSATA1.1, for this mapping example. To measure segregation between the mutation causing blue-colored flowers and the MSATA1.1 microsatellite locus, we need to have a subspecies of the same plant that is homozygous wild type (red flowers) and carries different alleles of the microsatellite loci than the mutant subspecies. Note that the allele names (e.g., 100) indicate the number of base pairs in the allele sequence. (B) A homozygous blue-flower parent plant was crossed to a homozygous red-flower (wild-type) parent plant, generating doubly heterozygous F1 plants (heterozygous at the loci for flower color, and the MSATA1.1 locus). DNA is extracted from the parent plants and the heterozygous F1 plants, PCR is used to amplify the microsatellite locus, and separation of the PCR products using agarose gel electrophoresis allows us to visualize the microsatellite genotypes in the form of gel banding patterns. (C) The heterozygous F1 are self-fertilized. We expect all combinations of flower-color phenotypes and microsatellite banding patterns to occur in the F2 population. As we are able to infer the genotype of the recessive blue-flower plants, we are able to classify the bands on a gel for the blue-flower plants as either “recombinant” or “parental,” representing the segregation that occurred in the F1. (D) If the microsatellite locus being tested is not genetically linked to the mutation causing blue flowers, we expect an equal proportion of parental and recombinant bands, indicating independent assortment. (E) If the microsatellite locus being tested is genetically linked to the mutation causing blue flowers, we expect a higher proportion of parental bands and a lower proportion of recombinant bands, compared to when there is independent assortment.
For simplicity, we focus the lesson on an example in which the new mutation is recessive to the wild-type allele. Mapping requires that we have access to two different subspecies (e.g., varieties; strains) for which a molecular marker map exists, indicating where the two subspecies possess different alleles for various microsatellite loci (Figure 2A). The unknown, homozygous mutation must occur in one subspecies (subspecies #1) that can be crossed with another subspecies (subspecies #2) that is wild-type and also carries different alleles of the microsatellites found in subspecies #1 (Figure 2A). This variation in phenotype and microsatellite alleles allows us to measure segregation between the microsatellite loci and the phenotype of interest.
Mapping typically begins with crossing two parental individuals, a homozygous mutant and a homozygous wild type, to generate a heterozygous F1 population (Figure 2B). The resulting F1 progeny are heterozygous for both the locus involved in the phenotype and for the microsatellite loci being used for mapping. The heterozygous F1 can be used in subsequent crosses to determine whether the newly identified phenotype (the new mutation) co-segregates with a particular microsatellite locus (Figure 2C). Since the phenotype of interest is caused by a recessive mutation in this scenario, we can infer the genotype of the F2 exhibiting the mutant phenotype to be homozygous for the new mutation. Based on our knowledge of which molecular marker alleles were carried by the homozygous parents, we can classify the microsatellite alleles (represented as bands on a gel) in the homozygous recessive F2 individuals as "parental" or "recombinant."
The frequency of parental and recombinant banding patterns will vary depending on whether or not the new mutation is genetically linked to the specific microsatellite locus. Most microsatellite markers will show independent assortment (i.e. they are not genetically linked to the mutation), resulting in an equal number of parental and recombinant microsatellite bands in the mutant phenotype F2 plants (Figure 2D). If a microsatellite marker is genetically linked to the new mutation, the F2 progeny that exhibit the recessive mutant phenotype will more frequently also inherit the microsatellite allele that was present in the parent mutant strain. That is, the new mutation is located on the same chromosome and close to that particular microsatellite locus, so that the mutation and the microsatellite do not independently assort (Figure 2E). Thus, by analyzing the molecular marker data, one can map the location of the gene controlling the new phenotype. Some examples of using molecular markers, such as microsatellites, to identify loci of interest include resistance to aphids in wheat(8), drought resistance in rice(9, 10)and corn(11), and polycystic kidney disease in humans(1)(2).
Intended Audience
The intended audience is sophomore-level undergraduate biological sciences majors in an introductory genetics class (i.e. at the 200 level).
Learning time
The lesson includes an in-class component with clicker questions and a group-based prediction activity with a handout, requiring about 110 minutes of in-class time. The in-class component is followed-up with a scaffolded practice problem and post-test. The follow-up problem and post-test are ideal for Teaching Assistant (TA)-led tutorials/discussion sessions/recitations that we will refer to as TA-led tutorials, but could also be used as homework. Our TA-led tutorials are small group (25 student), two-hour sessions facilitated by a graduate student TA. In these tutorials, students work in groups on a problem set related to the material they learned in class that week. See Table 1 for learning time and lesson components.
