Using Synthetic Biology and pClone Red for Authentic Research on Promoter Function: Genetics (analyzing mutant promoters)
Published online:
Abstract
Students often memorize the definition of a transcriptional promoter but fail to fully understand the critical role promoters play in gene expression. This laboratory lesson allows students to conduct original research by characterizing functional regions within known prokaryotic promoters. Students begin the lesson by learning the properties of transcriptional promoter DNA sequences. They design mutations for a constitutive promoter and discuss their designs as a class to choose which mutations to clone and characterize. This lesson provides an easy way for faculty with limited time and budgets to give their students access to real research in the context of traditional teaching labs that meet once a week for under three hours. The pClone Red Genetics lesson uses synthetic biology methods and makes cloning so simple that we have 100% success rates with sophomores taking Genetics. Students archive promoter sequences and their performances under standard conditions. The database permits synthetic biology researchers around the world to find a promoter that suits their needs and compare relative levels of transcription. The core methodology in this lesson is identical to the core methodology in the companion Introductory Biology Lesson by Campbell and Eckdahl. The methods are reproduced in both lessons for the benefit of readers. The two CourseSource lessons provide the detailed information needed to reproduce the pedagogical research results published in CBE – Life Sciences Education by Campbell et al., 2014.
Citation
Eckdahl, T.T. and Campbell, A.M. 2015. Using Synthetic Biology and pClone Red for Authentic Research on Promoter Function: Genetics (analyzing mutant promoters). CourseSource. https://doi.org/10.24918/cs.2015.5Society Learning Goals
Genetics
- Molecular biology of gene function
- How is genetic information expressed so it affects an organism's structure and function?
- Gene Expression and Regulation
- How do genes and genomes control changes in an organism's structure and function throughout its life cycle?
- Genetic variation
- How do different types of mutations affect genes and the corresponding mRNAs and proteins?
- Genetics of model organisms
- How do the results of molecular genetic studies in model organisms help us understand aspects of human genetics and genetic diseases?
- Methods & Tools in Genetics
- What experimental methods are commonly used to analyze gene structure and gene expression?
Science Process Skills
- Process of Science
- Pose testable questions and hypotheses to address gaps in knowledge
- Plan, evaluate, and implement scientific investigations
- Interpret, evaluate, and draw conclusions from data
- Construct explanations and make evidence-based arguments about the natural world
- Address novel questions through authentic research experiences
- Modeling/ Developing and Using Models
- Recognize the important roles that scientific models, of many different types (conceptual, mathematical, physical, etc.), play in predicting and communicating biological phenomena
- Make inferences and solve problems using models and simulations
- Build and evaluate models of biological systems
- Quantitative Reasoning/ Using Mathematics and Computational Thinking
- Apply the tools of graphing, statistics, and data science to analyze biological data
- Communication and Collaboration
- Share ideas, data, and findings with others clearly and accurately
Lesson Learning Goals
- Students understand how genes are regulated at the transcriptional level by internal and external conditions.
- Students will know how cells with the same genome can produce different proteins.
- Students will demonstrate how cell genomes can be manipulated through experimentation to alter function.
- Students will know how microorganisms serve as good model organisms for fundamental processes.
Lesson Learning Objectives
- Describe how cells can produce proteins at the right time and correct amount.
- Diagram a bacterial promoter with −35 and −10 elements and the transcription start site.
- Describe how mutational analysis can be used to study promoter sequence requirements.
- Develop a promoter mutation hypothesis and design an experiment to test it.
- Successfully and safely manipulate DNA and Escherichia coli for ligation and transformation experiments.
- Design an experiment to verify a mutated promoter has been cloned into a destination vector.
- Design an experiment to measure the strength of a promoter.
- Analyze data showing reporter protein produced and use the data to assess promoter strength.
- Define type IIs restriction enzymes.
- Distinguish between type II and type IIs restriction enzymes.
- Explain how Golden Gate Assembly (GGA) works.
- Measure the relative strength of a promoter compared to a standard promoter.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
One of the core concepts in biology is information storage and flow (1). To help students master this core concept, genetics courses typically cover how promoters serve as key genetic elements that determine the conditions under which a gene will be transcribed and how much mRNA will be produced. For more than twenty years, we have noticed that our students could recite the definition of a promoter, but many were unable to relate that definition to cell functions. For example, students were often surprised to learn that mutations in promoters can result in new phenotypes or diseases. To help students develop a deep understanding of the role of promoters in gene expression, we developed a new lab lesson that would give our students extensive, hands-on experience with promoters that they designed based on information from their textbook and material covered during class time. This lesson was designed for a genetics course and is closely associated with a similar CourseSource lesson in Introductory Biology.
In addition to learning about promoter structure and function, we also wanted our students to conduct real research rather than simply demonstrate a known outcome. Conducting research improves science education while also increasing the number and diversity of scientifically literate citizens (3-8). To accomplish these research goals, we drew on our years of experience of mentoring undergraduates in synthetic biology research (9-15). Synthetic biology is the use of engineering principles, molecular cloning methods, and mathematical modeling to design and construct biological parts, devices, and systems with applications in energy, medicine, and technology. Synthetic biology is a new area of research that is amenable to undergraduate and high school students (16-20). In the context of the recently redesigned AP Biology curriculum, the pClone Red Genetics lesson would also be appropriate for high school AP Biology courses (21, 22).
To fully understand promoters, we believe that students need to study a promoter in the lab. Specifically, our students design mutations in a known promoter and make predictions about how the promoter function will change as a result. They test their predictions by cloning their mutated promoter in the pClone Red expression plasmid, as described in detail later in this lesson. Because it is very difficult to measure promoters in absolute units (e.g., number of mRNA molecules per cell), we provide students with a positive control RFP expression device (number J04450) that uses a very well characterized promoter (Plac; part number R0010) and have them calculate the relative strength of their promoter compared to Plac which is 200 bp long. One benefit of using Plac is that most genetics classes also cover the lac operon. Using this familiar promoter in lab has the added benefit of reinforcing concepts learned during class time. For practical considerations, we limit choice of wildtype promoters that are no more than 60 basepairs (bp) long because the company that supplies our oligonucleotides charges more per base for synthesis over 60 bases. Students taking Genetics as sophomores at MWSU are charged with applying what they have learned in lecture and from their textbook about the properties of bacterial promoters to a research proposal they design. To start the process, students complete a take-home quiz (see S2) in which they propose two mutations to make in a consitutive promoter that would provide information about the promoter’s function. The students share their mutation plans with each other and discuss the merits of their designs during lab. Through this discussion, they arrive at a group decision about which two mutations to clone and study in each lab section. With three lab sections, the class as a whole can clone and study six promoter mutations. During the discussion, we emphasize that the students are engaging in authentic and often original research.
