Predicting and classifying effects of insertion and deletion mutations on protein coding regions
Published online:
Abstract
Mutations in genes can affect the encoded proteins in multiple ways, and some of these effects are counterintuitive. As for any other knowledge, students must create their own deep understanding of the Central Dogma. Students may not develop this understanding because they have limited opportunity to practice manipulating DNA sequences and classifying their effects. Such practice can improve student appreciation for the myriad possible effects of DNA change (mutation) on amino acid sequence. In this Lesson, a series of scaffolded exercises provides this opportunity. Students first identify gene sequences from an online database, create their own insertion/deletion mutations, and predict the effects. Students then use a web-based tool to translate and observe the effect of the mutation on protein sequence. Subsequent comparison of predicted and observed effects employs the chi-square test. Discussion of results with peers involves categorizing the types of possible effects. The lesson concludes with an exercise asking students to create a mutation with an intended effect on the protein. Together, the exercises integrate quantitative reasoning and statistical analysis, information literacy, and multiple Bloom's learning levels. Student progress is monitored using three formative and three summative assessments.
Citation
Ross, J.A. 2016. Predicting and classifying effects of insertion and deletion mutations on protein coding regions. CourseSource. https://doi.org/10.24918/cs.2016.18Society Learning Goals
Bioinformatics
- DNA - Information Storage [GENOMICS]
- Where are data about the genome found (e.g., nucleotide sequence, epigenomics) and how are they stored and accessed?
- How can bioinformatics tools be employed to analyze genetic information?
Genetics
- Transmission - Patterns of Inheritance
- How can one deduce information about genes, alleles, and gene functions from analysis of genetic crosses and patterns of inheritance?
- Genetic variation
- How do different types of mutations affect genes and the corresponding mRNAs and proteins?
- Methods & Tools in Genetics
- What experimental methods are commonly used to analyze gene structure and gene expression?
Science Process Skills
- Process of Science
- Pose testable questions and hypotheses to address gaps in knowledge
- Interpret, evaluate, and draw conclusions from data
- Construct explanations and make evidence-based arguments about the natural world
- Modeling/ Developing and Using Models
- Recognize the important roles that scientific models, of many different types (conceptual, mathematical, physical, etc.), play in predicting and communicating biological phenomena
- Make inferences and solve problems using models and simulations
- Build and evaluate models of biological systems
- Quantitative Reasoning/ Using Mathematics and Computational Thinking
- Apply the tools of graphing, statistics, and data science to analyze biological data
- Communication and Collaboration
- Share ideas, data, and findings with others clearly and accurately
Lesson Learning Goals
Students will understand how insertion/deletion mutations affect the reading frame of translation and the resulting protein sequence.Lesson Learning Objectives
Students will be able to:- accurately predict effects of frameshift mutations in protein coding regions
- conduct statistical analysis to compare expected and observed values
- become familiar with accessing and using DNA sequence databases and analysis tools
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
As part of the central dogma of biology, ribosomes translate messenger RNA (mRNA) molecules to produce proteins. The mRNA nucleotide triplets (codons) that base pair with transfer RNA anticodons specify the individual amino acids that become incorporated into the growing polypeptide chain. In eukaryotes, the reading frame of each mRNA is established by the location of the first AUG codon encountered by the ribosome as it scans the mRNA from the 5' end. The ribosome then interprets each successive codon according to base pair complementarity with a tRNA anticodon. In the classroom, the translation of the nucleic acid language into the amino acid language is facilitated by the codon table, which indicates (for most cases) how each mRNA codon corresponds to the amino acid that is incorporated into a protein.
Instruction on the central dogma is common in collegiate biology coursework, with many genetics textbooks devoting at least one chapter to each step (DNA replication, transcription, translation). In spite of this instruction, student misconceptions persist, especially in understanding how changes in DNA sequence affect protein sequence. In the analysis of the Genetics Concept Assessment (GCA), for example, student responses to nine of 25 questions on the instrument suggested the presence of alternative conceptions because greater than twenty percent of students answered one incorrect answer in the post-test (1). Four of those nine questions, where pervasive alternative conceptions hinder student success, involve the effects of DNA mutations on protein sequences (GCA questions 4, 11, 12 and 15). Of these four, the one that seems most intuitively difficult to address is question 12. Student responses to this question reveal that 34% of students think that insertion mutations cannot shorten the length of a protein. Such a result could occur, for example, by mutation resulting in an early termination of translation) (1). Although we know that it is critical for retention that students develop their own understanding of a desired concept (2), it can be difficult in practice to provide structured time for students to explore mutations on their own and to come to their own realization of the myriad (often counterintuitive) ways in which mutations can alter protein sequences. For example, deletion mutations can increase the length of proteins; insertion mutations can shorten proteins. Small insertion or deletion mutations can have severe effects on proteins, while larger mutations might not. Thus, I wished to employ active learning strategies, which improve conceptual learning (8,9). Thus, the Lesson I present here, which comprises a set of active-learning modules for the lecture course, fills an important niche.
It is important for students to understand how various types of nucleic acid mutations change the protein encoded by a mutated gene, in part because phenotypic differences between populations are typically due to genetic variants that alter protein sequences (e.g. 12). Students will work both individually and in groups. Individual work is optimal for helping students develop the relationships between existing and new knowledge (13,14), while group participation has been shown to improve academic success especially of underrepresented minorities (15) and to increase engagement and retention (16,17).
The framework for the exercises that comprise the Lesson involves each student first selecting a coding DNA sequence from a sequence database. Giving the student the ability to choose the substrate for the exercises to follow is important because student choice improves attitudes about their learning and is associated with improved performance compared to no-choice scenarios (18). This open-choice approach also enhances critical thinking (19). The instructor then introduces the class to some simple computer-based DNA sequence analyses to perform on their chosen genes. I introduced this component in part because the advent of the genomics era has led to broad need and advocacy for courses helping students learn how to access and analyze sequences (23).
Next, students mutate their sequence by making insertion or deletion mutations of their choosing, However, before observing the effect of the mutation on protein sequence, the instructor first asks them to make predictions to compare to the empirical outcomes they will produce. Discrepancies are discussed in small groups and then in the full class. I incorporated the step of making a prediction because identifying the expected outcome is a critical component of the scientific method and because student academic success is enhanced by having students make predictions (24).
I designed a number of formative and summative assessment activities to incorporate through the Lesson, in order to help the instructor and the students gauge and respond to student achievement and progress.
