A clicker-based case study that untangles student thinking about the processes in the central dogma
Published online:
Abstract
The central dogma of biology is a foundational concept that provides a scaffold to understand how genetic information flows in biological systems. Despite its importance, undergraduate students often poorly understand central dogma processes (DNA replication, transcription, and translation), how information is encoded and used in each of these processes, and the relationships between them. To help students overcome these conceptual difficulties, we designed a clicker-based activity focused on two brothers who have multiple nucleotide differences in their dystrophin gene sequence, resulting in one who has Duchenne muscular dystrophy (DMD) and one who does not. This activity asks students to predict the effects of various types of mutations on DNA replication, transcription, and translation. To determine the effectiveness of this activity, we taught it in ten large-enrollment courses at five different institutions and assessed its effect by evaluating student responses to pre/post short answer questions, clicker questions, and multiple-choice exam questions. Students showed learning gains from the pre to the post on the short answer questions and performed highly on end-of-unit exam questions targeting similar concepts. This activity can be presented at various points during the semester (e.g., when discussing the central dogma, mutations, or disease) and has been used successfully in a variety of courses ranging from non-majors introductory biology to advanced upper level biology.
Citation
Pelletreau, K.N., Andrews, T., Armstrong, N., Bedell, M.A., Dastoor, F., Dean, N., Erster, S., Fata-Hartly, C., Guild, N., Greig, H., Hall, D., Knight, J.K., Koslowsky, D., Lemons, P.P., Martin, J., McCourt, J., Merrill, J., Moscarella, R., Nehm, R., Northington, R., Olsen, B., Prevost, L., Stoltzfus, J., Urban-Lurain, M., and Smith, M.K. 2016. A clicker-based study that untangles student thinking about the processes in the central dogma. CourseSource. https://doi.org/10.24918/cs.2016.15Society Learning Goals
Genetics
- Molecular biology of gene function
- How is genetic information expressed so it affects an organism's structure and function?
- Genetic variation
- How do different types of mutations affect genes and the corresponding mRNAs and proteins?
Lesson Learning Goals
- Students should understand how different types of mutations affect alleles, and the corresponding mRNAs and proteins.
- Students should understand how genetic information is expressed, and how changes in DNA can affect an organism's phenotype.
Lesson Learning Objectives
Students will be able to:- explain the differences between silent (no change in the resulting amino acid sequence), missense (a change in the amino acid sequence), and nonsense (a change resulting in a premature stop codon) mutations.
- differentiate between how information is encoded during DNA replication, transcription, and translation.
- evaluate how different types of mutations (silent, missense, and nonsense) and the location of those mutations (intron, exon, and promoter) differentially affect the processes in the central dogma.
- predict the molecular (DNA size, mRNA length, mRNA abundance, and protein length) and/or phenotypic consequences of mutations.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
The central dogma of molecular biology describes the transfer of sequence information among DNA, RNA, and proteins (1,2). In undergraduate biology courses, central dogma units often include three primary processes: DNA replication, transcription, and translation (3,4). These three processes illustrate the storage and flow of genetic information, one of the five core concepts identified by the American Association for the Advancement of Science (AAAS) Vision and Change report (5) and a disciplinary core idea in the Next Generation Science Standards (6) for K-12 students.
Despite the importance of the central dogma, the relationships between gene, mRNA, protein, and phenotype remain difficult concepts for undergraduate students (7-11). For example, an exploration of student mental models using concept maps and information gathered during student interviews revealed that many students: 1) make inappropriate connections between DNA and RNA or omit them altogether, and 2) fail to connect mRNA with translation (10). Furthermore, open-ended interviews revealed that students were comfortable using technical terms such as "transcribe" even when their underlying mental models of the terms were incorrect (10).
Asking students to predict the outcomes of mutations, in particular stop codon mutations, on central dogma processes has also been used to assess student understanding of the central dogma. Student answers to a multiple-choice question developed for the Genetics Concept Assessment (GCA, 12) revealed that even after taking a genetics course, nearly half of the students incorrectly identified that a stop codon mutation will stop transcription (9). Furthermore, nearly one-third of the students failed to identify that a frameshift mutation can result in a premature stop codon that in turn leads to the production of a shorter protein.
More recently, student thinking about the differential effects of a stop codon mutation on the processes in the central dogma have been explored using constructed-response short-answer questions (13). Undergraduate students from multiple institutions were asked to answer a three-part short-answer question (modified from the GCA) on the effect of a premature stop codon on replication, transcription, and translation (Figure 1).

Figure 1. Pre/post short answer questions about the role of the stop codon in the central dogma. The pretest questions were answered after the instructors covered the central dogma through translation, but before the in-class activity was used. The posttest questions were answered 7-10 days after the activity.
Student responses were scored as correct, incorrect, or irrelevant/incomplete (e.g., they failed to address the question, and/or it was unclear which process they were talking about because of vague language) using tools from the Automated Analysis of Constructed Response project (AACR; http://www.msu.edu/~aacr). Student answers revealed variability in their understanding about DNA replication, transcription, and translation; moreover they often conflated two or more of the processes in their explanation (13). For example, students would incorrectly describe the effect of a premature stop codon on DNA replication and transcription (often writing that a premature stop codon will stop replication and transcription early), but go on to correctly describe the effects on translation.
One way to address mixed models of student thinking about the central dogma is to develop in-class activities that target these difficulties. Although lessons targeting student thinking about transcription and translation have been developed (e.g., 14,15), these do not directly address students' tangled understanding of when and to what extent changes in DNA will affect replication, transcription, and translation. Therefore, we developed an interactive classroom activity to clarify student thinking about how mutations, with particular emphasis on premature stop codon mutations, affect the individual processes in the central dogma (Supporting File S1: Untangling the central dogma - Lecture slides). This 50-minute clicker-based activity is centered on a case study of two brothers, one of whom has X-linked Duchenne muscular dystrophy (DMD). The activity follows the exploratory and experimental steps a researcher would use to determine which of five nucleotide differences in the brothers' dystrophin gene may be the cause of one brother having the disease phenotype. Specifically, students are asked to predict how different types of mutations (silent, missense, nonsense, promoter, and intron) affect replication, transcription, and translation and how that outcome relates to the disease phenotype.