Once the students have designed the mutant promoters, they design complementary oligonucleotides containing the mutant promoter sequence. We order their oligonucleotides, which the company returns within three business days. The students clone their mutant promoters into the pClone Red plasmid (part number J119137), which we designed and built (Figure 1; (23)).
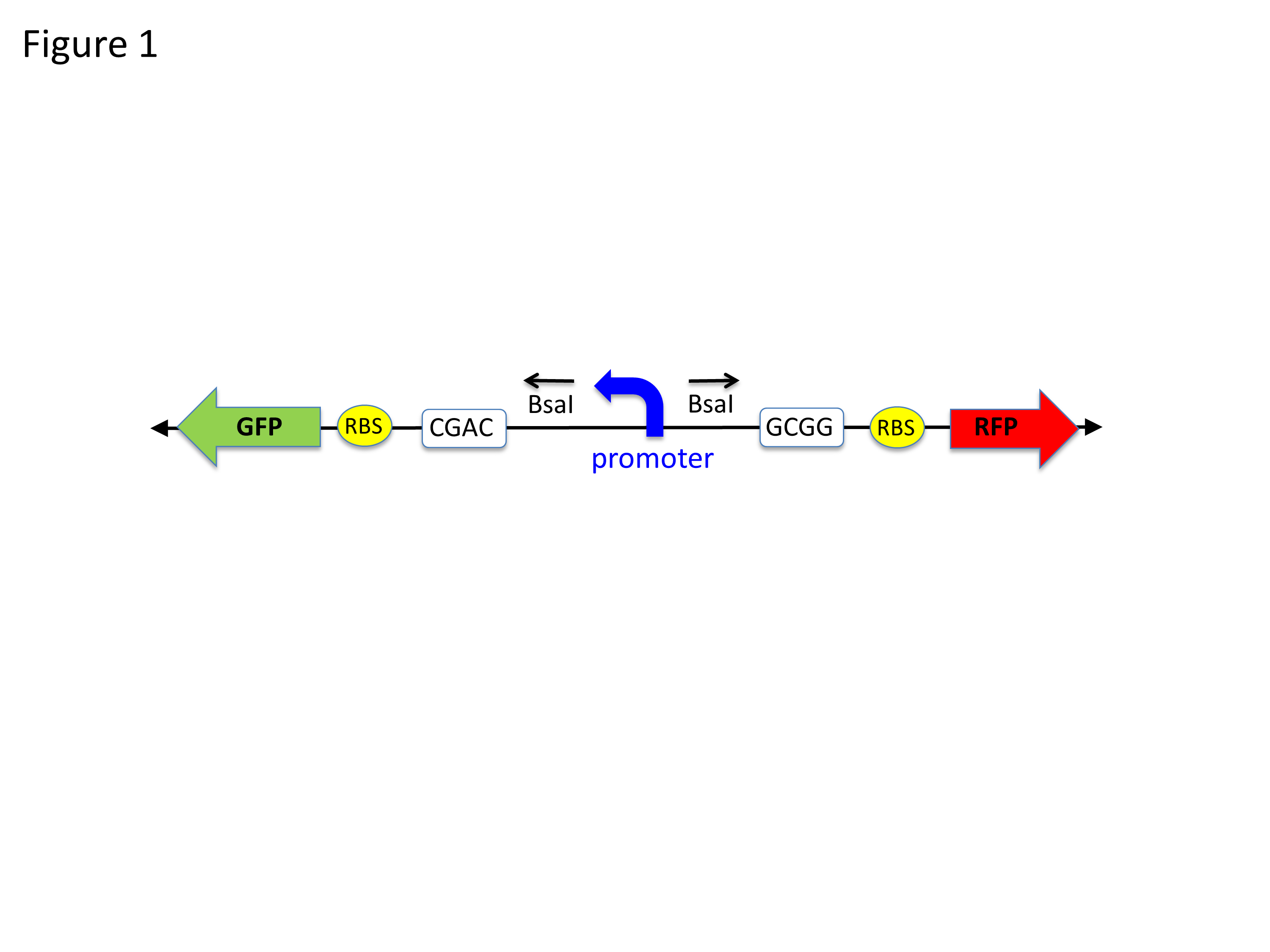
Figure 1. Diagram showing the key parts of the receiving plasmid pClone Red. A “backwards” GFP gene (promoter, RBS, and GFP coding) is oriented from right to left. The backwards PlacIQ1 promoter is flanked by outwards-facing Bsa I restriction sites. Digestion by BsaI removes the promoter and generates unique sticky ends (white boxes) that allow a mutant “forward” promoter to be ligated in the proper orientation to initiate transcription of RFP, to the right of RBS.
The pClone Red plasmid is the key to the success of this lesson. “Red” refers to the common reporter red fluorescent protein (RFP) that is included in the plasmid. If unaltered pClone Red is transformed into cells, the cells appear green because they contain a GFP expression cassette of a “backward” green fluorescent protein (GFP) gene and a “backward” (right to left in Figure 1) promoter (PlacIQ1; part number K091112). This backward promoter is located between GFP and the “forward” (left to right in Figure 1) RFP coding sequences on the plasmid. If the students’ mutant promoter is functional, it will initiate transcription of RFP (Figure 2) and the transformed cells will appear red if they produce sufficient amounts of RFP.