In all, the Lesson addresses student alternative conceptions about the effects of mutations on protein sequences. One strength of the series of exercises is that I have integrated a number of Core Competencies from Vision and Change, including quantitative reasoning, the process of science, and the relationship between science and society, as well as the Core Concepts of information flow and evolution (25).
INTENDED AUDIENCE
I designed this Lesson for upper-division biology majors and use it currently in a core undergraduate genetics course, with enrollment from 60 to 90 students. This Lesson could also be relevant and accessible to sophomore genetics classes and to honors first-year undergraduate biology classes.
REQUIRED LEARNING TIME
The Lesson comprises seven sequential, fifty-minute course meetings. Because I am involved with the California State University, Fresno initiative for instruction using tablet computers (the DISCOVERe program), I teach a genetics class in which each of my students brings a web-accessible computer to class every day. As a result, the integration of classroom activities with web-based analyses, as described in the Lesson, is feasible. However, this Lesson can easily be adapted for less device-rich situations by assigning web-based steps to be completed by the subsequent class period. For example, slides 11-21 could be distributed as homework for students to look over, and request that each student identify and bring their own CDS as a paper printout to the subsequent class period. Such modifications to the Lesson can also shorten the amount of required in-class time.
PRE-REQUISITE STUDENT KNOWLEDGE
Ideally, this Lesson will be used at the end of a typical genetics textbook chapter on eukaryotic translation, after students have encountered the concepts of: eukaryotic DNA replication, mutation and variation; transcription; translation initiation, elongation, and termination. Students might meet this prerequisite by reading a chapter on translation prior to the first class period of this Lesson. Specifically, students should have been exposed to (but not necessarily have mastered) the concepts of:
- Genetic variation, including terminology like mutation, allele, single nucleotide polymorphism
- Transcription, including
- the polarity of mRNA molecules
- the removal of introns by splicing
- Translation, including
- the CDS (coding DNA sequence) as the portion of the mRNA molecule that is translated (flanked at the 5' end by a start codon and the 3' end by a stop codon)
- the codon
- eukaryotic initiation proceeding by a ribosome scanning a mRNA starting at the 5' end and searching for the first AUG (methionine, start) codon, establishing the "reading frame"
- the ribosome moving one codon (three nucleotides, non-overlapping) at a time from 5' to 3' translating each codon and incorporating the cognate amino acid into the growing polypeptide chain (protein)
- Translation terminating at the first in-frame stop codon encountered by the translating ribosome
- using a codon table to translate an RNA codon sequence into the amino acid encoded by that codon
- Statistics, including
- the chi square test
- its use (to assess whether expected and observed values are different)
- how to perform the chi-square calculation
- how to interpret the chi square test statistic, along with the number of degrees of freedom, to estimate a p value (26)
- p values < 0.05 indicate that the observed and expected values are significantly different
- p >= 0.05 indicates that the observed and expected values are not different
SCIENTIFIC TEACHING THEMES
Active Learning
The Lesson extensively incorporates active learning approaches, alternating instructor-driven examples with exercises combining individual and group activities. Employing group discussion and consensus building drives peer instruction, with feedback from the instructor. Hands-on in silico identification of a DNA sequence selected by each student and subsequent manipulation requires the student to make decisions about how to modify the sequence and to make predictions about outcomes. This choice also motivates active engagement and resists the potential for some students to rely on peers to achieve group objectives.
Assessment
Three pairs of formative and summative assessments, formatted as printed exams and exercises, are provided as Supporting Files S3 and S4; S6 and S7; S10 and S11. Additional opportunities exist for the instructor to poll the class to obtain oral feedback on student comprehension, and regular student group work provides opportunities for students to perform self-assessment.
Inclusive Teaching
The Lesson addresses diversity in at least five ways. First, by engaging students in think-pair-share and group discussion as well as by using volunteer-response questions, the Lesson accommodates both introverts and extroverts. Having both student-selected groups as well as groups formed randomly also lets all the students access the varying expertise of peers that they might not normally choose to interact with. Third, the topic of the Lesson itself, how mutation affects protein sequence, is inextricably coupled with appreciation for the basis of diversity: DNA sequence changes leading to protein sequence changes via the central dogma. The Lesson also promotes diversity by allowing students to select a gene sequence of their own choosing for analysis, rather than every student working on the same gene selected by the instructor. Such an approach allows the Lesson to show case genetic diversity and to allow the students to draw broad conclusions about similarities and differences resulting from related types of mutations. Finally, the example used in the first class to motivate the Lesson, the genetic basis of human earwax type, provokes questions about the genetic differences between human populations and potential forces promoting those differences.
LESSON PLAN
In this Lesson, which I deliver over seven, fifty-minute course periods, between 60 and 95 biology major students complete the seven following activities:
- Identify a coding DNA sequence (CDS) available in a public sequence database (wormbase.org).
- Perform basic sequence analyses on the CDS by calculating nucleotide frequencies and using a chi square test to compare this observation to a null hypothesis.
- Conduct in silico translation of the CDS.
- Identify the observed location of in-frame stop codons in the CDS and using a chi-square test to compare this observation to a null hypothesis.
- Perform stochastic in silico mutagenesis of the CDS and predicting the effect on the encoded protein.
- Translate the mutant CDS; categorizing and discussing the outcomes of the effect of the mutation on the protein compared to the mutations of other students (representing mutations of various lengths and types).
- Design mutations with the intention of creating specific, desired effects on protein sequence.
These activities are interspersed with assessments and interactive lecture segments.
The activities and materials needed are summarized in Table 1.
