An Introduction to Eukaryotic Genome Analysis in Non-model Species for Undergraduates: A tutorial from the Genome Consortium for Active Teaching
Published online:
Abstract
Advances in Next Generation Sequencing (NGS) technology have led to development of many research methods for analyzing genomic data. Students studying biology at the undergraduate level must hone their skills at genomic analysis to participate in productive research during and after their undergraduate studies. To counteract the barriers such as a lack of funding, equipment, or bioinformatics experience at many undergraduate institutions, the GCAT-SEEK network has produced a tutorial designed to train biology and related majors about tools used for contemporary eukaryotic genomic analysis. This lesson allows students to work through relevant chapters including, but not limited to, quality filtering and trimming of short sequence reads, genome assembly, annotation, variant detection and genome visualization, with an emphasis on the Linux operating system. The tutorial has been used in a variety of settings including a completely independent self-paced tutorial for research students, a partially independent "flipped" classroom, a heavily supported semester-long laboratory, and as the basis for an 18-hour faculty-development workshop. After completion of these lessons, students should be able to demonstrate skills related to bioinformatics in the context of genomic data, as well as understand biological concepts involved in data quality, genome assembly, annotation and variant detection.
Citation
Buonaccorsi, V.P., Hamlin, D., Fowler, B., Sullivan, C., and Sickler, A. 2017. An Introduction to Eukaryotic Genome Analysis in Non-model Species for Undergraduates: A tutorial from the Genome Consortium for Active Teaching. CourseSource. https://doi.org/10.24918/cs.2017.1Society Learning Goals
Bioinformatics
- Computation in the life sciences
- What is the role of computation in hypothesis-driven discovery processes within the life sciences?
- What computational concepts are important in bioinformatics?
- What statistical concepts are important in bioinformatics?
- DNA - Information Storage [GENOMICS]
- How can bioinformatics tools be employed to analyze genetic information?
- Computational Skills
- What higher-level computational skills can be used in bioinformatics research?
Genetics
- Methods & Tools in Genetics
- What experimental methods are commonly used to analyze gene structure and gene expression?
Science Process Skills
- Process of Science
- Interpret, evaluate, and draw conclusions from data
- Quantitative Reasoning/ Using Mathematics and Computational Thinking
- Apply the tools of graphing, statistics, and data science to analyze biological data
Lesson Learning Goals
Students will:- Understand the steps of genome assembly, annotation, and variant discovery.
- Appreciate the role of computers in data analysis.
- Understand basic statistical concepts.
- Understand tools to explore and manage massive datasets.
- Learn the utility of command line based computer programming and its applications to biology.
Lesson Learning Objectives
At the end of the activity, students will be able to:- Explain the steps involved in genome assembly, annotation, and variant detection to other students and instructors.
- Create meaningful visualizations of their data using the integrated genome viewer.
- Use the Linux command line and web-based tools to answer research questions.
- Produce annotated genomes and call variants from raw sequencing reads in non-model species.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Continued development of Next Generation Sequencing (NGS) technologies has significantly affected both research and traditional instruction of the biological sciences (1). These technologies have decreased the cost of sequencing, enabled data to be generated faster than it can be analyzed, and encouraged improvements of analytical tools. As a result, genomic analysis provides increased opportunities to introduce authentic research experiences to undergraduates. Huge repositories of publicly available data like the NCBI short read archive, gene expression omnibus, and MG-RAST database for metagenomics data (2) can provide opportunities for students to engage in the process of science. The chapters presented below introduce instructors of differing backgrounds to the applications of genomics to molecular evolution and biochemistry using questions in a non-model marine fish as the basis for exploration (3-5).
Difficulties related to up-front equipment costs and training in advanced computer skills can impede engagement in genome-scale investigation for both faculty and students. To help ameliorate these difficulties the Genome Consortium for Active Teaching using Next Generation Sequencing (GCAT-SEEK; 6-8) seeks to design and disseminate educational materials to train both undergraduates and faculty teaching undergraduates about concepts and basic computing skills in the Linux environment that are necessary for eukaryotic genomic analysis, and to facilitate analysis of members' novel data with NGS platforms. Here, we present a seven-chapter tutorial that trains students and/or faculty in the use of key tools and techniques relevant to analyzing eukaryotic genomes [Supporting Materials S1; Tables 1 & 2].