INTENDED AUDIENCE
This activity has been used in ten large-enrollment undergraduate biology courses at five different institutions including: first-year introductory biology, introductory biology for biology majors, introductory biology for cell and molecular biology majors, genetics, and biology for non-majors.
REQUIRED LEARNING TIME
This activity was designed to fit into a 50-minute class period.
PREREQUISITE STUDENT KNOWLEDGE
This lesson is intended for use at any point in the semester after students received instruction on the central dogma. Students should be: able to define silent, missense and nonsense mutations; familiar with the processes in the central dogma, including DNA replication, transcription, and translation; comfortable using codon tables; and capable of transcribing DNA sequences into mRNA, and translating mRNA sequences into protein.
PREREQUISITE TEACHER KNOWLEDGE
The instructor should have some familiarity with DMD and the dystrophin gene, and basic understanding of the central dogma and mutations. Information about DMD is included in the notes section of the activity slides. A list of relevant resources explaining DMD and the mechanisms of the disease have been compiled in Table 1.
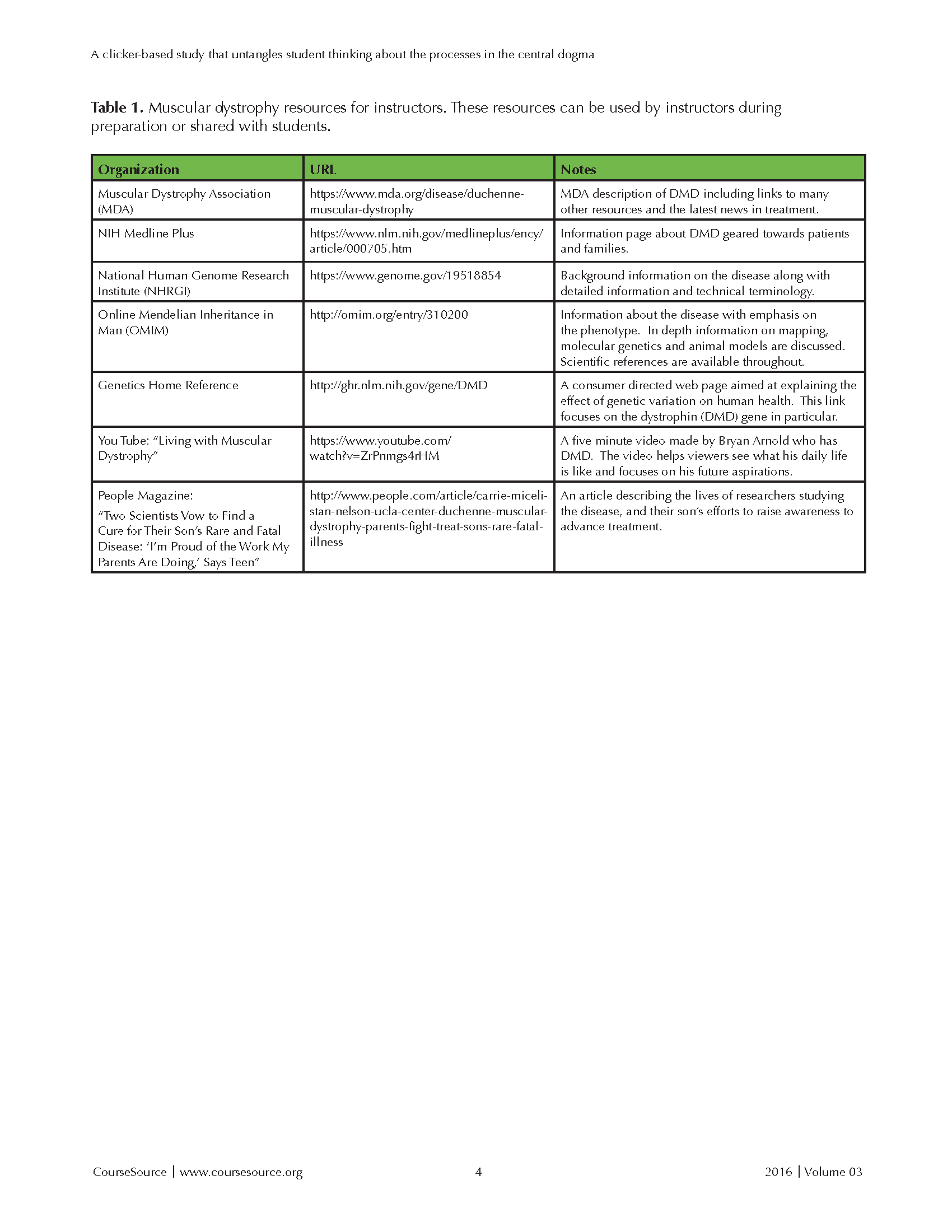
Table 1. Muscular dystrophy resources for instructors. These resources can be used by instructors during preparation or shared with students.
SCIENTIFIC TEACHING THEMES
ACTIVE LEARNING
The activity includes short answer discussion questions and multiple-choice questions with real-time student participation (i.e. clickers questions), think-pair-share (16), and whole class discussions. In whole class discussions, dialogue is shared across the classroom as the instructor facilitates moving the conversation from student to student. This dynamic can be achieved with or without clickers. Often a student will answer a question posed by the instructor and another student may support or disagree with the first student. For example, an instructor can request an explanation for a possible answer to the multiple-choice clicker question after peer discussion. When a student provides an answer, the instructor then asks other students to support or disagree with the answer and provide evidence. Having multiple students respond directly to one another enables students to hear diverse perspectives, requires students to verbalize their thoughts for a larger audience, and provides opportunity for interaction with a different subset of peers.
ASSESSMENT
Assessment of student understanding came from three sources: pre and post-activity responses to the AACR stop codon question (Figure 1), clicker questions during the classroom activity (Supporting File S2: Untangling the central dogma- Clicker questions and responses), and multiple-choice exam questions (Figure 2).