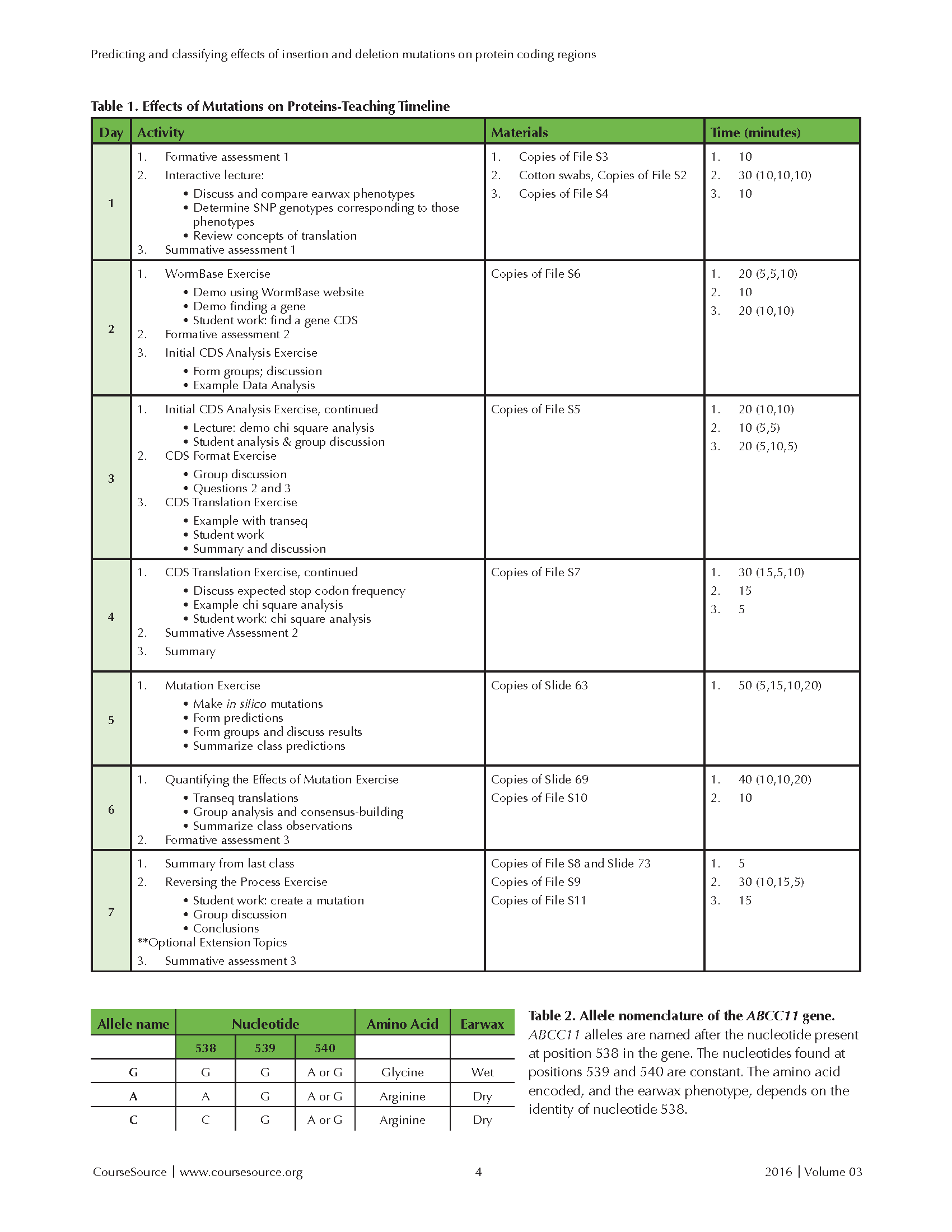
Tables 1 and 2
DAY 1
At the time of this Lesson, the 60 to 90 students enrolled in my upper-division biology majors' course in genetics have recently completed class sessions on DNA replication, mutation and transcription. They just read the textbook chapter on translation in preparation for the first day of this Lesson.
Each of the class meetings for this Lesson will require a computer and projector (or overhead transparencies and projector, or document camera and printouts of PowerPoint slides) for displaying Supporting File S1 (Effects of Mutations on Proteins - Presentations.pptx). Ideally, students will ideally always bring an internet-accessible laptop or tablet computer (or at least one device to share per group of 3-4 students).
Before today's class, prepare by adding photos of earwax phenotypes to Slide 3. For each student, bring: one cotton swab (optional) and a photocopy of (1) Supporting File S2 (a codon table; ask students to bring the codon table back to each class); (2) Supporting File S3 (Formative Assessment 1); and (3) Supporting File S4 (Summative Assessment 1).
FORMATIVE ASSESSMENT 1 (SUPPORTING FILE S3; KEY AVAILABLE IN SUPPORTING FILE S12) - 10 MIN.
INTERACTIVE LECTURE (SUPPORTING FILE S1) - 30 MIN.
To begin instruction, distribute one cotton swab to each student, keeping one for yourself. Then, direct everybody to clean their ears. Tell the students to keep their swabs, and then proceed to the lecture slides.
Slide 3 (Supporting File S1): Tell the class that there is a mutation known to correlate with earwax type ITE.DATA (11), making this example useful for determining one's genotype from one's phenotype.
Slide 4: After the students have identified their earwax types, this slide shows the frequencies of two alleles of ABCC11. The alleles are named A and G, after two nucleotides (adenine and guanine) that can be found at position 538 in the ABCC11 gene. Refer to Table 2 for a summary (pictured with Table 1). The A allele (recessive, white in the figure's pie charts) causes production of dry earwax; the G allele (dominant, black in the pie charts) produces wet earwax. After asking students to identify whether any geographic trends in the allele frequencies appear to exist, you can ask them whether their self-identified earwax phenotype correlates with their ethnicity based on the allele frequency figures for different worldwide ancestral populations. Now, students have experienced an example of how a mutation can cause a phenotypic change. To improve their appreciation for the vast range of ways that mutations affect protein sequences, it is next important to discuss the specific change involved in the ABCC11 gene that underlies the difference in human earwax type. This discussion usually requires five to seven minutes.
Slide 5: introduces a third ABCC11 allele, in which a C (cytosine) exists at nucleotide 538 (like the A allele, the C allele is also recessive and produces dry earwax). Let students read the information, ask clarifying questions of you if needed, and then think-pair-share, or use clickers or other polling approach, to have students answer the question at the bottom of the slide. Distribute File S2 copies for the students to use in this and subsequent classes. The only way that a GGN (N = aNy amino acid) codon, all four of which encode glycine, can mutate from a glycine codon to an arginine is if a mutation occurs in the first codon position. G-to-A mutations at this position will change GGA or GGG (glycine codons) to AGA or AGG (arginine codons). G-to-C mutations at the first position will change GGA or GGG (glycine) to CGA or CGG (also encoding arginine). Conclude with the point that this one amino acid change, caused by a difference in a single nucleotide between different human populations, causes an observable difference in phenotype: earwax consistency.
Slide 6: This introduction to the principle of DNA mutation affecting the encoded protein (the central dogma) motivates the next few days of class: exploring how mutations affect proteins.
Slide 7: Review these three points with the students. This review can be achieved by asking students to volunteer answers or through think-pair-share. I prefer to have students work in groups of three to four and then report out to the class, which takes about ten minutes of class time: three to five minutes for group discussion and the remainder for class discussion. It is important for students to recall that the first AUG codon from the 5' end of the mRNA is where a ribosome initiates translation and that translation terminates at the first in-frame stop codon (UAA, UAG, UGA). The third question is posed to make students consciously explore how protein length is determined: the class consensus should be that the number of codons between the start codon and the first in-frame stop codon dictates the number of codons and therefore the length of the encoded polypeptide.