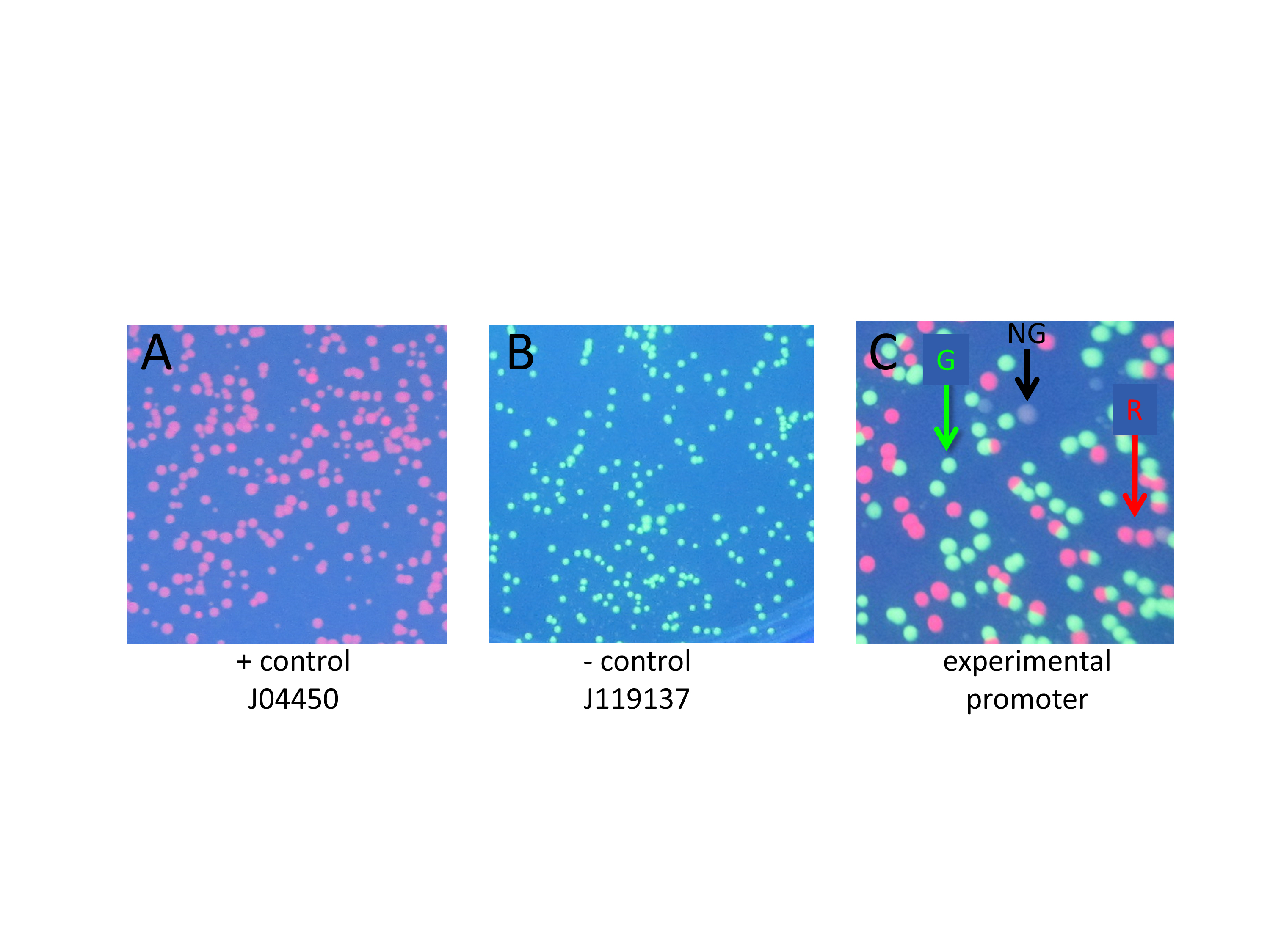
Figure 2. Close up photographs showing positive control, negative control, and experimental transformation plates. A) Positive control transformation of part J04450: colonies contain RFP and transcription is driven by Plac, producing only red colonies; no GGA performed. B) Negative control transformation: colonies contain pClone Red after GGA in the absence of mutant promoter oligonucleotides. The green colonies contain an intact pClone Red plasmid with the backwards promoter, and a few colorless colonies that contain pClone Red that religated after removal by BsaI of the Plac promoter. C) Experimental transformation: pClone Red after GGA in the presence of mutant promoter sequences. The plate shows all three colony colors as indicated by colored arrows (R = red, G = green, NG = not green). Red colonies contain a mutant promoter that drives RFP transcription; green colonies contain unaltered pClone Red; white colonies have a non-functional mutant promoter that cannot drive sufficient RFP transcription.
Students who clone a promoter can see three possible colors of colonies when their tranformation plates are viewed under UV light. Green colonies still contain the original backwards Plac promoter. Red colonies contain a functional mutated promoter. Colorless colonies contain a mutant promoter that is unable to drive sufficient expression of RFP to be visible in UV light.
To accomplish all of the cloning steps within the constraints of the ~ 3 hr lab periods, students use a new cloning method that is extremely easy, called Golden Gate Assembly (GGA; (24, 25). GGA involves a one-tube reaction containing the type IIS restriction enzyme BsaI and DNA ligase. The restriction enzyme removes the backward PlacIQ1 promoter and generate distinct sticky ends for directional cloning of the mutant promoter. The DNA ligase directionally inserts the double stranded mutant forward promoter into the gap. Thus, GGA eliminates GFP transcription driven by the backward PlacIQ1, while simultaneously cloning the “forward” mutant promoter (left to right in Figure 1). The removal of the constitutive backward PlacIQ1 promoter and cloning of the mutant forward promoter with GGA takes place in a single tube in one hour. Transformation of E. coli takes about 15 minutes. Students can easily perform the entire process in a normal laboratory session. After students have transformed and plated the bacterial cells, each lab group prepares to quantify the RFP produced by the transformed cells using a fluorometer (23).
Intended Audience
Genetics laboratory course for potential majors, sophomores
Learning Time
Minimum of four weeks in lab, plus time for oral presentations.
Pre-requisite Student Knowledge
Students need to be able to pipet small volumes and work sterilely with bacteria. They need to know DNA base pairing and have learned about transcription. This laboratory lesson goes hand-in-hand with classroom material and they reinforce each other.
SCIENTIFIC TEACHING THEMES
Active Learning
- Student lab groups decide mutations they want to make to the promoter and formulate a testable hypothesis about the function of their mutated promoter.
- Students use their textbook and understanding of how promoters work to design a mutation they want to make and test in the promoter.
- Students perform the laboratory procedures of cloning a promoter, transforming cells, and identifying colonies that contain the mutated promoter.
- Students test whether their mutated promoter performs as they had predicted.
- Students interpret the quantitative data they produce.
- Students generate graphical representations of their data and conclusions.
- Students communicate their findings in oral presentations.
Assessment
- Students will assess whether their results support their hypothesis.
- Students present their results and interpretations in graded oral lab reports.
- Students take pre- and post-surveys to measure comprehension of core concepts.
- Students submit their promoter designs and results to the Registry of Functional Promoters (RFP) database.
Inclusive Teaching
- Student lab groups include individuals with different learning styles and prior knowledge. They learn how to work together through cooperation.
- Students must communicate, collaborate, and self-organize at many points throughout the laboratory sessions. Each person must contribute at multiple steps every week.