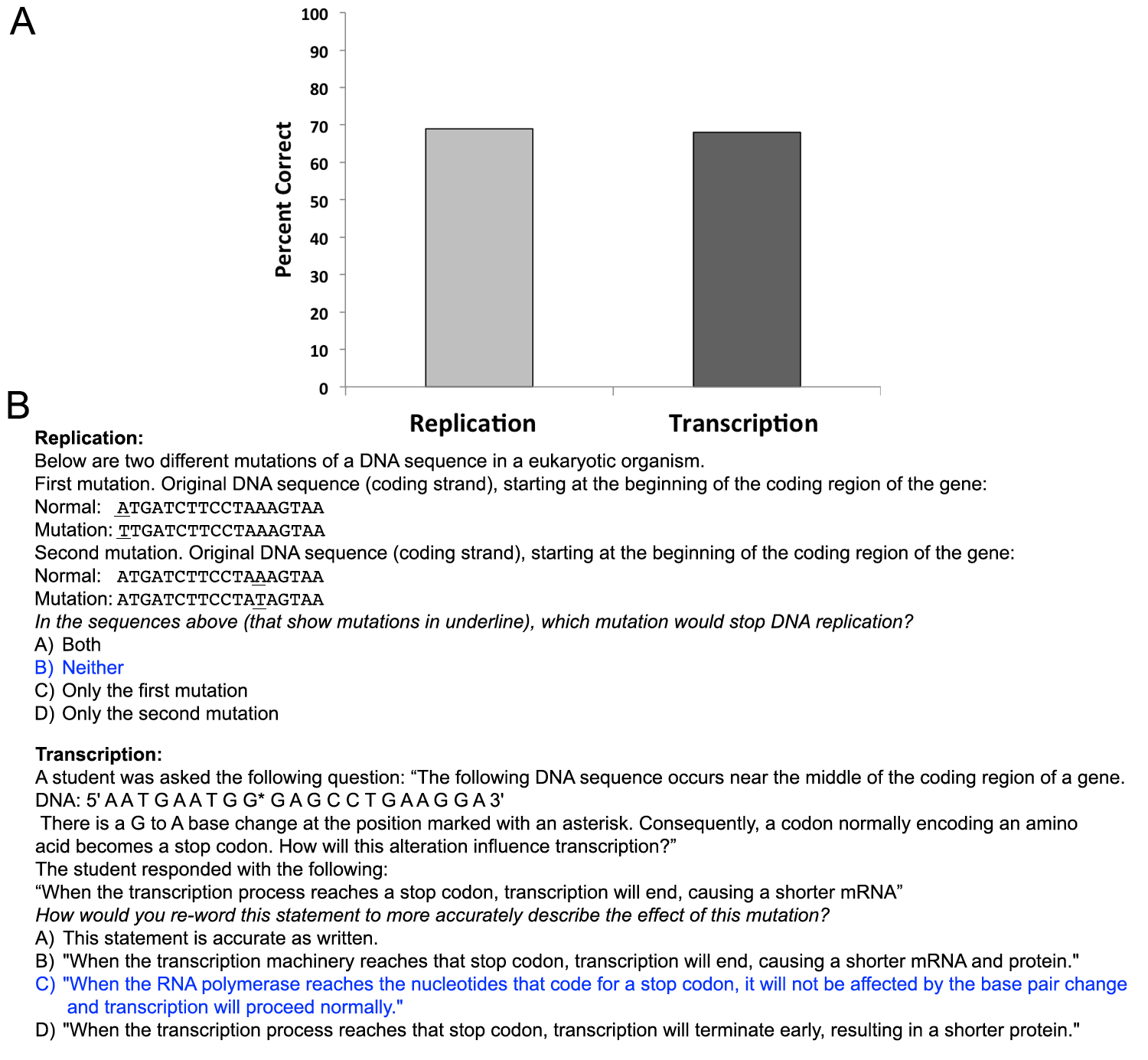
Figure 2. Student performance on end of unit exam questions addressing the role of stop codons on replication and transcription. (A) Weighted averages representing student responses from 8 of the classes. (B) Exam questions asked to students, correct answers are indicated in blue font.
After instruction on the central dogma but before the activity, students answered three open response pretest questions about the effect of a premature stop codon mutation on DNA replication, transcription, and translation (Figure 1). Student responses were categorized into correct, incorrect, or irrelevant/incomplete using tools from the AACR project (http://www.msu.edu/~aacr; 13). Students then answered the same three open response questions as a posttest 7-10 days after the activity.
In-class formative assessment of student understanding was measured using clicker questions. Student responses to clicker questions are included in the lesson plan and in Supporting File 2 (Supporting File S2: Clicker questions and responses). Summative exam questions were used to evaluate student retention of the material (Figure 2).
INCLUSIVE TEACHING
In this activity, students are asked to place themselves in the role of a research scientist and investigate open research questions. The activity includes animations, diagrams, and verbal descriptions. In addition, students answer clicker questions individually, discuss the questions with their neighbors and answer them again, and then participate in a whole class discussion (17,18). The engagement of students using active learning techniques such as think-pair-share during clicker questions has been shown to increase student learning and decrease drop out rate (19), especially for underrepresented students (20).
LESSON PLAN
This activity is designed for a 50-minute class period after the basic steps of the central dogma through translation have been covered. The progression of the clicker-based activity is outlined with estimated timing in the teaching timeline (Table 2).