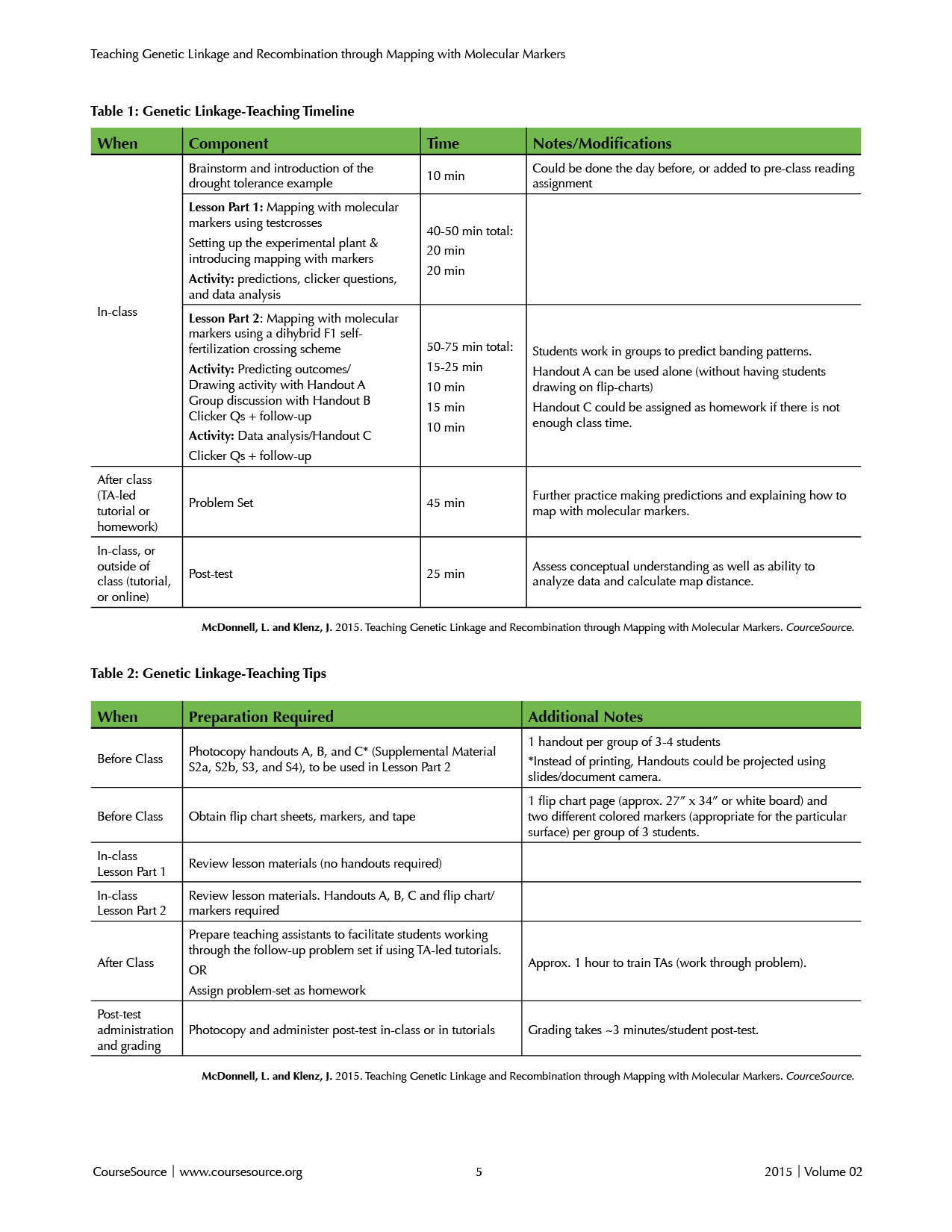
Table 1. Lesson times. Pre-class, in-class, and tutorial/recitation activities with estimated time required.
Table 2. Preparation required for the lesson, tutorial, and post-test.
Pre-requisite student knowledge
- Student pre-requisite knowledge for this activity includes the ability to:
- Explain what these are: locus, gene, and allele.
- Diagram meiosis and meiotic crossing over
- Describe the consequences of crossing over between two loci on gamete genotypes and frequencies, compared to when crossing over does not happen.
- Describe the difference between parental and recombinant allele combinations/genotypes
- Describe how genes that are close together on a chromosome are most often inherited together.
- Predict the outcomes of a dihybrid testcross (heterozygote x homozygous recessive) and a dihybrid x dihybrid cross when genes are assorting independently.
- Understand how PCR can be used to identify different alleles of molecular markers such as microsatellites.
- Interpret bands on a gel (separated using gel electrophoresis) and be able to explain how bands on a gel are similar or different (in terms of size and amount).
SCIENTIFIC TEACHING THEMES
ACTIVE LEARNING
In-class: Students will make predictions, comparisons, and teach each other during the group activity. Any confusion that is not identified and cleared up by peer group discussion will be revealed using the follow-up clicker questions and feedback from instructors.
After-class: The follow-up problem-set (ideally done in TA-led tutorials or as homework) will provide additional opportunities for students to engage in deliberate practice of applying their knowledge, making predictions, and analyzing data. If the follow-up problem-set is completed during a TA-led tutorial session, students will be able to receive immediate feedback on their thinking and further engage in peer discussion about the problem.
ASSESSMENT
Post-assessment: We assessed student learning via an open-ended (short answer) question set that tests students' conceptual understanding of the use of molecular markers for mapping, how banding patterns can be used to determine linkage, and their ability to analyze banding patterns to create a linkage map. Results of this post-test provide valuable information to instructors about student understanding, and formative feedback to students. Discussion of the post-test results can also provide additional opportunities for students and instructors to reflect on their learning.
INCLUSIVE TEACHING
We believe that the framing story used in the lesson about drought tolerant crops can be interesting to a variety of students, as most of them have encountered information about climate change, drought, and the impact of climate change on food crops. However, the framing story can be modified depending on the interests of the student audience, such as choosing genes and phenotypes related to diseases.
This lesson allows students to engage in the material in a variety of ways: individual clicker questions, peer discussion, drawing, and writing. Clicker questions, which students answer individually for the first vote, allow all students, regardless of ability, to engage in thinking and applying their knowledge, and receiving feedback (as the clicker questions are always followed-up with feedback from the instructor). We do not grade the clicker questions for correctness but rather use them for instructor and student feedback, creating a feedback-rich, low-stakes learning opportunity. Students are allowed to self-select their peer groups or to work individually (although in our experience almost all students participated in the group work portion of the class). The combination of clicker questions, peer discussion, and in-class problem solving with handouts allows for the variety of student abilities and learning preferences to be satisfied, while maintaining a high level of engagement.
LESSON PLAN
As presented, the activity requires approximately 110-160 minutes of class time, which can be split across multiple sessions depending on class length. This lesson has two parts: Part 1 introduces mapping with molecular markers using a testcross scenario whereby a dihybrid strain (heterozygous at two loci) is crossed to a homozygous recessive strain; Part 2 extends mapping with molecular markers through predicting and analyzing results from a dihybrid self-cross. If students are familiar with the basics of genetic linkage, i.e. predicting gamete genotypes when there is genetic linkage versus independent assortment, then Part 1 takes approximately 50 minutes of in-class time. However, if they are new to predicting the effects of genetic linkage, Part 1 could take nearly double that time (100 minutes). Part 2 takes approximately 50-80 minutes of in-class time. Our time estimates reflect our experience running the lesson and activities with a group of students who are very familiar with in-class group work. Classes that are less familiar with group work may require extra time for group assembly and for students to engage in group work.
Table 1 summarizes the lesson components and Table 2 provides tips for instructor preparation. Detailed background information and teaching notes are provided in the text. Slide-by-slide instructor notes and a suggested script are provided in the notes section of the Class Slides (S1a) and as a stand-alone document (S1b).
CLASS PREPARATION AND REQUIRED MATERIALS
Review and edit the Class Slides (S1a) as needed. Work through the student questions and worksheets, including drawing out labeled chromosomes.