- Oral presentations include questions and answers where some students thrive more than others do.
LESSON PLAN
The Timeline for this lesson is available in Supporting File S1.
Instructor Preparation before class
Instructors can purchase the pClone Red plasmid from Carolina Biological starting in the fall of 2015. Once they have the plasmid, they can use the reagents supplied in the Carolina Biological kit, or assemble their own reagents and produce ample plasmid from a single miniprep. Details of the ligation preparation procedure are available in the kit or from the supplemental file entitled “Golden Gate Assembly with pClone Red.” We have found that students more fully understand GGA when they perform the paper activity during the GGA reaction. Instructors will want to print enough copies for each student to have one copy (see supplemental PowerPoint file) as well as access to scissors and clear tape. We have provided several additional PowerPoint files that instructors can use to present and overview of GGA and experimental details. Instructors will want to familiarize themselves with the synthetic biology registry of standardized DNA parts (http://parts.igem.org/Main_Page). All part numbers (such as J119137) are described in detail in the registry.
Lab Sessions
General Information
Because this is a four-week wet-lab module, we start slowly to allow time in the classroom to cover DNA structure and gene regulation. Then, we cover how cloning works in general and GGA with pClone in particular (see S3). Students visit the two DNA registries they will be using so they realize their work will be part of a publicly accessible database of promoters and their functions. Knowing that their research will be part of a global research enterprise helps students recognize they are doing real science and not just reproducing outcomes that are already known.
Students take a pre-survey the first day of class to capture what they know prior to any instruction (see S4 and S5). There is no simple way into the literature for beginning students, so they are taught how to search for papers that describe the function of bacterial promoters and contain promoter DNA sequences. Students do not need to understand the entire paper and should focus on the promoter’s function in a prokaryote and whether it is positively or negatively regulated, or constitutive (always functions). From this starting point, they can develop a hypothesis about how their promoter design will function in E coli. To limit our costs and increase cloning success rates, the total length of the mutant promoter cannot exceed 60 bp. Our DNA provider, www.idtdna.com, has a cost inflection point at 60 bases (about $7 for each of the two oligos), so none of the student mutant promoters can be longer than 60 bp long. This size restriction will accommodate most constitutive bacterial promoters.
Designing a Mutated Promoter and Generating Promoter Oligonucleotides - Week 1
The first lab period in this sequence begins with a summary presentation to students about how pClone Red makes promoter research accessible (see S6). After question and answers, students begin the design phase of their research process. Students refer to their completed Promoter Quiz, in which they propose two mutations in a consitutive promoter that would provide information about the promoter’s function (see S2). The students share their mutation plans in lab and discuss the merits of each idea. Through this discussion, the group chooses which mutations to clone and test. Sometimes the discussion generates new ideas that combine different student proposals. We encourage the students to be specific about predicting the effect of their mutations on promoter function, based on the structure/function relationships they have learned. Student often ask their instructor what the effect of their mutation is going to be and are empowered when the instructor does not know the answer and adds that perhaps no one knows the answer.
The lab session continues with an instructor presentation showing the details of how pClone works (see S7). The main outcome of the Week 1 lab is to design the two oligonucleotides that will form the double-stranded DNA (dsDNA) with the mutant promoter sequence that will be cloned into the pClone Red plasmid (see S8). Students will need to add one sticky end to each oligonucleotide, so that the resulting dsDNA will be correctly oriented to drive RFP expression in the plasmid. Students must submit their promoter sequences to the Registry of Standard Parts (26). Students submit their sequence to the registry and the default designation is “planning.” Once the students clone and test their promoter, they will provide functional information and convert the status from “planning” to “works” or “failed.” This is the normal procedure for all parts submitted to the Registry.
Cloning the Promoter - Week 2
For each of three lab sections, two mutation plans are developed. The six mutations are cloned and studied in each of the lab sections. The afternoon before lab, at least one student from each lab group comes to lab to mix their oligos, boil them for 5 minutes, and allow them to anneal overnight (see S9). During their regular lab meeting the next day, students dilute the cooled oligos in preparation for the GGA reaction. After diluting the annealed oligos, students add them to the GGA mixture, which contains the pClone Red plasmid, type IIS restriction enzyme BsaI, DNA ligase, ATP, and a buffer (see S10). The instructor prepares aliquots of the GGA mixture in advance; the aliquots are stored at -20° C. The students prepare a negative control GGA reaction that lacks the mutant promoter oligonucleotides. Later, they will transform their positive control plasmid (part J04450) that uses the constitutive promoter Plac to drive RFP transcription.
The GGA reaction tubes are placed into a thermocycler for at least 20 rounds of a two-step cycle: 1 minute at 37° C and 1 minute at 16° C. (More rounds produce higher success rates but take longer.) The temperature cycles reflect optimal temperatures of the enzymes in the GGA reaction: the restriction enzyme BsaI works optimally at 37° C and DNA ligase works well at 16° C. After the cycles, a final step of 15 minutes of 37° C is added to reduce the number of undigested pClone Red plasmids. Once GGA is completed, students do three transformations with different DNA samples: 1) negative control of pClone Red after GGA, without addition of mutant promoter oligonucleotides that produce only green colonies; 2) the experimental GGA reaction containing their mutated promoter and 3) positive control plasmid with Plac promoter driving transcription of RFP (part number J04450; see S11). The students plate each transformation onto separate LB + ampicillin plates, which are allowed to grow about 16 hours at 37° C before photographing under UV light.
While the GGA reaction is in the thermocycler, students perform the paper GGA exercise to better understand what is happening to pClone Red and their oligonucleotides (see S12).
Characterizing the Promoter - Week 3
The afternoon prior to lab, at least one student from each group picks three colonies from each plate and grows them overnight at 37° C in 2 ml of LB + ampicillin broth (see S13). To select colonies, students place the transformation plates colony side down a UV light box, which allows students to distinguish red, green, and colorless colonies. Each group picks three green colonies from the negative control plate, three red colonies from the positive control plate, and three non-green colonies (red or colorless) from the experimental colonies. No matter how few in number, red colonies indicate the mutated promoter is working. If no red colonies appear, then the mutated promoter is not strong enough to drive sufficient RFP transcription to produce a visible red color, or does not work at all.