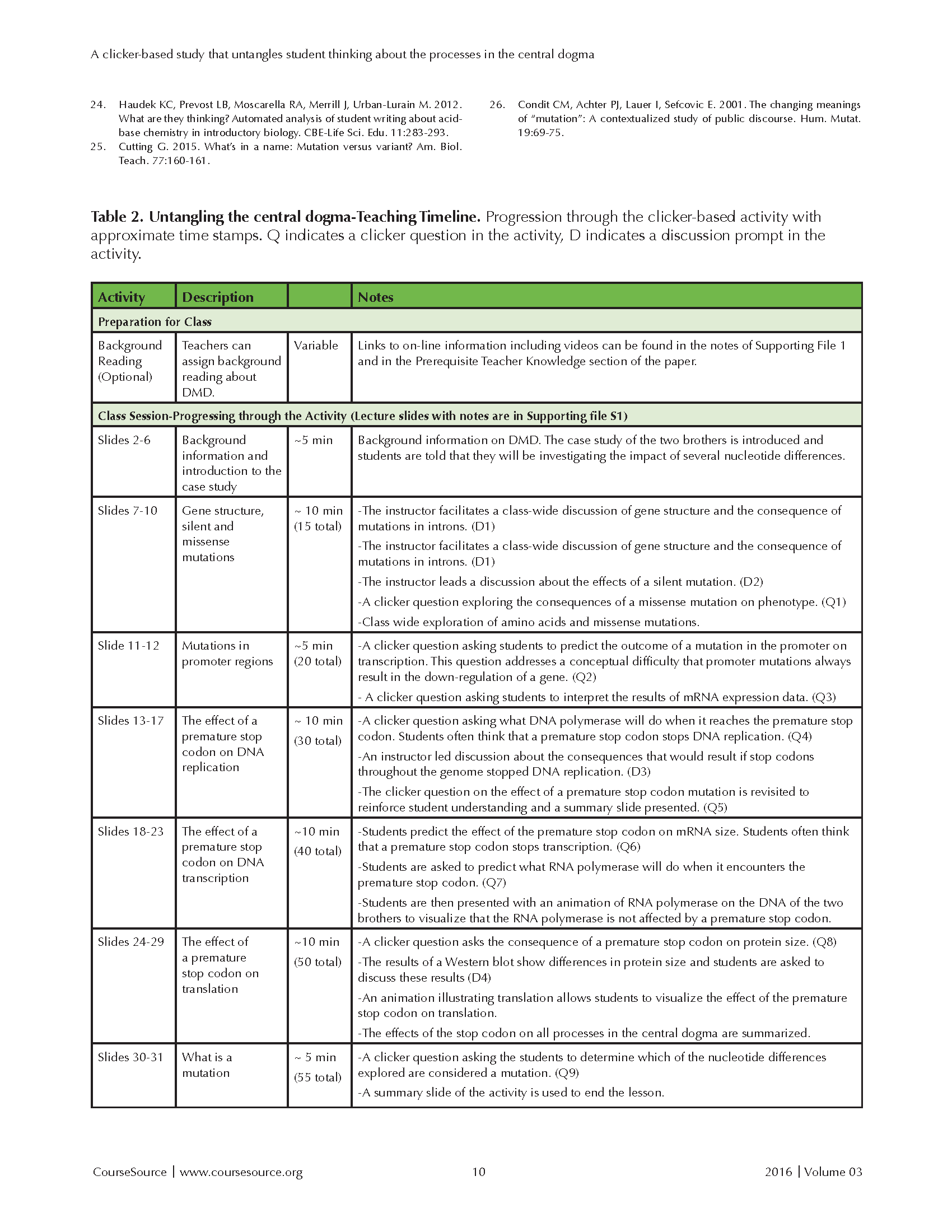
Table 2. Untangling the central dogma-Teaching Timeline. Progression through the clicker-based activity with approximate time stamps. Q indicates a clicker question in the activity, D indicates a discussion prompt in the activity.
PRE-CLASS PREPARATION
Instructors are encouraged to familiarize themselves with DMD and the role and function of the dystrophin protein. Background information about the disease including websites and optional videos are listed in Table 1 and in the notes section of the classroom presentation file. We encourage instructors to reflect on how they can best use the discussion prompts and clicker questions at a level appropriate for their individual classrooms.
USE OF CLICKER QUESTIONS
This activity is designed for the peer instruction model of clicker use, which includes think-pair-share (16,17). Briefly, students are presented with a clicker question, answer it on their own, turn to their neighbor to discuss the question, and vote again. The instructor then leads a discussion with the entire class explaining both the correct and incorrect answers. Typically, students are not shown their responses until after the peer discussion, subsequent vote, and class discussion are complete. This combination of student voting followed by an instructor explanation improves student learning (21).
Transitions through the activity are important for setting up clicker questions and whole class discussion questions (suggested transitions are included in the notes section of each slide, Supporting File S1: Untangling the central dogma - Lecture slides). Instructors are encouraged to read through the slides and notes prior to teaching to understand where these transitions are needed, and what patterns of student thinking have been commonly observed during the activity. The clicker questions for the activity, along with the student responses after peer discussion observed from eight classrooms at five different institutions are provided in Supporting File S2: Untangling the central dogma - Clicker questions and responses.
PROGRESSING THROUGH THE ACTIVITY
The clicker-based activity begins with background information on DMD, the dystrophin gene (mutations in this gene cause DMD), and information about two brothers: Liam who has DMD and Elijah who does not have the disease (Supporting File S1: Untangling the central dogma - Lecture slides, slides 2-5). Students are then presented with DNA sequence information for Liam and Elijah, and are told that the brothers have five nucleotide differences in the dystrophin gene (Supporting File S1: Lecture slides, slide 6). Throughout the activity the term "nucleotide difference" is recommended rather than mutation. This word choice was deliberate, because students often think of mutations only as those changes that affect phenotype (9).
Students start the activity by investigating nucleotide difference #4, which is located in an intron (Supporting File S1: Lecture slides, slide 7). Students are asked to discuss whether a nucleotide difference in an intron is more or less likely to result in a DMD phenotype. The instructor facilitates a class-wide discussion about gene structure and the students conclude that a nucleotide difference in an intron is not likely to result in a muscular dystrophy phenotype because introns are removed during RNA splicing. Instructors may choose to state that an intron is the "least likely" cause of mutation based on gene structure alone and move to the next slide. Alternatively, instructors may take this opportunity to discuss with students when and how splice site mutations affect mRNA transcripts. Several references describing the effects of splice site mutations on transcription are provided in the notes sections of slide 7 (Supporting File S1: Lecture slides).
Next students are asked to consider nucleotide difference #2, a silent mutation that does not alter the amino acid sequence of the protein (Supporting File S1: Lecture slides, slide 8). The instructor leads a class-wide discussion and students conclude that this difference is not likely to result in DMD. As nucleotide differences #4 and #2 are not likely candidates for causing disease, they are marked with the symbol "✖".