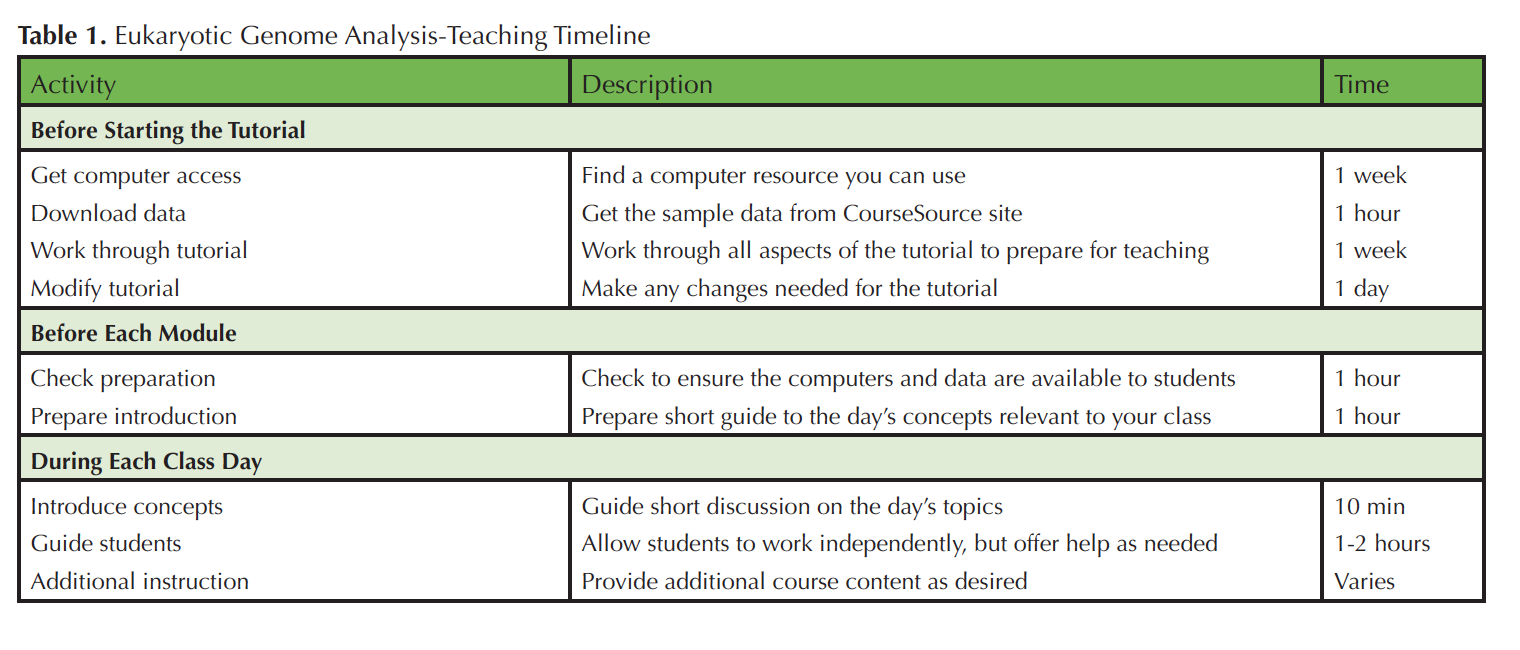
Table 1. Eukaryotic Genome Analysis-Teaching Timeline.