When lab starts, students photograph the plates and tubes of cells before using a 96-well plate reader to measure RFP fluorescence (585 nm excitation and 615 nm emission) and light absorbance (590 nm) to quantify cell density. Students calculate the fluorescence to absorbance ratio to normalize the amount of RFP signal to the number of cells present (see Options to Extend the Lesson to measure promoter strength in lieu of a fluorometer). Fluorescence and absorbance do not have absolute units so they are measured in arbitrary units. Students spend their remaining time interpreting their results and generating graphical representations of their data. They also submit their results to the two DNA registries, including fold induction when comparing the Plac promoter to their mutant promoter (Figure 3). Students submit the information as prompted by the Registry of Functional Promoters (27). Students also return the Registry of Standard DNA Parts and update their promoters with the “experience” data so other users can see how well the promoter works, if it works at all.
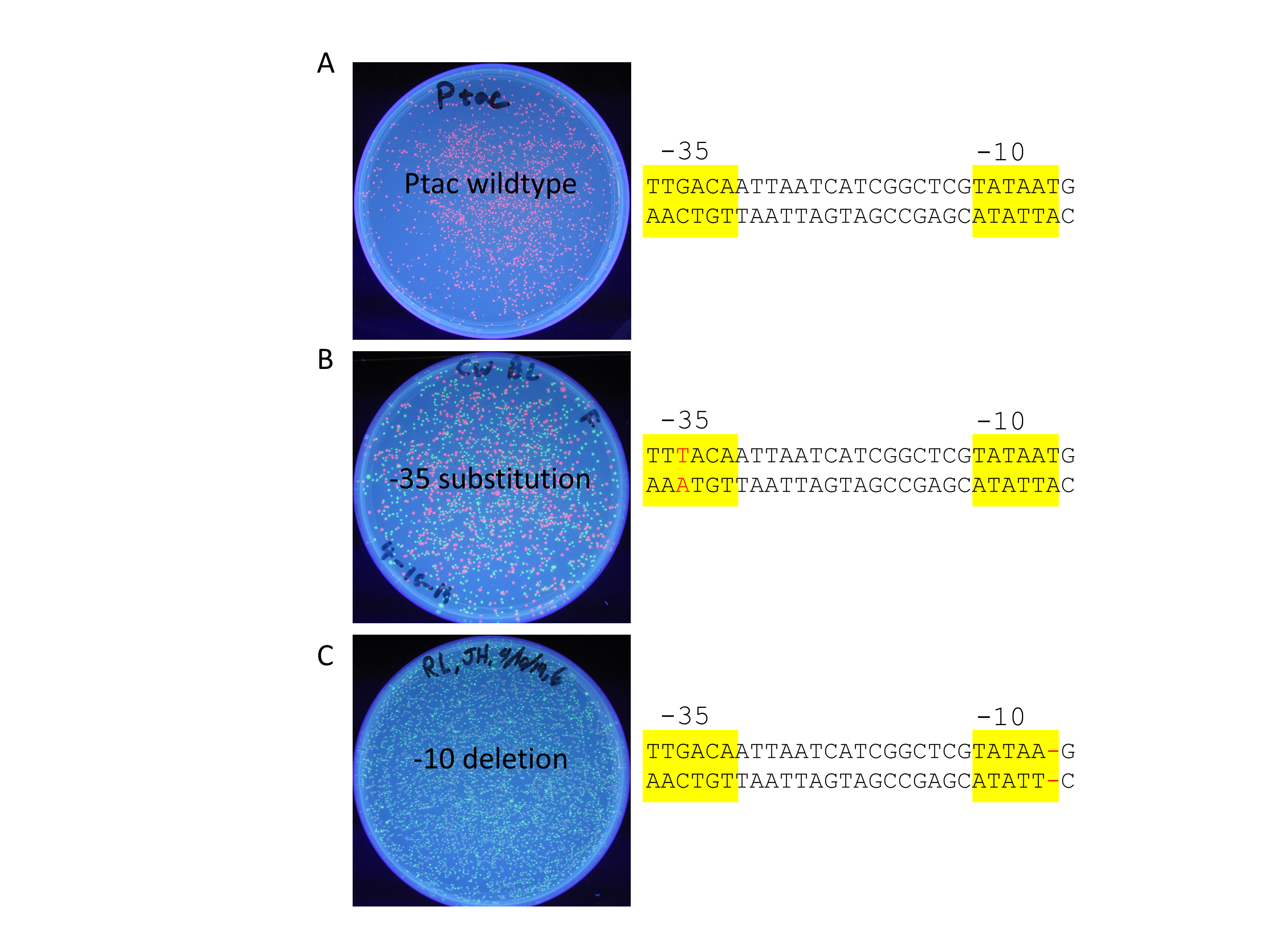
Figure 3. Student data comparing wild-type and two mutant promoters. A) Wild-type Ptac promoter driving the production of RFP. B) Mutant promoter with a base pair change from G to T (red font) on the top strand. C) Mutant promoter with a one base deletion (red dash) in the -10 box.
Presenting the Results - Week 4
Outside of lab, students prepare and practice their oral presentations. Each presentation is limited to ten minutes and divided into four sections – introduction, methods, results, and discussion. Students work together to prepare their presentations and are encouraged to share the workload and the presentation time. The instructor provides students with a graded evaluation of their presentation that counts for one quiz grade.
Options to Extend the Lesson
The pClone lesson has four possible extensions, all of which are appropriate for genetics students.
Extension 1: Quantifying without a fluorometer
If a fluorometer is not available, students can take a photograph of colonies and then use the free software ImageJ to quantify the redness of the colonies (28). This photographic method is not as accurate as using a fluorometer, but it will give quantitative data that students can use to assess relative promoter function. The details for using ImageJ are in the supplemental materials (see S14, (23)). It is worth noting that, unlike GFP, RFP is easily visualized with normal room lighting.
Extension 2: Quantifying without a UV light box
If you do not have a UV light box, or want to use a different reporter, you can use pClone Blue (part J119313) that has amilCP Blue as the reporter gene instead of RFP (29). pClone Blue is paired with a positive control plasmid (part J119128) that contains the well-characterized promoter P5. As with pClone Red, pClone Blue can be quantified by photographs of colonies analyzed by ImageJ. There is a second method to collect qualitative data using pClone Blue. You can grow the positive control blue cells and produce a serial dilution. From this dilution series, you can measure absorbance at 400 nm and generate a standard curve against which you can compare any mutant promoters (see S15).
Extension 3: Dissect function of bases in known promoter
Rather than mutating a known promoter, students can identify a new promoter from the literature for cloning and testing. This extension is described in detail in the companion CourseSource Introductory Biology Lesson by Campbell and Eckdahl. Analysis of a new promoter could be combined with the one described in the current lesson by having student introduce mutations into promoters that are expected to be under positive or negative control.