Students then consider a missense mutation, nucleotide difference #5 (Supporting File S1: Lecture slides, slide 9, Q1). In Elijah, the codon codes for aspartate and in Liam (affected) the codon codes for glutamate. Students are asked a clicker question about whether nucleotide difference #5 is a possible cause of DMD in Liam. In our classes, the majority of the students voted yes (Supporting File S2: Untangling the central dogma- Clicker questions and responses, Q1, weighted average (WA) = 85%). Students are then given additional information that glutamate and aspartate are both structurally and biochemically similar (Supporting File S1: Lecture slides, slide 10). At this point, the class can explore protein structure and function. The instructor then states that nucleotide difference #5 might be a cause of DMD and that some scientists might decide to pursue the difference further, but many researchers would explore the remaining options first to see if there is a more likely candidate. A "?" appears above nucleotide difference #5 to indicate that this nucleotide difference has not been ruled out as causal.
Students are asked to shift their thinking from the coding region to regulatory regions as they investigate nucleotide difference #1 (Supporting File S1: Lecture slides, slide 11, Q2), which is located in the promoter. Students are asked: "If difference #1 caused DMD, we would predict the mRNA levels in Liam to be __________ the mRNA levels in Elijah" and are given the choices a) the same as, b) higher than, c) lower than, or d) different in some way from (Elijah). Emphasis is placed on the word "If" because in subsequent slides data will be presented contrary to this prediction. The correct answer is d) but in our classes on average 65% of students chose c) lower than (Supporting File S2: Clicker questions and responses, Q2). These clicker results suggest that students often think that a mutation in the promoter will cause the promoter to become nonfunctional. The whole-class discussion following the clicker vote provides an opportunity for the instructor to emphasize that mutations in a promoter region can result in either an increase, decrease, or no change in the regulation of transcription, and that without additional information the outcome is not certain. More information about promoter mutations can be found in de Vooght et al, 2009 (22) and in the notes section of slide 11.
Students are then presented with mRNA expression data that show the normalized dystrophin mRNA levels in muscle tissue from Elijah and Liam are equal (Supporting File S1: Lecture slides, slide 12, Q3). Given these data, the students are then asked a clicker question to reflect on whether difference #1 is a likely cause of DMD in Liam. In our classes the majority of students selected "no" (Supporting File S2: Clicker questions and responses, Q3, WA = 90%). Nucleotide difference #1 is then eliminated as a candidate for causing disease and is marked with the symbol "✖".
The activity now shifts to explore the influence of a premature stop codon on the processes in the central dogma starting with DNA replication. To orient the students, the instructor presents a slide that shows the steps of the central dogma (Supporting File S1: Lecture slides, slide 13), with DNA replication circled. Students are told that nucleotide difference #2 results in a premature stop codon in Liam (affected) compared to a glutamine codon in Elijah (Supporting File S1: Lecture slides, slide 14, Q4). Students are asked a clicker question about what DNA polymerase will do when it reaches this stop codon: a) stop when it reaches the first nucleotide encoding the premature stop codon, b) stop when it reaches the last nucleotide encoding the premature stop codon, or c) not be affected by this base change and will continue to read through the nucleotide difference. In our classes on average 73% of students incorrectly answered either a) or b) (Supporting File S2: Clicker questions and responses, Q4), suggesting a tendency for students to associate the term "stop codon" with stopping DNA replication. This response pattern provides an opportunity for the instructor to discuss the process of DNA replication and the fact that codons have no meaning to DNA polymerase. Alternatively, the instructor can choose to discuss this point after the students have worked through the two subsequent slides that explore the mechanisms at work during DNA replication.
The next slide (Supporting File S1: Lecture slides, slide 15) presents an image of DNA with red boxes indicating regions of the DNA that could encode stop codons. Students are asked to think about the consequence on DNA replication if DNA polymerases recognized any, or all of these stop codons. Students are then re-asked the clicker question about what a DNA polymerase will do when it reaches the stop codon (Supporting File S1: Lecture slides, slide 16, Q5). At this point in our classes, 75% of students correctly answered c) not be affected by this base change and will continue to read through the nucleotide difference (Supporting File S2: Clicker questions and responses, Q5). The instructor then presents the next slide (Supporting File S1: Lecture slides, slide 17) that states: "Polymerases read only one base at a time, not triplet codons. The DNA polymerase would not be affected by this base change in Liam's DNA and replication would proceed normally."
Following DNA replication, the activity explores the effect of a premature stop codon on transcription. The instructor presents a slide to reinforce that the premature stop codon will have no effect on DNA replication and reminds students of the position of transcription in the central dogma (Supporting File S1: Lecture slides, slide 18). Students are asked to predict the effect of the nucleotide difference #3, the premature stop codon in Liam (affected), on mRNA size using Northern blot data. Specifically, students are asked whether Liam's mRNA will be a) shorter, b) longer, or c) the same size as Elijah's mRNA (Supporting File S1: Lecture slides, slide 19, Q6). In our classes, 49% of the students incorrectly answered a) shorter (Supporting File S2: Clicker questions and responses, Q6). Even though many students may be incorrect, the instructor is encouraged to proceed onto the next slide without discussing why c) is the correct answer ("That's interesting, let's see what else we can find out").
The next slide asks what the RNA polymerase will do when it reaches the nucleotides encoding the stop codon: a) stop when it reaches the first nucleotide encoding the premature stop codon, b) stop when it reaches the last nucleotide encoding the premature stop codon, or c) not be affected by this base change and will continue to read through the nucleotide difference (correct answer) (Supporting File S1: Lecture slides, slide 20, Q7). Although students had just been asked a similar question about DNA polymerase, in our classes a subset of students (Supporting File S2: Clicker questions and responses, Q7, WA = 33%) still incorrectly answered either a) or b). The incorrect understanding of the effect of a premature stop codon thus persists for many students when thinking about transcription, regardless of the fact that a very similar question about DNA polymerases was previously asked (Supporting File S1: Lecture slides, slides 14, 16). The question about RNA polymerase aims to reinforce the similar functionality of DNA and RNA polymerases. Students are then presented with the results of the Northern blot experiment and can be encouraged to speculate why there is no difference in the size of the mRNA molecules for Elijah and Liam (Supporting File S1: Lecture slides, slide 21).