SUMMATIVE ASSESSMENT 1 (SUPPORTING FILE S4; KEY AVAILABLE IN SUPPORTING FILE S13) - 10 MIN.
*This assessment should occupy about 10 minutes of class time. All assessments in this Lesson are meant to be conducted on individual students, not in groups.
DAY 2
For each student, bring: a photocopy of Supporting File S6 (Formative Assessment 2).
WORMBASE EXERCISE - 20 MIN.
Slides 11-20: introduce students to one method for accessing an online nucleotide sequence database to retrieve gene sequences. I use the WormBase.org website mainly because of its efficient user interface; it requires fewer steps to get students from the entry webpage to being able to look at gene sequences than human genome websites. Present slides 9-16 first, as an example of how to use the WormBase.org website. These screen shots and presenter notes walk through the process of using WormBase.org to access coding DNA sequence (CDS) entries for genes.
Not only is WormBase efficient to use, but I also use this opportunity to discuss with students the use of model organisms in science. WormBase.org was created to share information about the nematode Caenorhabditis elegans. This species of worm is studied by thousands of scientists around the world, in fields like development, genetics, neurobiology, behavior, pharmacology, and molecular biology. It has the same key traits as other model species, like a small physical size (about 1 mm per adult), simple and inexpensive husbandry, large numbers of offspring (a single hermaphrodite, one of the two sexes in this species, can produce over 200 offspring over its roughly two week lifespan), and a completely sequenced genome. The genome is much smaller than the human genome, and contains fewer genes, but many of the genes in C. elegans have homologs in humans. Thus, by studying C. elegans, we have learned quite a bit about genetics that is directly applicable to humans.
At the end of the WormBase Exercise, each student should have obtained the CDS of the gene he or she has chosen to use. From slide 18, tell the students that, before you release them for about ten minutes to access the WormBase.org website and retrieve a gene sequence, you will first show them one way to find gene names for performing the sequence database query (slides 19 and 20). Students can also enter search terms in the WormBase search window to find genes related to those terms (e.g. "neuron," "paralysis," "sleep." Then release the students to obtain a CDS and display slide 21, which summarizes the workflow you just walked them through, while they complete this exercise. Genes that have been used include daf-1, which is involved in chemosensation, egl-2, which is involved in egg-laying defects in nematodes, and unc-3, which has neuronal function. Some students might happen to select noncoding genes (such as rRNA and tRNA genes). If this occurs, you should gently suggest that the student pick a protein-coding gene, as this is essential for this Lesson. Some genes (of the thousands in the C. elegans genome) that could be useful to suggest to any students who have issues with selecting a gene include sxl-1, tra-1, tra-2, fem-1, fem-2, fem-3, for example.
PERFORM FORMATIVE ASSESSMENT 2 (SUPPORTING FILE S6; KEY AVAILABLE IN SUPPORTING FILE S14) - 10 MIN.
INITIAL CDS ANALYSIS EXERCISE - 20 MIN.
Slide 23: now that each student has identified a CDS to analyze, students should again form groups of three to four and then discuss how to efficiently determine the number of nucleotides and the nucleotide frequencies in the CDS. Allow students about ten minutes to explore options, then suggest that they use the "Find/Replace" tool of their favorite text editor or word processing program to find the number of instances of each of the nucleotides. No group should initially resort to counting by hand. If they do, set them onto the idea that they should be able to devise a much more efficient approach.
Slide 24 contains a demonstration sequence, the CDS of the nuo-1 gene, for you to use in showing students how to perform the following analyses. The gene nuo-1 produces a protein involved in mitochondrial oxidative phosphorylation, the process that produces cellular energy in the form of the molecule ATP. I selected this gene because I am a mitochondrial geneticist and am interested in understanding how mitochondrial mutations can impact organismal fitness. This gene is homologous to a gene that we (humans) also have. Mutations in the human homolog can cause metabolic dysfunction.
Slide 25 contains the nucleotide frequencies and CDS length for the instructor's example CDS (the nuo-1 gene, first seen as the example gene in the previous course period). After displaying this example, and discussing whether any groups had vastly different percentages for their CDSs, ask the class whether the nuo-1 nucleotide composition (and whether the composition of each of their own CDS) seems to comprise a random sequence of nucleotides. It is useful to introduce the value of this point to students by noting that you will soon be asking questions about how mutations impact the amino acid compositions and lengths of proteins. It is important first to know whether the nucleotide composition of the gene is biased toward particular nucleotides. After a brief discussion, ask each student group to arrive at the expected nucleotide frequency of nuo-1. After letting each group discuss for one to two minutes, bring the groups back together and discuss whether a consensus is reached: that if there are four nucleotides (A, T, G, C), then a random sequence should comprise equal numbers of each nucleotide (or 25% of each of the four).
Slide 26 asks each student to formalize the expectation for a specific case: nuo-1. With 1,440 nt in the CDS, 25% of 1,440 = 360, which is the expected number of A, T, G, and C nucleotides if this gene sequence is essentially random.
Slide 27 summarizes, in a structure that suggests an impending statistical test, our nuo-1 data to this point.
DAY 3
For each student, bring: a photocopy each of Supporting File S5 (chi-square table - ask the students to bring this copy back to each class meeting).
INITIAL CDS ANALYSIS EXERCISE, CONTINUED - 20 MIN.
Slide 30 should be used to remind the class quickly how to conduct a chi-square test, or you can ask each student to individually calculate the chi-square test statistic value using the formula displayed.
Slide 31 explicitly adds the filled-in chi-square formula to use.
Slide 32 then raises the point that, to interpret the chi-square test statistic, we need to know the number of degrees of freedom in the calculation.
Slide 33 adds detail about how to determine the number of degrees of freedom (df) for a chi-square calculation. A chi-square calculation comprises the sum of a number of arithmetical clauses of the form (O-E)/E, where O is an observed number of nucleotides, in this case, and E is the expected number. Each of the four nucleotides will be represented in one such clause, so the entire chi-square formula will be a sum of the four clauses. The df in the calculation is equal to the number of categories of data (in this case the four nucleotide frequencies), minus one, so 4-1 = 3 for nucleotide frequency tests. This slide also instructs in the interpretation of the chi-square test statistic value and the degrees of freedom using a chi-square table.