Extension 4: Verify cloning of non-functional promoter
One limitation of pClone results is that plasmids may contain no promoter at all; the original promoter was removed and the plasmid re-ligated without insertion of the mutant promoter into the plasmid. This plasmid religation produces colorless colonies, which is the same phenotype as a plasmid that contains a mutant promoter that does not work or works at a very low level. To distinguish these two possible genotypes, you could either have some plasmids sequenced, or use PCR to distinguish the longer original promoter (PlacIQ1 is 70 bp) from the shorter mutant promoters (see S16). For example, the P5 promoter is only 36 bp which is easily distinguished from PlacIQ1. We have used PCR screening with genetics students before, but stopped because it was not central to the learning goals.
TEACHING DISCUSSION
Technical Issues
Perhaps the most compelling aspect of this lesson is that 100% of all lab groups have successfully cloned their mutant promoters despite them being sophomores with no prior cloning experience. pClone Red is inexpensive and does not require any specialized equipment unless you want to quantify the RFP by fluorescence. Students have cloned and tested many different types of mutations including insertions, deletions, and substitutions. Less than half of these mutant promoters have worked as predicted. Students have various reactions to disproving their hypotheses. Some assume that they have made a mistake; others are frustrated that they did not get the results they thought they were supposed to get; and others realize that they may have discovered something new about how bacterial promoters function. Regardless of the nature of students’ reactions to the results, we help them learn that disproving hypotheses is a valuable part of science. Students realize that their hypotheses can be wrong even when they did everything correctly. We want them to realize that even a negative result is worth recording in the two Registries associated with this lesson. As in “real” science, unexpected results can lead students to the literature where they are looking for good explanations for their results (Figure 3B).
Student Comments
Students often start this lab lesson with a fair bit of confusion about what they are doing, but early confusion can be stimulate learning. As they progress into the semester, they learn more and more about promoters so their lab experiences make more sense with time. Students have repeatedly indicated that they like doing real promoter research rather than canned labs. Students often express surprise at how accessible authentic research on promoter function was made using the pClone Red lesson. These comments are consistent with the data discussed in Vision and Change (1) as well as BIO2010 (3). Cognitive psychologists have documented that students learn best when they construct their own knowledge. The concept of knowledge construction is consistent with students in our class moving from initial confusion to eventual clarity about how a promoter works and how their research with pClone Red tested the function of their promoter (30).
Learning Outcomes
Recently, Classroom Undergraduate Research Experiences (CUREs; (8, 31-36) have received a lot of attention. The pClone Red promoter research lesson provides an easy way to scale up CURE opportunities in all teaching lab sections. In our paper describing pClone Red (23), we showed that students accomplished seven of the nine genetics learning objectives. Interestingly, the students were not able to adequately describe the GGA method, even though they had performed it to clone their mutated promoters! Because of their confusion about GGA, we instituted the paper activity where students physically manipulate paper versions of DNA during GGA (see S12). This activity can be performed by students during the hour-long GGA reaction.
The pClone Red Genetics lesson addressed one of the core concepts and several core competencies outlined in Vision and Change (1). It provides faculty with a lost cost lab module that has allowed 100% of the student lab groups to clone a promoter they designed. The students evaluated their hypotheses about how their promoters would function. Students are actively engaged in constructing their own understanding about promoters while using many learning strategies. Feedback on course evaluations has been very favorable and students synthesize their knowledge in oral and written laboratory reports that confirm that many of the learning goals and objectives are accomplished.
A common misconception is that different cell types in a multicellular organism contain different genes rather the correct understanding that all cells containing the same genes that are differentially regulated at the transcriptional level. Working with promoters over an extended period and testing the strength of a promoter they designed helps students gain a greater appreciation for the central role promoters play in cell and molecular biology. Furthermore, students learn to clone using a new method that is substantially easier and more effective than the traditional approach that often requires gel purification and the need for sticky ends generated when type II restriction enzymes cut within their recognition sites. Students often think that the newest method is normal, rather than being as impressed as their instructors are by the innovation. Despite their inability to appreciate the advantages of the new methods, it is very common for students to share their excitement during lab that this synthetic biology promoter lab module is real research that other biologists can access and use in their own research. It should not be surprising that students prefer to do meaningful lab work, rather than busywork replicating previous results, or studying a new question that is of little interest by the scientific community. It is exciting to offer Genetics students a meaningful research experience that is inexpensive and allows every lab group to clone and test original mutation hypotheses. For four years, 100% of the student lab groups have successfully cloned their mutated promoters. Often the effect of the mutations on promoter function is surprising to students and the instructor. This produces a very rich learning environment when students can learn that unexpected results are normal in research.
This lesson spells out the details of how to implement a course-based, undergraduate research experience (CURE) within the budget and time constraints of a typical lab. In particular, the degree of difficulty and cost of the lab are consistent with a typical introductory biology course intended for majors and potential majors. In 2014, we published a detailed description of the learning objectives and student assessment data in CBE – Life Sciences Education (23).
SUPPORTING MATERIALS
- S1. pClone Genetics-Timeline of pClone Red Genetics Laboratory Lesson
- S2. pClone Genetics-Genetics Promoter Mutation Quiz
- S3. pClone Genetics-Links to Introduction to Synthetic Biology and Cloning
- S4. pClone Genetics-Pre- and Post-Assessment Questions, no answers
- S5. pClone Genetics-Pre- and Post-Assessment Questions, with answers
- S6. pClone Genetics-Overview of Golden Gate Assembly for Promoter Research
- S7. pClone Genetics-Details of Golden Gate Assembly for Promoter Research
- S8. pClone Genetics-Designing Mutant promoter
- S9. pClone Genetics-Annealing Promoter Oligonucleotides
- S10. pClone Genetics-Golden Gate Assembly with pClone Red
- S11. pClone Genetics-Transforming DNA into E. coli Cells
- S12. pClone Genetics-Paper Activity for GGA Cloning into pClone
- S13. pClone Genetics-Phenotype Analysis after Transformation
- S14. pClone Genetics-Quantifying pClone Red Promoter Strength with ImageJ (no fluorometer)
- S15. pClone Genetics-Using pClone Blue instead of pClone Red (no UV light)
- S16. pClone Genetics-pClone Genotyping to Verify Promoter Cloning
ACKNOWLEDGEMENTS
We thank Brian Cronk, Corinne Andresen, Paul Frederick, Samantha Huckuntod, Claire Shinneman, Annie Wacker, and Jason Yuan for their help in developing pClone Red. Support was provided by the National Science Foundation RUI grants MCB-1120558 to MWSU and MCB-1120578 to Davidson College; the MWSU Program of Research, Teaching, and Applied Learning (PORTAL); Howard Hughes Medical Institute grant 52006292 to Davidson College; and the James G. Martin Genomics Program at Davidson College. A.M.C. and T.E. are members of GCAT SynBio. pClone Red is being marketed by Carolina Biological and therefore the authors declare their conflict of interest.