Students then see an animation that illustrates how RNA polymerase interacts with DNA molecules (Supporting File S1: Lecture slides, slide 22). This animation is embedded in the presentation slides, but is also available as a separate file (Supporting File S3: Untangling the central dogma Transcription animation). The students observe the polymerase approach and continue to read through Liam's (affected) premature stop codon. The instructor then presents a slide (Supporting File S1: Lecture slides, slide 23) that states: "Polymerases read only one base at a time, not triplet codons. The RNA polymerase would not be affected by this base change in Liam's DNA and transcription would proceed normally."
A summary slide of the central dogma (Supporting File S1: Lecture slides, slide 24) is displayed reinforcing that a premature stop codon will have no effect on DNA replication and transcription, and students are asked to focus on translation. Students are asked a clicker question (Supporting File S1: Lecture slides, slide 25, Q8) to predict whether the effect of the premature stop codon in Liam (affected) will: a) result in a smaller dystrophin protein in Liam (correct), b) result in a larger dystrophin protein in Liam, or c) result in the same size protein in Liam and Elijah. The majority of students in our classes correctly answered a) (Supporting File S2: Clicker questions and responses, Q8, WA = 83%). Students are then shown the result of a Western blot experiment that shows that Liam (affected) produces a shorter protein when compared to Elijah (Supporting File S1: Lecture slides, slide 26). Students can be asked to explain why they are seeing these results. The results of the experiment are followed by a second animation (Supporting File S4: Untangling the central dogma - Translation animation) illustrating how the ribosome falls off the mRNA when it reaches Liam's (affected) premature stop codon (Supporting File S1: Lecture slides, slide 27). The instructor can then present information about how the translation machinery recognizes stop codons (Supporting File S1: Lecture slides, slide 28) and a summary slide of the role of stop codons in the central dogma (Supporting File S1: Lecture slides, slide 29).
In the final clicker question in the activity (Supporting File S1: Lecture slides, slide 30, Q9) students revisit all of the nucleotide differences and are asked which of the five nucleotide differences are mutations: a) all five are mutations (correct answer), b) the three in the exons (difference #2, #3, #5), c) the two in exons that change the amino acid sequence (differences #3, #5), or d) the one in the exon that causes Duchenne muscular dystrophy (difference #5). In our classes, even at the end of this activity, on average 41% of students selected one of these three common conceptual difficulties (b, c, or d) (Supporting File S2: Clicker questions and responses, Q9). This final clicker question provides an opportunity to reinforce that any permanent DNA sequence change is considered a mutation (correct answer a).
The activity is completed with a summary slide capturing the major points of the activity (Supporting File S1: Lecture slides, slide 31) and the conclusion that, in this case, the premature stop codon (nonsense mutation) was the most likely mutation to lead to the expressed phenotype of DMD in Liam because of the effect on translation and the subsequent truncated protein produced.
TEACHING DISCUSSION
STUDENT PERFORMANCE ON PRE-POST ASSESSMENT
We assessed the effect of this activity by having students answer three open response questions about the role of a nonsense mutation on DNA replication, transcription, and translation (13; http://create4stem.msu.edu/aacr/question/stop-codon; Figure 1). The students answered these questions twice (pre- and post-activity). The pre-activity time point was before the activity but after the instructors taught about the central dogma through translation. The post-activity time point was 7-10 days after the in-class activity. Student responses were classified as being correct, incorrect, or irrelevant/incomplete using tools from the AACR project (13).
Students from five institutions in classes ranging from introductory biology to advance molecular biology participated in the activity. To measure student thinking before and after the activity, we calculated average normalized learning gain
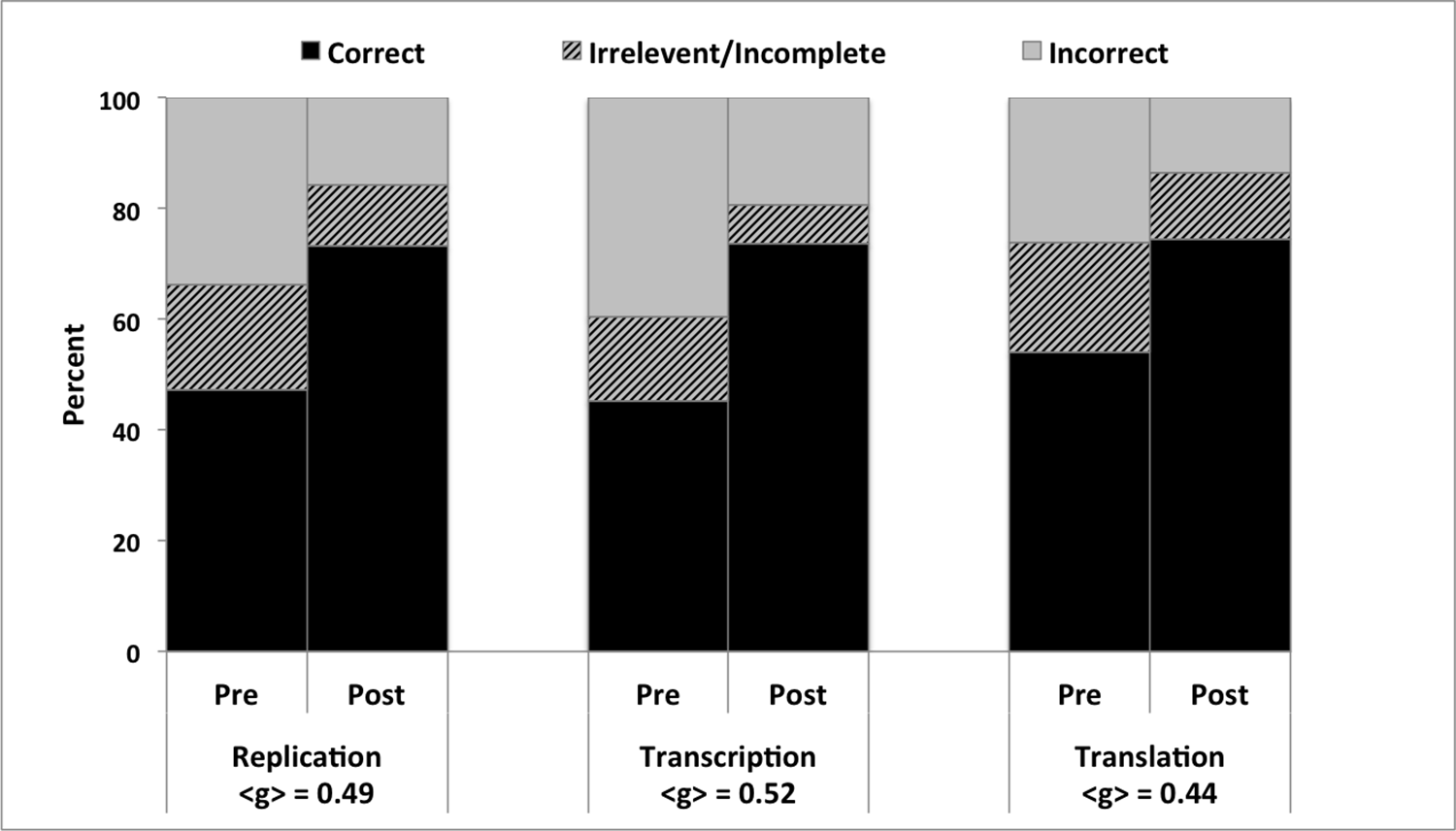
Figure 3. Aggregate student performance on pre/post short answer questions about the role of the stop codon in the central dogma asked in 10 different classes. Student responses were coded as correct, incorrect or irrelevant/incomplete. Normalized learning gains (<g> = (% of students who scored correct post – % of students who scored correct pre)/(100 – % of students who scored correct pre) are also reported (<g>, 23).
In addition, answers on shared end of unit exam questions show student performance on the effect of premature stop codons on replication and transcription at the end of the semester (69% and 68% correct respectively; Figure 2).