Slide 34 then details the process of using a chi-square table, with the test statistic and number of degrees of freedom, to ascertain the associated p value.
Slide 35 describes the interpretation. The p value associated with the nuo-1 nucleotide frequency chi-square test statistic and three degrees of freedom is between 0.05 and 0.10.
Slide 36 indicates that the observed and expected values are not statistically significantly different. In other words, the observed nucleotide frequencies match our prediction that there will be 25% of each nucleotide.
Slide 37: at this point, having walked through one example, have each student use the chi-square test to calculate the chi-square test statistic and interpret that value using a chi-square table to identify the associated p value for the sequence of their selected gene. In each group, students should work together to compare outcomes. There is no specific (intended) outcome for this exercise. It is likely that most genes will not differ from the expectation of 25% composition of each nucleotide. However, some might differ from the expectation, perhaps by chance, because some proteins happen to be composed of amino acids that tend to be encoded by more AT-rich or GC-rich codons. This is partly to do with the redundancy of the codon table. If a protein happens to have many glycines (GGN codons), prolines (CCN codons), or alanines (GCN codons), for example, then the CDS would be biased toward G and C nucleotides. Codon bias, the empirical observation that some degenerate codons are used more frequently than others, also impacts this expectation. The main purpose of conducting this analysis is to convince the class that they are working with genes of essentially random nucleotide composition. This point is critical for upcoming analysis of the observed vs. expected frequencies of particular codons (stop codons) in their CDSs.
CDS FORMAT EXERCISE - 10 MIN.
Slide 38: Students work in the same small groups to calculate the number of codons in each of their CDS. Because in the previous class each student calculated the number of nucleotides in their CDS, simply dividing this number by 3 (nucleotides/codon) will yield the number of codons.
Slide 39: To orient the students to the layout of the CDS, the next question to ask them is where the start and stop codons are located in each of their CDS. In their groups, each should rapidly arrive at the conclusion that each CDS begins with an AUG (the start codon) and ends with one of the three stop codons (UAG, UGA, or UAA). It is useful at this point to remind students that DNA contains thymine nucleotides (T), which are represented in RNA by uracil (U). Thus, the codon table contains U in place of T. However, CDS are, by definition, written as DNA sequence (containing T instead of U). Thus, the start codon found in a CDS is written ATG, and the stop codons are TAG, TGA, and TAA.
Slide 40 then shows the nuo-1 CDS, which reinforces the idea that the start and stop codon represent the start and the end of the CDS.
CDS TRANSLATION EXERCISE - 20 MIN.
Now that students have calculated the number of codons in their CDS, it is time to empirically determine the amino acid sequence of the CDS.
On Slide 41, the URL to the transeq (sequence translation) web tool is given, although other web-based translation tools might be as useful. Please note that such online services appreciate advance notice of courses making use of their servers, so please provide them the courtesy of giving that advance notice (http://www.ebi.ac.uk/support/sequences). Each student (or group) should navigate to the URL shown but not begin work yet.
Show Slides 42-44 to walk the class through the use of the transeq web tool. You should use the nuo-1 CDS sequence from Slide 24 (also provided in the notes field of Slide 42) to demonstrate this process.
While displaying Slide 45, ask each student to translate their CDS and then compare their calculation of the number of codons in their CDS (from PowerPoint slide 38, from the previous class meeting) with the results from the transeq translation, and share their results among their group-mates to reach consensus and resolve any issues.
Slide 46 poses a critical question for assessing the effects of mutations on protein sequences. Now is a good time to reiterate to the class that we've twice already agreed that the length of a protein is determined by the number of codons between the start codon and the first in-frame stop codon. So, it would be interesting to determine whether this means that all proteins are roughly the same length, with an average related to the frequency with which we expect by chance to observe stop codons in nucleotide sequences. In this context, this question is Socratic - the idea is to motivate the next exercise. An alternate approach is to assign this calculation as a homework problem for students to address and bring answers for discussion in the following class period.
DAY 4
For each student, bring one printout of Supporting File S7 (Summative Assessment 2, upon each of which you've hand-written a whole number between 1 and 150 in the space provided on the front page, attempting not to repeat a number and to roughly evenly span this range of values).
CDS TRANSLATION EXERCISE, CONT. - 30 MIN.
Slide 49: rephrases the question about the frequency of stop codons in a CDS in a format that should get the students thinking that another chi-square test is imminent.
With the codon table displayed on slide 50, groups should discuss the answer to the question of what the expected frequency of stop codons should be and come to a consensus on a prediction for this value. Before showing Slide 51, facilitate the class coming to consensus.
Slide 51 makes the concluding point that the expected frequency of stop codons should be, on average, every ~21 codons. This calculation results from analysis of the codon table: with 3/64 codons being stop codons, the expected frequency is 0.046.
Slide 52: The calculated frequency of stop codons gets us closer to comparing our observed frequency of stop codons in the nuo-1 CDS (1/480) with an expectation, but the expected value has to be an expected number of stop codons (not an expected frequency). Thus, 480/(3/64) = ~22.5 stop codons expected in a random DNA sequence of the same length as the nuo-1 CDS (480 amino acids).
Slide 53 incorporates this expectation into the full chi-square formula and arrives at a p value that is much smaller than 0.05.
Slide 54: Release students to perform the stop codon frequency analysis on their CDS and to discuss results in their groups before coming to a class consensus on the results. Of course, each CDS only contains a single stop codon (at its 3' end), so the general trend that will be observed is that students who happened to choose genes with longer CDS will be more significantly different from the expectation. Only very short CDS might not differ from the expectation. If practical, the instructor can tally the results of the class on a projected spreadsheet or on a white board, noting the number of codons in each student's CDS.
SUMMATIVE ASSESSMENT 2 (SUPPORTING FILE S7; KEY AVAILABLE IN SUPPORTING FILE S15) - 15 MIN.
SUMMARY - 5 MIN.
Slide 56: At the conclusion of this assessment, after the students have handed in their worksheets, spend a few minutes polling the class about their results. The structure of this exercise was to create the key to this assessment (i.e. Supporting File S15) distributed among the class by having each student calculate a single chi-square value and interpret its associated p-value. A brief discussion should suffice to arrive at a consensus that proteins above a certain length (117 codons) are significantly lacking stop codons.
Slide 57: summarizes the critical point that CDS overwhelmingly lack the expected frequency of stop codons. The vast majority of proteins in sequence databases are larger than 116 amino acids, so stop codons are truly underrepresented in CDS.