References
- AAAS. 2011. Vision and Change: A Call to Action, Final Report. http://visionandchange.org/finalreport/ Accessed November 13.
- Campbell AM, Eckdahl T. 2015. CourseSource pClone Intro Biology Lesson. http://coursesource.org/courses/using-synthetic-biology-and-pclone-red-for-authentic-research-on-promoter-function
- Council NR. 2003. BIO2010: Transforming Undergraduate Education for Future Research Biologists. National Academies Press, Washington, DC.
- Seymour EHA, Laursen SL, DeAntoni T. 2004. Establishing the benefits of undergraduate research for undergraduates in the sciences: first findings from a three-year study. Sci Educ 88:493-594.
- Lopatto D. 2006. Undergraduate research as a catalyst for liberal learning. Peer Rev 8:22-25.
- Laursen S, Hunter A, Seymour E, Thiry H. 2010. Undergraduate Research in the Sciences: Engaging Students in Real Science. Real Science San Francisco, Jossey-Bass:320.
- Slater SJ, Slater TF, Bailey, M. J. 2010. Discipline-Based Education Research: A scientist’s Guide, New York, New York.
- Harrison M, Dunbar D, Ratmansky L, Boyd K, Lopatto D. 2011. Classroom-Based Science Research at the Introductory Level: Changes in Career Choices and Attitude. CBE—Life Sciences Education 10:279–286.
- Haynes KA, Broderick ML, Brown AD, Butner TL, Dickson JO, Harden WL, Heard LH, Jessen EL, Malloy KJ, Ogden BJ, Rosemond S, Simpson S, Zwack E, Campbell AM, Eckdahl TT, Heyer LJ, Poet JL. 2008. Engineering bacteria to solve the burnt pancake problem. J Biol Eng 2:8.
- Baumgardner J, Acker K, Adefuye O, Crowley ST, DeLoache W, Dickson JO, Heard L, Martens AT, Morton N, Ritter M, Shoecraft A, Treece J, Unzicker M, Valencia A, Waters M, Campbell AM, Heyer LJ, Poet JL, Eckdahl TT. 2009. Solving a Hamiltonian Path Problem with a Bacterial Computer. J Biol Eng 3:11.
- Eckdahl TT, Campbell AM, Heyer LJ, Poet JL. 2010. Synthetic Biology as a new opportunity for multidisciplinary undergraduate research. CUR Quarterly 30:42-48.
- Wolyniak MJ, Alvarez CJ, Chandrasekaran V, M. GT, Holgada A, J. JC, W. MR, L. PA, J. S, M. WT, Y. Y. 2010. Building Better Scientists through Cross-Disciplinary Collaboration in Synthetic Biology: A Report from the Genome Consortium for Active Teaching Workshop 2010. CBE Life Sci Educ 9:399-404.
- Pearson B, K. H. Lau, A. Allen, J. Barron, R. Cool, K. Davis, W. DeLoache, E. Feeney, A. Gordon, J. Igo, A. Lewis, K. Muscalino, M. Parra, P. Penumetcha, V. G. Rinker, K. Roland, X. Zhu, J. L. Poet, T. T. Eckdahl, L. J. Heyer and A. M. Campbell. 2011. Bacterial Hash Function Using DNA-Based XOR Logic Reveals Unexpected Behavior of the LuxR Promoter Interdisciplinary BioCentral 3:10.
- Sawyer EM, C. Barta, R. Clemente, M. Conn, C. Davis, C. Doyle, M. Gearing, O. Ho-Shing, A. Mooney, J. Morton, S. Punjabi, A. Schnoor, S. Sun, S. Suresh, B. Szczepanik, D. L. Taylor, A. Temmink, W. Vernon, A. M. Campbell, L. J. Heyer, J. L. Poet and T. T. Eckdahl. 2012. Bacterial Logic Devices Reveal Unexpected Behavior of Frameshift Suppressor tRNAs. Interdisciplinary BioCentral 4:10.
- Eckdahl TT, Campbell AM, Heyer LJ, Poet JL, Blauch DN, Snyder NL, Atchley DT, Baker EJ, Brown M, Brunner EC, Callen SA, Campbell JS, Carr CJ, Carr DR, Chadinha SA, Chester GI, Chester J, Clarkson BR, Cochran KE, Doherty SE, Doyle C, Dwyer S, Edlin LM, Evans RA, Fluharty T, Frederick J, Galeota-Sprung J, Gammon BL, Grieshaber B, Gronniger J, Gutteridge K, Henningsen J, Isom B, Itell HL, Keffeler EC, Lantz AJ, Lim JN, McGuire EP, Moore AK, Morton J, Nakano M, Pearson SA, Perkins V, Parrish P, Pierson CE, Polpityaarachchige S, Quaney MJ, Slattery A, Smith KE, Spell J, Spencer M, Taye T, Trueblood K, Vrana CJ, Whitesides ET. 2015. Programmed evolution for optimization of orthogonal metabolic output in bacteria. PLoS One 10:e0118322.
- Biotechnology and Biological Sciences Research Council DSaTL, Engineering and Physical Science Research Council, Medical Research Council. 2011. Joint Synthetic Biology Initiative (JSBI). http://www.bbsrc.ac.uk/funding/grants/priorities/synthetic-bio/ Accessed November 13.
- Knight TF. 2003. Idempotent Vector Design for Standard Assembly of Biobricks. MIT Synthetic Biology Working Group.
- Campbell AM. 2005. Meeting report: synthetic biology Jamboree for undergraduates. Cell Biol Educ 4:19-23.
- Khalil AS, Collins JJ. 2010. Synthetic biology: applications come of age. Nat Rev Genet 11:367-379.
- Kahl LJ, Endy D. 2013. A survey of enabling technologies in synthetic biology. J Biol Eng 7:13.
- Wood WB, Mathematics NRCCoPfASo, Science in American High S. 2002. Advanced high school biology in an era of rapid change: a summary of the biology panel report from the NRC Committee on Programs for Advanced Study of Mathematics and Science in American High Schools. Cell Biol Educ 1:123-127.