All of the information needed for Part 1 is included in the Class Slides. However, the drawing and prediction activities in Lesson Part 2 require some materials. During Part 2, three-person student groups draw gametes and predicted banding patterns on large flip-chart sheets (27" x 34"). They affix these sheets to the wall around the classroom with masking tape. For a class of 60 students (20 groups), we brought 40, fat-tip Crayola(C) coloring markers that the students could use to do their drawings. Students spread their papers out on the floor or taped them on the wall to work on them. The Crayola(C) markers we used did not bleed through and mark the wall; if one were concerned about this possibility, students could be given two sheets of paper. If you have a smaller class with white boards or chalk boards, student groups could draw directly on the boards, eliminating the need for the flip chart paper.
We chose to have students draw on large sheets of paper because the size encouraged more collaboration and critiquing, compared to when students do their independent drawings on individual 8.5" x 11" pieces of paper. We also felt that having more space to capture their ideas would encourage students to create more elaborate drawings that included meiosis and banding patterns. However, in a class with 200 students, we found it too challenging to manage the activity with flip chart sheets. We still had students work in small groups to make their predictions but each student did their own drawing on individual copies of Handout A (S2a and S2b). A more detailed description of how to manage this activity is included below in the during class section.
Lesson Part 2 also requires three handouts: A, B, and C (S2 (both "a" and "b" forms), S3 and S4). These handouts will need to be photocopied ahead of time if you choose to distribute them to students in class (vs. projecting the handout on the overhead projector or document camera). You will need to make the following for a class of size "X": 1/3X copies of Handout A1; 1/3X copies of Handout A2; 1/6X copies of Handout B; 1/3X copies of Handout C.
DURING CLASS
LESSON PART 1: MAPPING WITH MOLECULAR MARKERS USING A TESTCROSS (S1A AND S1B, LESSON SLIDES 2-28)
1.A. BRAINSTORM: WHAT IS MAPPING GOOD FOR?
The instructor uses the first four slides to set the background for the activity. Slide 5 poses three brainstorm questions for students to answer in self-selected groups.
If we know the location of some genes in a genome, how can we use that information to determine what DNA sequences cause specific traits of interest? (Answer: we expect genes that are linked together on a chromosome will be inherited together frequently. Therefore, alleles of those linked genes will tend to be inherited together - so we'll expect to see traits of linked genes segregate together more often than they would if they were not linked, i.e. located on different chromosomes or far apart on the same chromosome.
What are some examples of traits for which you (or someone) might want to know the location of the gene(s) in the genome that controls that trait? (Examples: genes involved in diseases; genes that give a desired in crop plants such as increased growth or disease resistance)
Why would you want to know the location of a gene? (Answer: once we know the location of the gene, we can do more analysis of the gene: e.g., sequence the gene, identify promoters, identify mutations that cause the trait, or clone the gene to do further manipulations and experiments.)
Each group writes down their combined answers and hands them in after the brainstorming time. (We do not assign grades for this activity, but use it as an incentive for students to complete the activity and to give the instructor information about student thinking).
The purpose of this brainstorm is to encourage students to (1) get into the research frame of mind; (2) to propose traits in plants and animals that various research groups might be interested in mapping; and (3) to consider how and why people might want to determine the location of genes within the genome. We give the students 3-5 minutes to brainstorm and write down their answers on a piece of notebook paper. This activity is particularly useful if done during the class period just prior to the lesson because the instructor can then review student responses and summarize briefly at the start of the mapping with molecular markers class.
1.B. INTERACTIVE LECTURE ABOUT USING MOLECULAR MARKERS TO MAP A GENE CONFERRING DROUGHT TOLERANCE
This lesson focuses on mapping a gene for drought tolerance in a crop plant. Fleshing out the potential local impacts of drought can provide an opportunity for students to connect to the scenario.
Background information for instructors: In this lesson, we are considering a locus involved in a drought tolerance phenotype, DR(T), (wild type is drought sensitive, DR(S)). We start our hypothetical mapping experiment using a microsatellite locus called "G" to map the location of the DR locus. Alleles of the G locus vary by the number of repeat units, and hence the size of the PCR amplified G locus can vary depending on the allele. The homozygous drought tolerant plant carries the G(2)(00) allele (DR(T )G(2)(00) / DR(T) G(2)(00)) and the homozygous drought sensitive plant carries the G(4)(00) allele (DR(S) G(40)0 /DR(S) G(400)). (The superscript numbers refer to the number of repeats of the given allele, and )therefore reflects the relative size of the PCR product (band) separated and visualized using gel electrophoresis. Figure 3 illustrates a testcross for this scenario and the expected banding patterns.
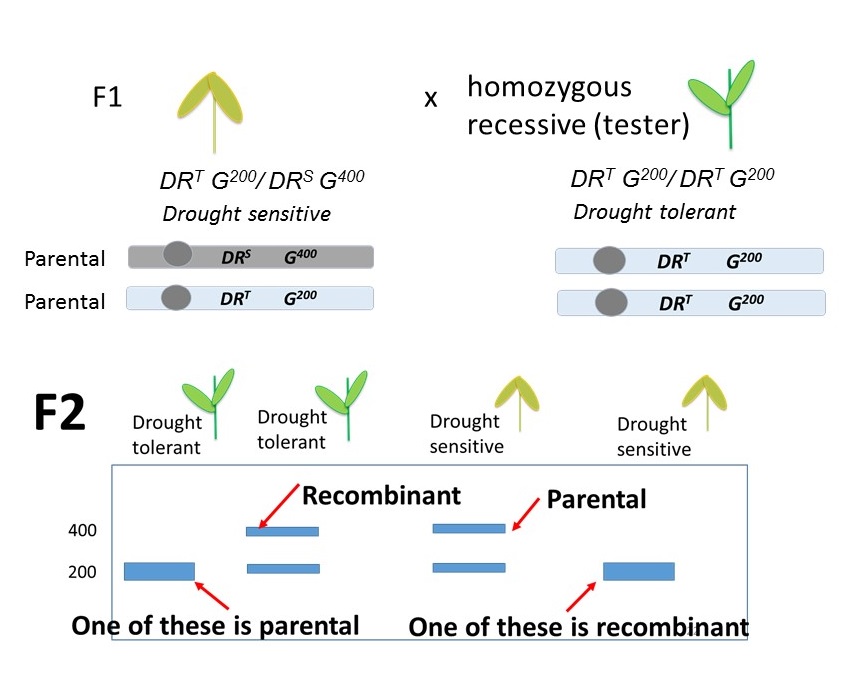
Figure 3. Predicting banding patterns from a testcross and identifying parental and recombinant bands. Each F2 receives one microsatellite allele from the homozygous recessive tester plant, so only one band in each plant can be classified as either parental or recombinant, representing the segregation that occurred in the F1.