DAY 5
For each student, bring a printout of slide 63 (Mutation Exercise - Summary of Predictions).
MUTATION EXERCISE - 50 MIN.
Slide 60 gives students the rules (just for this exercise - these are not biologically relevant rules, and it is important to make this point) of how you would like them to alter their CDS before analysis. Students should first choose whether they will insert new nucleotides into their CDS or delete existing nucleotides from it. For those who will insert nucleotides, they should write out between one and twelve nucleotides to insert (a sequence of their choosing) and choose a spot in the CDS where they will insert this sequence. For students who will make a deletion, they should choose between one and twelve consecutive nucleotides that they will remove from the CDS. My observation is that students tend to prefer to make changes near the near the 5' end, and students also seem to prefer to delete existing nucleotides over inserting them. Thus, I now actively urge students also to choose a seemingly random location in the CDS to place their mutation, and poll the class by hand-raising to ensure that I'm achieving a roughly 1:1 ratio of insertions to deletions.
Slide 61: Now that mutation has occurred, it is time to observe the result, but students should first predict what the effect of their insertion or deletion will be. The predictions should be structured as follows. Each student will predict whether their mutation will lengthen, shorten, or not change the length of the protein; they will also predict whether their mutation will severely, somewhat, or barely change the sequence of the protein.
Slide 62 asks students to form new groups. The compositions of the groups are dictated by the characteristics of the CDS mutations that they chose. Each group should ideally contain three to five students and comprise only students who made:
Group Name Mutation Type Size Change
I1 Insertion 1, 4, 7, 10 nucleotides
I2 Insertion 2, 5, 8, 11 nucleotides
I3 Insertion 3, 6, 9, 12 nucleotides
D1 Deletion 1, 4, 7, 10 nucleotides
D2 Deletion 2, 5, 8, 11 nucleotides
D3 Deletion 3, 6, 9, 12 nucleotides
Depending on the distribution of students into these groups, you might break some larger groups up into smaller groups of three to five, as long as the composition of each group remains homogeneous.
Provide up to ten minutes to form groups and for each group to debate and reach consensus on its predictions. Common predictions here involve the alternative conceptions that insertions necessarily make the resulting protein longer and that deletions necessarily make the resulting protein shorter. Predictions about the effect of the mutation on the protein sequence will typically be more variable, but there is often a component of the idea that increasing size of insertion/deletion corresponds to more qualitatively severe changes to protein sequence.
Slide 63: Once groups have reached consensus, facilitate a twenty-minute class discussion to reach a class consensus on predictions for each of the six types of mutation. These results can be tabulated on slide 63, or on printouts of this slide distributed to each group or each student. The presenter notes for this slide contains a summary of most likely predictions for each group.
DAY 6
For each student, bring one printout of Supporting File S10 (Formative Assessment 3) and of Slide 69 (Quantifying the Effects of Mutation Exercise: Summary of Observed Effects).
QUANTIFYING THE EFFECTS OF MUTATION EXERCISE - 40 MIN.
Slide 66: After clear predictions have been established, the students will use the transeq web tool to translate their mutant CDS and make observations about how their random insertion/deletion mutations affected the protein by comparison with the earlier translation of the wild-type CDS.
Slide 67: Groups should briefly discuss how to quantify the protein sequence characteristics related to the predictions that were just made: did their protein increase, decrease, or stay the same length; did the protein sequence severely, somewhat, or barely change?
Slide 68: After standardizing their analyses, each group member should quantify these characteristics for their mutant CDS and discuss with the rest of the group to see if a general conclusion can be reached on the effect of that group's type of mutation on protein length and on protein sequence.
After groups reach consensus, this is an optimal time to change the composition of groups, if time permits, in a jigsaw (forming new groups in which there is at least one member from each of the different original groups). Students representing different types of mutations (I1, I2, I3, D1, D2, D3) can share their observations.
Slide 69: Each group reports out to the class, to compare how different lengths and types of mutations were found to affect protein sequences. You can fill out the table on a projector or using a document camera, or each student can fill out an individual copy.
FORMATIVE ASSESSMENT 3 (SUPPORTING FILE S10; NO KEY OR RUBRIC PROVIDED) - 10 MIN.
DAY 7
For each student, bring (1) a printout of slide 73 (Quantifying the Effects of Mutation Exercise -Summary of Likely Observed Effects, Key); (2) Supporting File S8 (Example Mutation Results.docx); (3) Supporting File S9 (Designer Mutation Exercise.docx); and (4) Supporting File S11 (Summative Assessment 3).
SUMMARY FROM PREVIOUS CLASS - 5 MIN.
First, display Slide 73 to the class. If this slide displays a pattern that is unlike what the class concluded from their own observations (Slide 69 from the previous class), then it is more useful to show this slide to initiate a discussion about why the discrepancy might have arisen. This slide contains a visual key to a number of example mutations made to a fictitious CDS that is short enough to be represented on one line of print (see Supporting File S8 "Example Mutation Results.docx"). These mutations satisfy many of the possible outcomes of the effects of mutations on protein sequence. The most salient features of this table are that:
- Most or all mutation types can increase or decrease protein length; it is much less likely to have mutations that leave the protein the same length.
- Only in-frame mutations (multiples of 3) will likely produce mutations that will barely affect protein sequence (i.e. in-frame deletions or insertions of small numbers of codons).
- It should be rare to find a mutation that does not change the protein sequence at all.
REVERSING THE PROCESS EXERCISE - 30 MIN.
The observed results of this Lesson should naturally raise questions in class, such as: Is there any cell in the results table for which it is impossible to devise a mutation to occupy? This question is posed on Slide 74. Have students work alone or in small groups to specifically design a mutation, using Supporting File S9 that will fill any one of the blank cells from Slide 69. Conclude the class with Slide 75, summarizing the most important points from this series of exercises. If time remains, you might assign the extension assignment on Slide 76.
SUMMATIVE ASSESSMENT 3 (SUPPORTING FILE S11; KEY AVAILABLE IN SUPPORTING FILE S16) - 15 MIN.
TEACHING DISCUSSION
COURSE MODE AND PHILOSOPHY
I have observed that the investment of time required to implement the Lesson (five 50-minute class sessions) results in improved student intellectual investment in learning and in their approach to answering questions related to the effects of mutations on proteins. I generally employ a pedagogical approach of blending learning (27), specifically using a flipped classroom in which students often access material prior to the in-class period where we practice employing the material in context (28). Thus, I try to develop a classroom atmosphere that engages students in activities developed to produce deep understanding of key concepts, rather than broadly surveying the discipline of genetics.