PERSISTENT CONCEPTUAL DIFFICULTIES
In addition to using clicker questions with think-pair-share and whole-class discussion, this activity uses animations and targets word association to help students overcome conceptual difficulties associated with the central dogma.
Animations
In developing the activity, we noted that students were having trouble visualizing how DNA and RNA polymerases interacted with DNA and hypothesized that this issue was contributing to student difficulties with determining the role of stop codons on replication and transcription. Therefore, we added an animation to illustrate how the RNA polymerase would continue to transcribe through the stop codon in Liam (affected) and the non-mutant sequence in Elijah (unaffected) (Supporting File S1: Lecture slides, slide 22). Later in the lesson we added a second animation that showed that the ribosome would release the mRNA when reaching the premature stop codon in Liam (affected) and continue to synthesize the polypeptide chain in Elijah (unaffected) (Supporting File S1: Lecture slides, slide 27). These animations are intended to provide a clear visual representation showing polymerases interacting with one nucleotide at a time, and ribosomes interacting with triplet nucleotides (codons).
Targeting word association
This activity directly addresses how students think about the different processes in the central dogma by comparing and contrasting the role of mutations (nucleotide differences) during DNA replication, transcription, and translation. A student's inability to successfully understand the effect of these mutations may be compounded by incorrect word association such as: "stop codons stop everything" (13,24). For example, early in the activity students are asked what will happen to the DNA polymerase when it encounters a premature stop codon (Supporting File S1: Lecture slides, slide 14, Q4). The majority of our students incorrectly answered that the DNA polymerase will stop (Supporting File S2: Clicker questions and responses, Q4, WA=73%) and then participated in a class-wide discussion about the consequence of a DNA polymerase stopping every time it reached a stop codon in the genome. When students were then re-asked this clicker question (Supporting File S1: Lecture slides, slide 16, Q5), on average 24% of our students answered this question incorrectly (Supporting File S2: Clicker questions and responses, Q5). Several slides later (Supporting File S1: Lecture slides, slide 20, Q7), students are asked what will happen to the RNA polymerase when it encounters a premature stop codon. Despite the similarity of the questions, on average 33% of our students still answered that the RNA polymerase will stop (Supporting File S2: Clicker questions and responses, Q7). The answer patterns to and difficulty of these clicker questions indicate the need to discuss how DNA and RNA polymerases use DNA as a template at multiple points in the lesson.
In addition, students often define the word mutation as a change that has a phenotypic outcome (10,25,26). Therefore, another goal of this activity is to help students think about the definition of mutation. Even though the lesson exposes students to silent, missense, nonsense, and regulatory mutations, at the end of the activity students are confused about which of the five nucleotide differences is a mutation. On clicker question Q9, many students (Supporting File S2: Clicker questions and responses, WA=41%) selected that mutations must be in exons, change the amino acid sequence, or influence phenotype (Supporting File S1: Lecture slides, slide 30). This clicker question provides an opportunity for instructors to talk to students about the fact that any DNA change that is replicated is a mutation, which is a concept often misunderstood even at the end of a genetics course (10).
ADDITIONAL SUGGESTIONS - to enhance student learning while using this activity
In addition to designing the activity, faculty have developed several instructional extensions including in-class worksheets, pre-class assignments, and post-class assignments. Examples of these extensions can be found in Supporting File S5: Untangling the central dogma- Extensions on learning.
In-Class Worksheets: Students can also use a codon table to determine what amino acids the codons encode, expand on consequences of other types of mutations (frameshift, deletions), and/or model (draw out) the biological processes. Worksheets can also be used when an instructor does not teach with clicker questions.
Pre-class assignments: Students can complete assignments where they learn more about DMD. These assignments may include watching videos of people coping with the disease (for helpful webpages and video see the URLs in the Prerequisite Teacher Knowledge section of the paper).
Post-class assignments: Students can also evaluate their learning by answering a set of similar homework questions; sample assessment questions are included in the supplemental file.