Using the Class Slides, the instructor walks students through the logic behind a mapping experiment:
- Review how genetic crosses can determine that the phenotype (drought tolerance) results from a recessive mutation in a single gene. (Slides 1-9)
- Help students realize that knowing that the phenotype results from a recessive mutation does not provide any information about the specific gene involved or why this particular recessive mutation in this unknown gene leads to drought tolerance. (Slides 10-11)
- Explain how microsatellite molecular markers can be used in genetic crosses to map the physical location of a gene that produces a phenotype (i.e. drought tolerance). (Slides 12-23)
- Have students analyze microsatellite data to determine linkage and map distance. (Slides 24-28)
As the students work through this example, they must draw out the chromosomes involved, predict the phenotypes of progeny, and interpret results of gels showing microsatellite-banding patterns. The Class Slides and Lecture Script contain details about each step in the lesson.
LESSON PART 2: MAPPING WITH MOLECULAR MARKERS USING F1 X F1 DATA (S1A AND S1B, SLIDES 29-46)
After students have considered in Lesson Part 1 how to map a gene using a test cross, they now practice their skills on a new cross. The goal here is to push students to connect the events in meiosis to what we see in the F2 population. In addition, we want them to understand that, when assessing for linkage using microsatellites, we are looking at the relationship between the phenotype of interest (drought tolerance or sensitivity) and the banding patterns of the microsatellites in plants.
BACKGROUND INFORMATION FOR INSTRUCTORS
The F1 x F1 dihybrid cross (DR(T )G(200) / DR(S) G(400)( )X DR(T )G(200) / DR(S) G(400)), which could also be called an F1 self-cross, will produce an F2 population that contains both drought-tolerant and drought-sensitive plants. The only way to assess linkage between the drought tolerant phenotype and the G microsatellite is to determine the proportion of F2 progeny that carry a parental or recombinant combination of DR and G alleles. To determine parental versus recombinant, we must be able to infer the genotype with respect to the DR locus. In the offspring of this dihybrid cross, we can only infer the genotypes of the offspring with the recessive phenotype, i.e. the drought tolerant offspring.
In the progeny from the F1 self-cross, we know that the drought-tolerant plants must be homozygous recessive (DR(T)/DR(T)). However, a drought-sensitive plant could be either homozygous (DR(S)/DR(S)) or heterozygous (DR(S)/DR(T)). Because we cannot be sure of the genotype for the DR locus in the drought-sensitive F2 plants, we cannot use drought-sensitive plants to assess linkage. We can only use the drought-tolerant F2 plants to examine linkage between DR and G alleles. Consequently, we will examine the banding patterns among only the drought-tolerant F2 plants, once again looking for less than 50% recombinant types.
Contrast this situation with the testcross from Lesson Part 1, whereby a dihybrid F1 (DR(T) G(200)/ DR(S) G(400)) was crossed to a homozygous drought tolerant plant (DR(T) G(200)/ DR(T) G(200)). In the testcross, we could infer the genotype of both the drought-tolerant and the drought-sensitive offspring: all drought-tolerant progeny were homozygous for the DR(T) allele and all drought-sensitive progeny were heterozygous, i.e. DR(T)/DR(S).
Given that we would get information from each progeny produced from a test cross, but only 1/4 of the progeny produced from an F1 self-cross, why would anyone choose to study a self-cross? Practically speaking, it can be very laborious to force crosses between plants. Thus, just allowing the F1s to self-fertilize can be easier for the researcher. For this reason, we are going to explore how to determine if two loci are genetically linked using the results of a dihybrid F1 self-cross.
JIGSAW ACTIVITY: PREDICTING GENOTYPE AND PHENOTYPES FOR LINKED VERSUS UNLINKED GENES IN THE F2 PROGENY FROM AN F1 SELF CROSS
Using the class slides, provide the F1 x F1 scenario to students and then move on to the jigsaw activity(1)(4). Students should self-assemble into teams of six, and then each team of six should break into two, three-person groups. One group in the team will work on Prediction #1: predict the F2 genotypes, phenotypes, and banding patterns if the DR locus is independently assorting from the G microsatellite locus. The other group in the team will work on Prediction #2: predict the F2 genotypes, phenotypes, and banding patterns if the DR locus is genetically linked to the G microsatellite locus.
We recommend that each team of six assembles together in one area of the classroom. After you introduce the group goals, ask the groups to split from their team of six (i.e. all groups working on Prediction #1 could work on one side of the classroom and all groups working on Prediction #2 on the other side of the classroom). Provide each group with a sheet of flip chart paper (about 27" x 34"), a couple of colored flipchart markers, and either Handout A1 (predicting results if the loci are linked, S2a) or Handout A2 (predicting results if the loci are not linked, S2b). Encourage each group to draw the gametes, the chromosomes labeled with alleles, Punnett Squares, and the expected bands on a gel for the F2 population. (If you do not have the space, you can have groups work directly on the handout, omitting the flip-chart component.)
Groups can complete their predictions in approximately 10-15 minutes. We recommend that instructors and/or teaching assistants circulate around the room, asking questions and soliciting explanations about the groups' drawings. The biggest barrier to completing this task in the time allotted is often student's hesitation to begin. In such a case, we recommend that you demonstrate how to start on your own flip chart page or the document camera by drawing chromosomes labelled with alleles. Also, encourage groups to complete the questions on Handout A, as these questions will be needed for the second phase of the activity, when groups reassemble into their team of six to teach one another about their predictions.
In our experience, many students have a hard time determining which banding patterns will increase and decrease if genetic linkage is occurring. The key is to look at the gametes produced by the F1 and identify which are parental and recombinant. We can then adjust the frequencies of each gamete type according to our scenario. If there is no genetic linkage between the phenotype and the microsatellite, each recombinant gamete type should occur at a frequency of 25% (total recombinants of 50%). If there is genetic linkage between the phenotype and the microsatellite, the frequency of each recombinant gamete will be less than 25%. Figure 4 shows an example of a Punnett Square drawing used to make predictions. Slides 35-37 describe the answers to Handout A questions (S1a, Class Slides).