- CollegeBoard. 2011. AP Biology Curriculum Framework 2012-2013. New York.
- Campbell AM, Todd Eckdahl, Brian Cronk, Corinne Andresen, Paul Frederick, Samantha Huckuntod, Claire Shinneman, Annie Wacker, and Jason Yuan. 2014. pClone: Synthetic Biology Tool Makes Promoter Research Accessible to Beginning Biology Students. CBE Life Sciences Education 13:285-296.
- Weber E, Gruetzner R, Werner S, Engler C, Marillonnet S. 2011. Assembly of designer TAL effectors by Golden Gate cloning. PLoS One 6:e19722.
- Werner S, C. Engler, E. Weber, R. Gruetzner, Marillonnet, S. 2012. Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioeng Bugs 3:38-43.
- Group MW. 2013. Registry of Standard Biological Parts http://partsregistry.org/Main_Page Accessed March 11.
- Campbell AM, Hatfield, B., Heyer, L.J., Eckdahl, T.T. . 2013. Registry of Functional Promoters. http://gcat.davidson.edu/RFP/ Accessed March 11.
- Rasband WS. 1997-2012. ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA. http://imagej.nih.gov/ij/. Accessed March 11.
- iGEM TU-S. 2011. amilCP, blue chromoprotein. http://parts.igem.org/Part:BBa_K592009 Accessed November 13.
- Council NR. 2000. How People Learn: Brain, Mind, Experience, and School Expanded Edition. National Academies Press, Washington, DC.
- Lopatto D. 2007. Undergraduate research experiences support science career decisions and active learning. CBE Life Sci Educ 6:297-306.
- Wei CA, Woodin T. 2011. Undergraduate research experiences in biology: alternatives to the apprenticeship model. CBE Life Sci Educ 10:123-131.
- David Lopatto CH, Christopher J. Jones, Don Paetkau, Vidya Chandrasekaran, David Dunbar, Christy MacKinnon, Joyce Stamm, Consuelo Alvarez, Daron Barnard, James E. J. Bedard, April E. Bednarski, Satish Bhalla, John M Braverman, Martin Burg, Hui-Min Chung, Randall J. DeJong, Justin R. DiAngelo, Chunguang Du, Todd T. Eckdahl, Julia Emerson, Amy Frary, Donald Frohlich, Anya L. Goodman, Yuying Gosser, Shubha Govind, Adam Haberman, Amy T. Hark, Arlene Hoogewerf, Diana Johnson, Lisa Kadlec, Marian Kaehler, S. Catherine Silver Key, Nighat Kokan, Olga R. Kopp, Gary A. Kuleck, Jane Lopilato, Juan C. Martinez-Cruzado, Gerard McNeil, Stephanie Mel, Alexis Nagengast, Paul J. Overvoorde, Susan Parrish, Mary Preuss, Laura D. Reed, E. Gloria Regisford, Dennis Revie, Srebrenka Robic, Jennifer A. Roecklien-Canfield, Anne G. Rosenwald, Michael R. Rubin, Kenneth Saville, Stephanie Schroeder, Karim Sharif, Mary Shaw, Gary Skuse, Christopher D Smith, Mary Smith, Sheryl T. Smith, Eric P. Spana, Mary Spratt, Aparna Sreenivasan, Jeffrey S. Thompson, Matthew Wawersik, Michael Wolyniak, James Youngblom, Leming Zhou, Jeremy Buhler, Elaine Mardis, Wilson Leung, Christopher D. Shaffer, Jennifer Threlfall, Sarah C. R. Elgin. 2014. A Central Support System Can Facilitate Implementation and Sustainability of a Classroom-Based Undergraduate Research Experience (CURE) in Genomics. CBE Life Sci Educ 13:711-723.
- Hanauer, DI and Dolan EL. 2014. The Project Ownership Survey: Measuring Differences in Scientific Inquiry Experiences. CBE Life Sci Educ 13:149-158.
- Corwin LA, Graham MJ, Dolan EL. 2015. Modeling Course-Based Undergraduate Research Experiences: An Agenda for Future Research and Evaluation. CBE Life Sci Educ 14:1-13.
- Russell JE, D’Costa AR, Runck C, Barnes DW, Barrera AL, Hurst-Kennedy J, Sudduth EB, Quinlan EL, Schlueter M. 2015. Bridging the Undergraduate Curriculum Using an Integrated Course-Embedded Undergraduate Research Experience (ICURE). CBE Life Sci Educ 14.
Article Files
Login to access supporting documents
Eckdahl-Using Synthetic Biology and pClone Red for Authentic Research on Promoter Function- Introductory Biology analyzing mutant promoters_3.pdf(PDF | 728 KB)
S1. pClone Genetics-Timeline of pClone Red Genetics Laboratory Lesson.pdf(PDF | 40 KB)
S2. pClone Genetics-Genetics Promoter Mutation Quiz.docx(DOCX | 17 KB)
S3. pClone Genetics-Links to Introduction to Synthetic Biology and Cloning.pptx(PPTX | 1 MB)
S4. pClone Genetics-Pre- and Post-Assessment Questions no answers.docx(DOCX | 23 KB)
S5. pClone Genetics-Pre- and Post-Assessment Questions with answers.docx(DOCX | 23 KB)
S6. pClone Genetics-Overview of Golden Gate Assembly for Promoter Research.pptx(PPTX | 748 KB)
S7. pClone Genetics-Details of Golden Gate Assembly for Promoter Research.pptx(PPTX | 5 MB)
S8. pClone Genetics-Designing Mutant promoter.docx(DOCX | 179 KB)
S9. pClone Genetics-Annealing Promoter Oligonucleotides.docx(DOCX | 22 KB)
S10. pClone Genetics-Golden Gate Assembly with pClone Red.docx(DOCX | 25 KB)
S11. pClone Genetics-Transforming DNA into E. coli Cells.docx(DOCX | 23 KB)
S12. pClone Genetics-Paper Activity for GGA Cloning into pClone.pptx(PPTX | 69 KB)
S13. pClone Genetics-Phenotype Analysis after Transformation.docx(DOCX | 627 KB)
S14. pClone Genetics-Quantifying pClone Red Promoter Strength with ImageJ no fluorometer.docx(DOCX | 143 KB)
S15. pClone Genetics-Using pClone Blue instead of pClone Red no UV light.docx(DOCX | 793 KB)
S16. pClone Genetics-pClone Genotyping to Verify Promoter Cloning.docx(DOCX | 302 KB)
- License terms

Comments
Comments
There are no comments on this resource.