STUDENT ATTITUDES AND ASSESSMENT RESULTS
As expected, my class assessments that I conduct prior to using this Lesson reveal that my students share widespread, common alternative conceptions about the effects of mutations on proteins. For example, for students who create nucleotide deletions in their CDSs, 37/41 students in one semester predicted that the encoded protein would be shorter; 14/21 students who created insertion mutations predicted that these would product longer proteins.
Student responses demonstrate the effectiveness of the design of the activities comprising this Lesson. For example, students have reported appreciating the repeated use of the chi-square test in different scenarios. Additionally, students have responded favorably to the approach of working independently on DNA sequences and "playing with" (mutating) the sequences to observe the effect of the mutation on the protein. Expressions of surprise at how the same types of mutations (e.g. insertions of multiples of three nucleotides) can cause a diversity of effects on protein sequences are common. Students reported appreciating the opportunity to create their own summary of the vast variety of unexpected types of effects on proteins that different types of mutations can have and discovering how certain types of mutations are more likely than others to produce particular effects on the protein length/sequence. High-achieving students also particularly liked the challenge posed at the end of the Lesson to design a mutation to satisfy a particular protein requirement.
After completion of the Lesson, students improved in avoiding common alternative conceptions related to the effects of mutations on proteins, particularly with respect to the alternative conception that insertions necessarily make proteins longer and deletions make them shorter. Students consciously understood that more details must be considered before making a conclusion about the effect of a mutation on a protein sequence. For example, one student response to Formative Assessment 3 (File S10) was, "The groups that inserted one nucleotide predicted increase in amino acid sequence. Today's exercise showed a decrease in amino acid sequence because premature stop codons appeared." Also, the best improvement on Genetics Concept Assessment (GCA) questions 4, 11 and 12 that I have observed in one semester included 62% to 71%, 28% to 100%, and 35% to 43% correct responses (pre-test to post-test), respectively.
LESSON ADAPTATIONS AND EXTENSIONS
The Lesson offers opportunities for multiple adaptations. First, instead of using the earwax example to motivate the Lesson, an easy substitution is to employ phenylthiocarbamide test strips, which allow students to phenotype themselves for the ability to taste or not to taste phenylthiocarbamide. The genetic basis for this phenotype also involves known single nucleotide polymorphisms (29). Exploring the topic of the genetic structure of human populations following the earwax activity can be extended by discussing the use of microsatellite markers (DNA fingerprinting) in forensic science in a statistical approach to determining the likely ethnicity of forensic DNA samples (e.g. 30).
If you have already introduced specific genes during instruction of your course, then it would be appropriate to direct students to access the CDS of a list of genes you provide, along with the URL for the sequence database you wish them to use. Likewise, if your course has introduced human medical conditions, you should consider requiring students to use the CDS of a gene that is implicated in development of hereditary human disorders. One of the most useful resources for accomplishing this is the Online Mendelian Inheritance in Man (OMIM) website (http://www.omim.org), where students can search for a heritable human disease and read a curated description of studies that have explored the genetic basis for that disease. The entry for each disease also includes the names of genes implicated in the disease.
Finally, an important best practice to emphasize is that the layouts of web interfaces used (i.e. WormBase.org, transeq) might change over time. The screen shots incorporated in the class presentation slides are current as of 9 September 2015, but it is always important to perform a run-through of the analyses before in-class presentation to ensure that the URLs for the tools have not changed and that the screen shots in the slides do not need to be updated.
SUPPORTING MATERIALS FILES
- S1. Effects of Mutations on Proteins – Presentations.pptx
- S2. Effects of Mutations on Proteins – Codon Table.png
- S3. Effects of Mutations on Proteins – Formative Assessment 1.docx
- S4. Effects of Mutations on Proteins – Summative Assessment 1.docx
- S5. Effects of Mutations on Proteins – Chi Square Table.png
- S6. Effects of Mutations on Proteins – Formative Assessment 2.docx
- S7. Effects of Mutations on Proteins – Summative Assessment 2.docx
- S8. Effects of Mutations on Proteins – Example Mutation Results.docx
- S9. Effects of Mutations on Proteins – Designer Mutation Exercise.docx
- S10. Effects of Mutations on Proteins – Formative Assessment 3.docx
- S11. Effects of Mutations on Proteins – Summative Assessment 3.docx
- S12. Effects of Mutations on Proteins – Formative Assessment 1 Key.docx
- S13. Effects of Mutations on Proteins – Summative Assessment 1 Key.docx
- S14. Effects of Mutations on Proteins – Formative Assessment 2 Key.docx
- S15. Effects of Mutations on Proteins – Summative Assessment 2 Key.xlsx
- S16. Effects of Mutations on Proteins – Summative Assessment 3 Key.docx
ACKNOWLEDGMENTS
I offer my sincere thanks to a number of colleagues and mentors who have provided opportunities, inspiration, support, and encouragement for my scholarly efforts in pedagogy, including: Joseph Castro, Lynnette Zelezny, Susan Elrod, Andrew Lawson, Rudy Sanchez, Angel Sanchez, Chris Vieira, Mike Pronovost, Sue Yang, JoLynne Blake, Mary Bennett, and the DISCOVERe faculty fellows.
References
- Smith MK, Knight JK. 2012. Using the Genetics Concept Assessment to document persistent conceptual difficulties in undergraduate genetics courses. Genetics 191:21-32.
- McDaniel MA, Butler AC. 2011. A contextual framework for understanding when difficulties are desirable, p 560. In Benjamin A (ed), Successful remembering and successful forgetting: A festschrift in honor of Robert A Bjork. Psychology Press (Taylor and Francis), New York, NY.
- Branford J, Brown A, Cocking R. 2000. How people learn: Brain, mind, experiences and school. National Academies Press, Washington, D.C.
- Fox M, N H. 2003. Evaluating and improving undergraduate teaching in sciences, technology, engineering, and mathematics. Press NA, Washington, D.C.
- Kuh G, Kinzie J, Schuh J, Whitt W. 2005. Student success in college: creating conditions that matter. Jossey-Bass, San Francisco.
- McKeachie W. 2007. Good teaching makes a difference - and we know what it is, p 457-474. In Perry R, Smart J (ed), The scholarship of teaching and learning in higher education: An evidence-based perspective. Springer, Dordrecht, Netherlands.