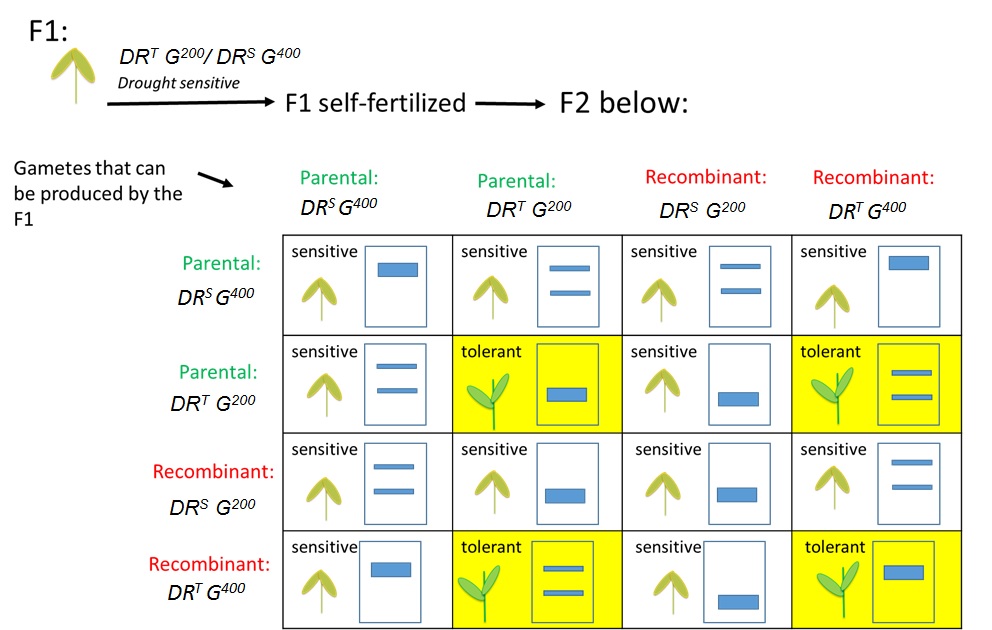
Figure 4. Predicting the F2 phenotypes and banding patterns from a dihybrid F1 self-fertilization. Labeling gametes as parental or recombinant allows for a quick assessment of which banding patterns represent recombinant and/or parental gamete genotypes.
Once the groups have completed their predictions, ask them to re-assemble into their original team of six and tape their posters up on the wall. Distribute a copy of Handout B (S3) to each team of six. They should now take the next 5-10 minutes to teach each other about their predictions by working through the questions on Handout B. As an incentive to complete the questions, we tell each group that they must hand in a completed Handout B, with all six team member names and student numbers on the worksheet, at the end of class. Once groups have completed their discussion, ask students to return to their seats.
CLICKER QUESTION SEQUENCE: F1 SELF-CROSS PREDICTIONS AND DATA ANALYSIS
The students will review the answers to some of the questions on Handout A and B by going through the clicker questions on slide 36-38, with feedback provided in response to clicker question results. When working through these clicker questions, we ask students to answer them individually for the first vote. If the responses indicate enough confusion to warrant peer instruction (less than 70% of the class select the correct answer), students could be cued to discuss their answer choice and reasoning with their peers before re-voting.
The question on Slide 36 asks students to recognize that each banding pattern in drought-sensitive F2 plants could represent a recombinant (question 1 from Handout B). Recall that drought sensitive is the dominant phenotype in this scenario. In our experience, only 33% of students selected the correct answer on the first vote. Students still struggle to select the correct answer, even after engaging in the group discussion activity with Handout B. Approximately 50% of students did not select the 200/200 banding pattern as one that could be represent a recombinant gamete genotype.
In a drought-sensitive F2 plant, the 200/200 band could be the result of a parental gamete (DR(T) G(200)) and recombinant gamete (DR(S) G(200)) combining (DR(T) G(200)/ DR(S) G(200)), or it could be the result of two recombinant gametes combining (DR(S) G(200)/ DR(S) G(200)). When only 33% of students selected the correct answer on the first vote, we asked students to consider what combination of gametes (parental and recombinant) could make drought-sensitive F2 plants and to discuss the answers with their neighbor, working systematically to decide if each banding pattern could represent a recombinant gamete. Students were given a few minutes to discuss and then revote, which resulted in 80% selecting the correct answer. After the second vote, we used the document camera to draw the crossing-over events in the F1 at the chromosomal level, indicate the resulting gamete genotypes, label the genotypes as parental and recombinant, and predict the resulting banding patterns generated from the union of various gamete types.
The clicker question on Slide 37 asks students to identify recombinant bands in the drought-tolerant (recessive, mutant phenotype) F2 plants (Question 2 of Handout B). Now, students are fairly successful at identifying the correct answer (74% correct on the first vote, Figure 5C and 5D). The clicker question on Slide 38 asks students to select which population they would choose for testing genetic linkage between the drought tolerance locus and the microsatellite in question, using F1 self-crossing (Question 3 of Handout B): the entire F2 population, drought-tolerant plants only, or drought-sensitive plants only. After discussions from the previous two questions, most students select the correct answer on the first vote (the drought-tolerant F2 only).