CONCLUSIONS
This clicker-based case-study activity is an engaging, adaptable lesson that effectively clarifies and re-enforces the role of mutations on phenotype as it explores the consequences of various mutations on DNA replication, transcription, and translation. The activity helps students make connections among topics that often appear disconnected and unrelated. Using clicker questions with think-pair-share, group discussions, animations, and predictions, students are able to explore how mutations affect genes, and the corresponding mRNA and proteins.
SUPPORTING MATERIALS
- S1. Untangling the central dogma-The clicker-based stop codon activity slides with notes.
- S2. Untangling the central dogma-Clicker questions used in the activity, distribution of student answers, and explanations for the range of student answers.
- S3. Untangling the central dogma-Animation file of transcription.
- S4. Untangling the central dogma-Animation file of translation.
- S5. Untangling the central dogma-Example extensions (worksheets and homework questions) that can be used along with the activity.
ACKNOWLEDGMENTS
We would like to thank Matthew Steele, and Andrea Bierema, Michigan State University, for their work on the AACR stop codon question algorithm and rubric respectively. We also would like to thank the students for their participation and contribution to this project.
References
- Crick F. 1970. Central dogma of molecular biology. Nature 227:561-563.
- Crick F. 1958 On protein synthesis. Symp. Soc. Exp. Biol. 12: 138-163.
- Reece J, Urry LA, Meyers N, Cain ML, Wasserman SA, Minorsky PV, Jackson RB, Cooke BN. 2011. Campbell Biology. Pearson, Boston.
- Morris JR. 2013. Biology: How Life Works. Macmillan Higher Education.
- American Association for the Advancement of Science 2011. Vision and change in undergraduate biology education: A call to action. American Association for the Advancement of Science, Washington, DC.
- NGSS Lead States. 2013. Next Generation Science Standards: For states, by states. National Academies Press, Washington, DC.
- Lewis J, Wood-Robinson C. 2000. Genes, chromosomes, cell division and inheritance - do students see any relationship? Int. J. Sci. Edu. 22:177-195.
- Marbach-Ad G. 2001. Attempting to break the code in student comprehension of genetic concepts. J Biol. Edu. 35:183-189.
- Smith MK, Knight JK. 2012. Using the Genetics Concept Assessment to document persistent conceptual difficulties in undergraduate genetics courses. Genetics 191:21-32.
- Wright LK, Fisk JN, Newman DL. 2014. DNA -> RNA: What do students think the arrow means? Cell Bio. Edu. 13:338-348.
- Newman DL, Snyder CW, Fisk JN, Wright LK. 2016. Development of the Central Dogma Concept Inventory (CDCI) assessment tool. Cell Bio. Edu. 15:ar9-ar9. 10.1187/cbe.15-06-0124.
- Smith MK, Wood WB, Knight JK. 2008. The Genetics Concept Assessment: a new concept inventory for gauging student understanding of genetics. Cell Bio. Edu. 7:422-430.
- Prevost LB, Smith MK, Knight JK. Using student writing and lexical analysis to reveal student thinking about the role of stop codons in the central dogma. 10.1187/cbe.15-12-0267 CBE Life Sci Educ vol. 15 no. 4 ar65
- Ross JA. 2016. Predicting and classifying effects of insertion and deletion mutations on protein coding regions. CourseSource.
- Zhou J and Brickman P. 2011. FOXP2 and Speech: A gene expression case. National Center for Case Study Teaching in Science, University at Buffalo, Buffalo New York.
- Lyman F. 1987. Think-Pair-Share: An expanding teaching technique: MAA-CIE Cooperative News. 1:1-2
- Mazur E. 2013. Peer Instruction: A User's Manual, 2nd ed. Pearson Higher Education, Upper Saddle River.
- Smith MK, Wood WB, Adams WK, Wieman C, Knight JK, Guild N, Su TT. 2009. Why peer discussion improves student performance on in-class concept questions. Science 323:122-124.
- Freeman S, Eddy SL, McDonough M, Smith MK, Okoroafor N, Jordt H, Wenderoth MP. 2014. Active learning increases student performance in science, engineering, and mathematics. Proc. Nat. Acad. Sci. U.S.A. 111:8410-8415.
- Eddy SL, Hogan KA. 2014. Getting under the hood: How and for whom does increasing course structure work? Cell Bio. Edu. 13:453-468.
- Smith MK, Wood WB, Krauter K, Knight JK. 2011. Combining peer discussion with instructor explanation increases student learning from in-class concept questions. Cell Bio. Edu. 10:55-63.
- de Vooght KMK, van Wijk R, van Solinge WW. 2009. Management of gene promoter mutations in molecular diagnostics. Clin. Chem. 55:698-708.
- Hake R. 1998. Interactive-engagement versus traditional methods: a six-thousand-student survey of mechanics test data for introductory physics courses. Am. J. Phy. 66:64-74.
- Haudek KC, Prevost LB, Moscarella RA, Merrill J, Urban-Lurain M. 2012. What are they thinking? Automated analysis of student writing about acid-base chemistry in introductory biology. CBE-Life Sci. Edu. 11:283-293.
- Cutting G. 2015. What's in a name: Mutation versus variant? Am. Biol. Teach. 77:160-161.
- Condit CM, Achter PJ, Lauer I, Sefcovic E. 2001. The changing meanings of "mutation": A contextualized study of public discourse. Hum. Mutat. 19:69-75.
Article Files
Login to access supporting documents
A clicker-based case study that untangles student thinking about the processes in the central dogma(PDF | 505 KB)
S1. Untangling the central dogma- Lecture Presentation Slides.pptx(PPTX | 4 MB)
S2. Untangling the central dogma- Clicker questions used in the activity distribution of student answers and explanations for the range of student answers.docx(DOCX | 154 KB)
S3. Untangling the central dogma- Animation file on transcription.avi(AVI | 289 KB)
S4. Untangling the central dogma- Animation file on translation.avi(AVI | 92 KB)
S5. Untangling the central dogma- Extensions on learning samples worksheets homework and exam questions.docx(DOCX | 255 KB)
- License terms

Comments
Comments
There are no comments on this resource.