- Resnick LB. 1991. Shared cognition, p 1-20. In Resnick L, Levine J, Teasley S (ed), Perspectives on socially shared cognition. American Psychological Association, Washington, D.C.
- Campbell AM, Eckdahl TT. Using Synthetic Biology and pClone Red for Authentic Research on Promoter Function: Introductory Biology (identifying new promoters). CourseSource.
- Eckdahl TT, Campbell AM. Using Synthetic Biology and pClone Red for Authentic Research on Promoter Function: Genetics (analyzing mutant promoters). CourseSource.
- Colosimo PF, Peichel CL, Nereng K, Blackman BK, Shapiro MD, Schluter D, Kingsley DM. 2004. The genetic architecture of parallel armor plate reduction in threespine sticklebacks. PLoS Biol 2:E109.
- Yoshiura K, Kinoshita A, Ishida T, Ninokata A, Ishikawa T, Kaname T, Bannai M, Tokunaga K, Sonoda S, Komaki R, Ihara M, Saenko VA, Alipov GK, Sekine I, Komatsu K, Takahashi H, Nakashima M, Sosonkina N, Mapendano CK, Ghadami M, Nomura M, Liang DS, Miwa N, Kim DK, Garidkhuu A, Natsume N, Ohta T, Tomita H, Kaneko A, Kikuchi M, Russomando G, Hirayama K, Ishibashi M, Takahashi A, Saitou N, Murray JC, Saito S, Nakamura Y, Niikawa N. 2006. A SNP in the ABCC11 gene is the determinant of human earwax type. Nat Genet 38:324-330.
- POGIL. Process Oriented Guided Inquiry Learning. http://www.pogil.org/. Accessed 11/17/2014.
- Alexander P, Murphy P. 1998. The research base for APA's learner-centered psychological principles, p 25-60. In Lambert N, McCombs B (ed), How Students Learn. American Psychological Association, Washington, D.C.
- Mayer R. 1998. Cognitive theory for education: What teachers need to know, p 353-377. In Lambert N, McCombs B (ed), How Students Learn. American Psychological Association, Washington, D.C.
- Fullilove RE, Treisman PU. 1990. Mathematics achievement among African American undergraduates of the University of California, Berkeley: An evaluation of the Mathematics Workshop Program. Journal of Negro Education 59:463-478.
- Rhodes T. 2010. Assessing outcomes and improving achievement: Tips and tools for using rubrics. Association of American Colleges and Universities, Washington, D.C.
- Treisman U. 1992. Studying students studying calculus: A look At the lives of minority mathematics students. College Mathematics Journal 23:362-372.
- von Mizener B, Williams R. 2009. The Effects of Student Choice on Academic Performance. Journal of Positive Behavior Interventions 11:110-128.
- Doyle T. 2011. Learner-centered teaching: Putting the research on learning into practice. Stylus, Sterling, VA.
- Maloney M, Parker J, Leblanc M, Woodward C, Glackin M, Hanrahan M. 2010. Bioinformatics and the undergraduate curriculum. CBE Life Sci Educ 9:172-174.
- Honts J. 2003. Evolving strategies for the incorporation of bioinformatics within the undergraduate cell biology curriculum. Cell Biology Education 2:233-247.
- Holtzclaw J, Eisen A, Whitney E, Penumetcha M, Hoey J, Kimbro K. 2006. Incorporating a new bioinformatics component into genetics at a historically black college: Outcomes and lessons. CBE Life Sci Educ 5:52-64.
- Campbell C, Nehm R. 2013. A critical analysis of assessment quality in genomics and bioinformatics education research. CBE Life Sci Educ 12:530-541.
- Crouch C, Fagen A, Callan J, Mazur E. 2004. Classroom demonstrations: Learning tools or entertainment? American Journal of Physics 72:835-838.
- American Association for the Advancement of Science. 2011. Vision and Change in Undergraduate Biology Education: A Call to Action. Washington, D.C.
- Bakewell MA, Wittkopp PJ. 2013. Basic Probability and Chi-Squared Tests. Genetics Society of America Peer-Reviewed Education Portal 2013.005.
- Bonk C, Graham C. 2006. The handbook of blended learning environments: Global perspectives, local designs. Pfeiffer (Wiley), San Francisco.
- Bergmann J, Sams A. 2012. Flip your classroom: reach every student in every class every day. International Society for Technology in Education, Washington, D.C.
- Kim U, Drayna D. 2004. Genetics of individual differences in bitter taste perception: lessons from the PTC gene. Clinical Genetics 67:275-280.
- Butler JM, Schoske R, Vallone PM, Redman JW, Kline MC. 2003. Allele frequencies for 15 autosomal STR loci on U.S. Caucasian, African American, and Hispanic populations. J Forensic Sci 48:908-911.
Article Files
Login to access supporting documents
Predicting and classifying effects of insertion and deletion mutations on protein coding regions(PDF | 194 KB)
S1. Effects of Mutations on Proteins Presentations.pptx(PPTX | 6 MB)
S2. Effects of Mutations on Proteins Codon Table.png(PNG | 252 KB)
S3. Effects of Mutations on Proteins Formative Assessment 1.docx(DOCX | 506 KB)
S4. Effects of Mutations on Proteins Summative Assessment 1.docx(DOCX | 356 KB)
S5. Effects of Mutations on Proteins Chi Square Table.png(PNG | 175 KB)
S6. Effects of Mutations on Proteins Formative Assessment 2.docx(DOCX | 42 KB)
S7. Effects of Mutations on Proteins Summative Assessment 2.docx(DOCX | 260 KB)
S8. Effects of Mutations on Proteins Example Mutation Results.docx(DOCX | 102 KB)
S9. Effects of Mutations on Proteins Designer Mutation Exercise.docx(DOCX | 90 KB)
S10. Effects of Mutations on Proteins Formative Assessment 3.docx(DOCX | 36 KB)
S11. Effects of Mutations on Proteins Summative Assessment 3.docx(DOCX | 556 KB)
S12. Effects of Mutations on Proteins Formative Assessment 1 Key.docx(DOCX | 78 KB)
S13. Effects of Mutations on Proteins Summative Assessment 1 Key.docx(DOCX | 36 KB)
S14. Effects of Mutations on Proteins Formative Assessment 2 Key.docx(DOCX | 78 KB)
S15. Effects of Mutations on Proteins Summative Assessment 2 Key.docx(DOCX | 77 KB)
S16. Effects of Mutations on Proteins Summative Assessment 3 Key.docx(DOCX | 64 KB)
- License terms

Comments
Comments
There are no comments on this resource.