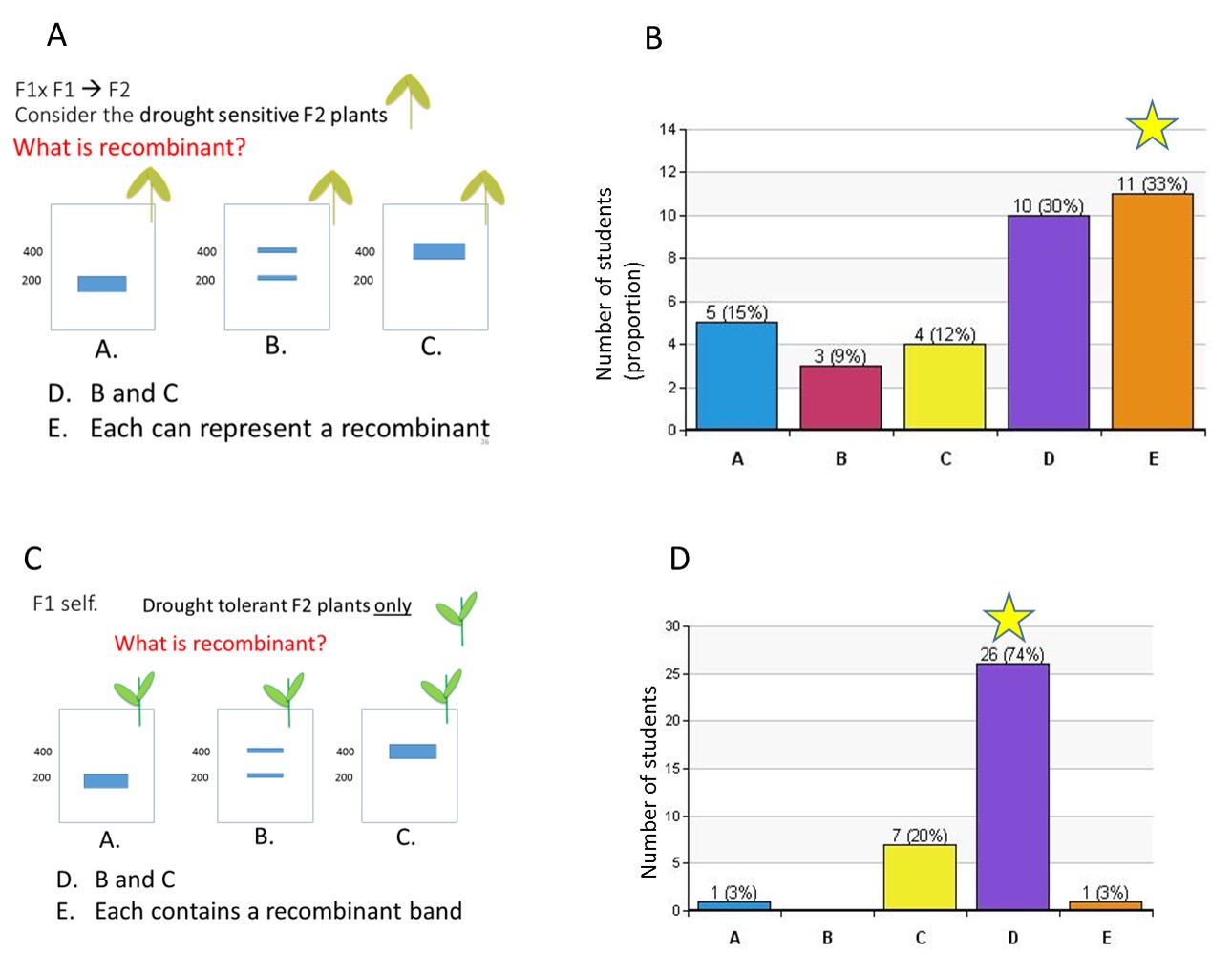
Figure 5. Clicker questions asked to check student understanding after they make predictions about F2 banding patterns. Questions were asked after student groups completed both the drawing activity (Handout A) and peer instruction (Handout B). (A) The first question asked students to identify bands on a gel as recombinant, which should also prompt them to consider which bands are parental (i.e. not recombinant). (B) Student responses indicate they may not be correctly inferring the genotype of the plant, and hence not recognizing all the bands could represent recombination. All three banding patterns can be the result of a recombinant and/or parental allele combination. It is for this reason that we cannot use the plants expressing the dominant phenotype to accurately assess linkage. Peer discussion after the first vote resulted in greater than 80% of students selecting the correct answer on a revote. (C) The second clicker question asked students to identify recombinant banding patterns when we are only examining the F2 plants that exhibit the recessive trait (i.e. Are drought tolerant and therefore we can infer they are homozygous for the recessive DRT allele?). (D) Student responses suggest improved understanding compared to the previous clicker question about which bands represent recombination events. The star indicates the correct answer.
The next clicker question sequence (Slides 39-40) leads students through calculating the map distance between the drought tolerance and microsatellite loci, using gel-banding patterns as their data set. Allow students to vote, and then share an explanation with them (see Lecture Script, S1b).
Note that, at the point in the lesson when we ask the question on Slide 39, we have intentionally not yet talked about how to calculate map distance in the F1 x F1 scenario, or that the method used for this cross differs from that used in the testcross scenario. In the drought tolerant F2 plants the 400 band is recombinant. If students apply the "rules" of calculating recombinant frequency and map distance that we learned in the testcross scenario, they will answer that the loci are linked and they are 30 map units apart, because 3/10 of the plants have the 400 band. In our experience, most students are likely to make this mistake, which we use as an opportunity to have a meaningful discussion about why we calculate map distance differently in the F1 x F1 scenario compared to the testcross. Slide 40 walks through the logic behind why simply counting plants that contain at least one recombinant chromosome is not the correct way to calculate map distance in this situation.
The emphasis of this discussion should be on the gamete genotypes that fuse to create the genome of the F2 plants. In the F1 x homozygous recessive testcross scenario shown on Slide 40 (DR(T) G(200)/ DR(S) G(400) x DR(T) G(200)/ DR(T) G(200) F2), only the gametes from the F1 dihybrid parent plant can be identified as parental or recombinant. In other words, only one of the DR-G allele combinations on one chromosome in the resulting F2 plants can be classified as parental or recombinant. Hence, we can classify the entire F2 plant as either a parental organism or a recombinant organism with respect to the two loci we are testing. To determine map distance we calculate the frequency of recombinant organisms in the total population analyzed.
In comparison, in the F1 x F1 cross (DR(T) G(200)/ DR(S) G(400)xDR(T) G(200)/ DR(S) G(400) F2), both gametes that make an F2 plant can be either parental or recombinant. As a result, we have to classify the DR-G allele combination on each chromosome that the F2 plant inherited as either parental or recombinant. With respect to the DR-G loci, one F2 plant may have no recombinant chromosomes, one recombinant chromosome, or two recombinant chromosomes. To determine recombination frequency we only analyze the plants that are drought tolerant (i.e. homozygous recessive, DR(T)/ DR(T)). So, in this F1 X F1 cross, to determine recombinant frequency (and, thus, map distance), we must count every recombinant band and divide by the total number of bands.
The question on Slide 41 can be used if time permits. The students are asked to analyze the banding patterns for drought-tolerant F2 plants produced by an F1 x F1 cross. All of the plants we analyze have the same homozygous 200/200 banding pattern. After the students vote, you can tell them: "In this case, all of the F2 plants have the same banding pattern (200/200), which indicates that all drought-tolerant F2 plants inherited the parental DR(T)G(200) and not the recombinant genotype DR(T)G(400). One reason for no recombinant genotypes showing up is that the DR and G loci are so tightly linked (i.e. very close to each other on the same chromosome), that little to no recombination occurs between them. It is possible that recombination does occur, but at such a low frequency that we do not see it in our sample size. You could extend this idea by asking the class "If no recombination occurs between the two loci, what genotypes and banding patterns do you expect in the drought-sensitive F2?" In this scenario, we predict the F2 from the F1 x F1 cross to be one quarter DR(T) G(200)/DR(T)G(200), one half DR(T) G(200)/DR(S)G(400), and one quarter DR(S)G(400)/DR(S)G(400). Among the drought-sensitive plants, two thirds of them will be 200/400 and one third will be 400/400.
OPTIONAL NEW SCENARIO: ASSESSING LINKAGE OF A BEETLE RESISTANCE GENE TO MULTIPLE MICROSATELLITES
This portion of the lesson involves students in assessing genetic linkage between molecular markers and a new trait of interest, resistance of potato plants to a pest, the potato beetle. After using the Class Slides to set up the scenario, ask students to work in groups of three on Handout C (S4). This handout shows the banding patters for six different microsatellites from progeny of an F1 X F1 cross. There are six different gels, each one showing the bands of a different microsatellite. This situation is much more challenging than the previous ones because the students have to classify multiple microsatellites as linked or unlinked to phenotype. However, it is also more like how an actual mapping experiment would be done. Tell the class that the first group to determine the correct genetic linkage, including the correct map distance, for all six markers wins the challenge. (We have sometimes rewarded the winning group with some chocolate bars, but have also simply awarded the winning team "bragging rights.") The group that finishes first can be encouraged to either raise their hand or come to the front of the room and explain their answer to the class, or to write their answer on a page projected by the document camera.
WHAT COMES AFTER MAPPING?
Slide 46 reminds students that crosses and linkage analysis are only a first step toward the larger goal of identifying the beetle (or drought) resistance locus. These microsatellite mapping experiments tell us that the gene of interest is located at a specific location on a specific chromosome. However, we still have work to do. We need to identify the specific gene and the specific mutation that are responsible for the resistant phenotype. Eventually, we would want to elucidate the function of the gene, as well as the mechanism by which this gene and gene product contribute to beetle resistance. Answering these questions can often take years.
AFTER CLASS
PROBLEM SET (TA-LED TUTORIAL OR HOMEWORK)
The tutorial problem set (S5a) was developed to give students additional practice with mapping using molecular markers. We usually have students work in groups of 3-4 in a TA-led tutorial session. Each person has an individual copy of the problem set, but students are encouraged to work as groups to help each other work through the problem. Alternatively, this problem set could be assigned as homework.
The first part of the problem set asks students to design a crossing scheme (i.e. true-breeding parents crossed to generate a heterozygote, followed by an F1 x F1 cross to generate an F2 population). After they design an experimental plan and make predictions about what the banding patterns will look like if there is genetic linkage, the teaching assistant can give each group a handout showing the gels to analyze (Handout D, S5b).
POST-TEST
The post-test asks students to explain the purpose of some steps in an experimental plan for mapping with molecular markers, to make predictions about banding patterns if two loci are assorting independently or are genetically linked, and to analyze data from a mapping experiment. Please see the S6 for the test and answer key. We have found this post-test to be very revealing in terms of student thinking and confusion related to linkage and mapping.
TEACHING DISCUSSION
The aim of this lesson plan was to help students connect the events of meiosis to the allele combinations found in gametes and offspring, while developing a stronger understanding of the effects of genetic linkage on both gamete and offspring genotypes and phenotypes. We chose to teach genetic linkage using mapping with molecular markers to provide a more modern context compared to traditional genetic linkage lessons and examples. The in-class activities are very amenable to adaptation to a tutorial format or as post-class homework assignments, although the in-class implementation lends itself very well to providing timely feedback to students. This activity would also be suitable as a pre-lab lesson for a molecular genetics lab that covers linkage or mapping. The examples of the lesson can easily be adapted to meet student and instructor needs. For example, if the F1 x F1 cross is too complex for the level of the class, one could teach only the testcross method and still have students predict the outcomes by drawing the expected banding patterns.
This activity was very successful at engaging students in using drawings of chromosomes to aid in making predictions. In addition, the group work and clicker questions allowed the students to benefit from peer and instructor feedback(15). There seemed to be just the right amount of struggle on the student's part: enough to stimulate learning, but not so much that they became disengaged. As instructors, we felt that the significant amount of drawing encouraged during this lesson was very important for student learning(16). In particular, it was rewarding for us to watch how the drawing and prediction activity forced students to confront their lack of awareness of the relationship between gametes, chromosome segregation in meiosis, and offspring phenotypes. During the whole lesson, students appeared to be very engaged and we observed multiple "ah ha" moments. There were many opportunities for us to interact with the students while they worked and discussed, making it a very enjoyable and rewarding lesson to deliver.
This lesson has been very effective at improving student's understanding of linkage. During one term of the course, students had a "traditional" lecture on mapping with molecular markers that did not include opportunities to predict banding patterns or discriminate between results of a testcross and an F2-self-fertilization cross. Post-test results of students in the traditional class revealed many misunderstandings about mapping with molecular markers (Figure 6). In particular, we noticed that the majority of students could detect genetic linkage correctly (i.e. linkage is occurring versus independent assortment); however, student explanations revealed that most of them had flawed logic for determining linkage. For example, many of them did not recognize how the bands on the gel could be used to determine if the gamete genotypes that created the F2 plant were parental or recombinant. When scoring the gel banding patterns from an F1 x F1 dihybrid cross, students frequently counted individual lanes rather than considering that each lane contains data from two chromosomes (i.e. a diploid plant). In comparison, when we delivered this very active lesson, they performed significantly better on the post-test (Figure 6). We feel that the practice and feedback provided by the in-class activities were critical factors that resulted in improved student understanding of genetic linkage, and mapping with molecular markers.

Figure 6. Student performance on one of the post-test items reveals improved understanding when we used this active in-class lesson. The test item required analysis of gel banding patterns to determine linkage between microsatellite markers and a locus of interest and a written explanation of how the data supports their conclusions. Students who engaged in the herein described active-learning based class, followed by the tutorial problem set, performed much better than students who received a more traditional (exposition focused) class followed by the tutorial problem set. Students from the active class were better at describing the linkage relationship correctly and correctly describing how the bands on the gel represented recombination. Students from the lecture-based class were much less likely to consider that each band represented a chromosome and a possible recombination (or parental) event.
This activity achieves our desired learning goals. Our in-class lesson is very active, providing students with multiple opportunities to directly engage with the material, make predictions, work in groups, and confront misunderstandings they may have. We hypothesize that requiring students to draw the chromosomes and connect those chromosomes to the banding patterns observed is one aspect of this lesson that helps students overcome some of the challenges to understanding meiosis and genetic linkage.
Use of this lesson and associated activities was a positive reminder for us that it is very important to allocate sufficient time for students to draw chromosomes when exploring meiosis and linkage. We strongly encourage others teaching genetic linkage to find new and engaging ways to teach this topic and get students deeply engaged in connecting chromosomes to gametes to alleles to phenotypes.
SUPPORTING MATERIALS
- S1a. Genetic Linkage-Lesson Power Point Slides
- S1b. Genetic Linkage-Lecture Script
- S2a. Genetic Linkage-Handout A1
- S2b. Genetic Linkage-Handout A2
- S3. Genetic Linkage-Handout B and Answers
- S4. Genetic Linkage-Handout C and Answers
- S5a. Genetic Linkage-Tutorial Problem Set and Answers
- S5b. Genetic Linkage-Tutorial Gels for Analysis
- S6. Genetic Linkage-Post-Test and Answers
ACKNOWLEDGMENTS
We would like to acknowledge the support we received from Dr. Craig Berezowsky who provided feedback on the tutorial activity, and helped administer the first iteration of the tutorial activity and the post-test. We thank our teaching assistants, who were instrumental in administering the tutorial activity and post-test. Thank you to all of the students who participated in class.
References
- CourseSource. Course Source and Genetics Society of America Learning Framework for the Course "Genetics". www.coursesource.org. Accessed July 16, 2014.
- Griffiths A, Wessler SR, Caroll SB, Doebley, J. 2012. Introduction to Genetic Analysis. New York, NY: W.H. Freeman.
- Mrazek J, Guo X, Shah A. 2007. Simple sequence repeats in prokaryotic genomes. Proceedings of the National Academy of Sciences USA 104: 8472-8477.
- Cosson P, Decroocq V, Revers F. 2014. Development and characterization of 96 microsatellite markers suitable for QTL mapping and accession control in an Arabidopsis core collection. Plant Methods 10:2-6
- Bell CJ, Ecker JR. 1994. Assignment of 30 Microsatellite Loci to the Linkage Map of Arabidopsis. Genomics 19: 137-144.
- Senior ML, Heun M. 1993. Mapping maize microsatellites and polymerase chain reaction confirmation of the targeted repeats using a CT primer. Genome 36: 884-9
- Remya KS, Joseph S, Lakshmi PK, Akhila S. 2010. Microsatellites in varied arenas of research. Journal of Pharmacy and BioAllied Sciences. 2: 141-143.
- Liu XM, Smith CM, Gill BS, Tolmay V. 2001. Microsatellite markers linked to six Russian wheat aphid resistance genes in wheat. Theoretical and Applied Genetics 102: 504-510
- Zhang J, Zheng HG, Aarti A, Pantuwan G, Nguyen TT, Tripathy JN, Sarial AK, Robin S, Babu RC, Nguyen BD, Sarakarung S, Blum A, Nguyen HT. 2001. Locating genomic regions associated with components of drought resistance in rice: comparative mapping within and across species. Theoretical and Applied Genetics 103: 19-29.
- Courtois B, McLaren G, Sinha PK, Prasad K, Yadav R, Shen L. 2000. Mapping QTLs associated with drought avoidance in upland rice. Molecular Breeding 6: 55-66.
- Rahman H, Pekic S, Lazic-Jancic V, Quarrie SA, Shah SMA, Pervez A, Shah MM. 2011. Molecular mapping of quantitative trait loci for drought tolerance in maize plants. Genetics and Molecular Research 10: 889-901.
- Harris PC, Thomas S, Ratcliffe PJ, Breuning MH, Coto E, Lopez-Larrea C. 1991. Rapid genetic analysis of families with polycystic kidney disease 1 by means of a microsatellite marker. The Lancet 338: 1484-1487.
- Padgette S, Goette J, Mazour C. 2012. Drought Tolerant Corn. Retrieved from http://www.monsanto.com/sitecollectiondocuments/whistlestop-drought-posters.pdf
- Aronson A. 1978. The Jigsaw classroom. Beverly Hills, CA: Sage.
- Smith MK, Wood WB, Krauter K, Knight JK. 2011. Combining peer discussion with instructor explanation increases student learning from in-class concept questions. CBE Life Sciences Education 10: 55-63.
- Bell JC. 2014. Visual Literacy Skills of Students in College-Level Biology: Learning Outcomes following Digital or Hand-Drawing Activities. The Canadian Journal for the Scholarship of Teaching and Learning 5: 6.
Article Files
Login to access supporting documents
Teaching Genetic Linkage and Recombination through Mapping with Molecular Markers(PDF | 1 MB)
S1a. Genetic Linkage-Lesson Power Point Slides.pptx(PPTX | 4 MB)
S1b. Genetic Linkage-Lecture Script.docx(DOCX | 40 KB)
S2a. Genetic Linkage-Handout A1.docx(DOCX | 129 KB)
S2b. Genetic Linkage-Handout A2.docx(DOCX | 130 KB)
S3. Genetic Linkage-Handout B and Answers.docx(DOCX | 209 KB)
S4. Genetic Linkage-Handout C and Answers.pptx(PPTX | 87 KB)
S5a. Genetic Linkage-Tutorial Problem Set and Answers.docx(DOCX | 152 KB)
S5b. Genetic Linkage-Tutorial Gels for Analysis.ppt(PPT | 1 MB)
S6. Genetic Linkage-Post-Test and Answers.docx(DOCX | 67 KB)
- License terms

Comments
Comments
There are no comments on this resource.