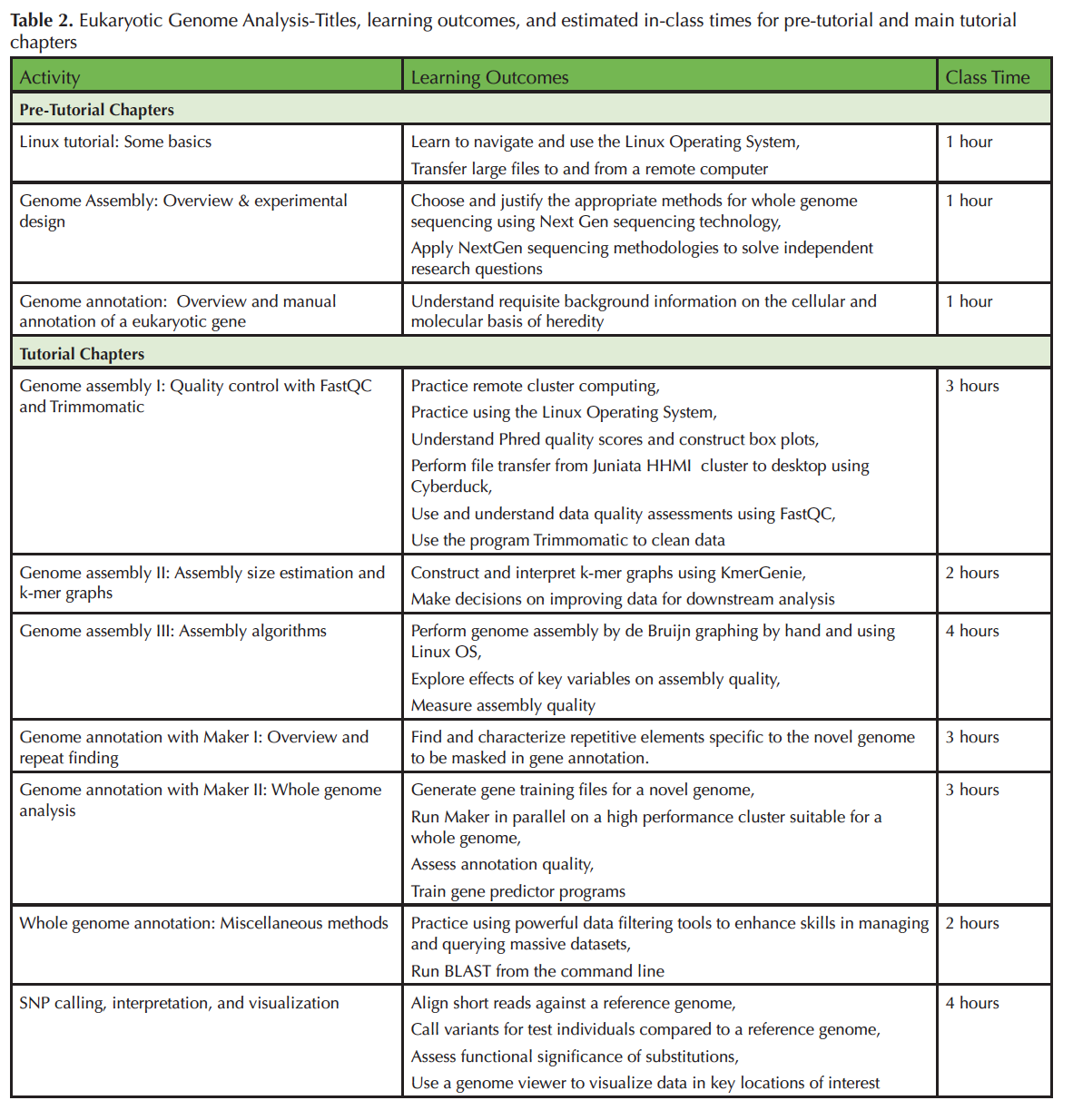
Table 2. Eukaryotic Genome Analysis-Titles, learning outcomes, and estimated in-class times for pre-tutorial and main tutorial chapters.
The selected protocols introduce modern bioinformatics research tools as well as specific research tools used for assembly, annotation and variant calling for non-model genomes. Given the declining cost of genome data, we anticipate increased sequencing of novel genomes at the undergraduate level, providing a basis for molecular and evolutionary inquiries within the undergraduate classroom. For $5k to $10k, genome core facilities are capable of deep sequencing gigabase-sized novel genomes and returning data within a few weeks of receiving genomic DNA. Faculty may wish to start an independent or course-based research experience with raw sequence reads on an organism important to their research program, and have students analyze the genomes themselves using methodology of their design. Faculty may want to perform less computationally intensive and less expensive re-sequencing analysis to characterize variants compared to a reference genome. The tutorial provided is meant for researchers working on the great majority of species that lack the kind of extensively developed databases and resources that characterize model species such as, for example, baker's yeast, Saccharomyces cerevisiae.
A tutorial from the GCAT-SEEK network detailing RNAseq analysis (8) and a series of educational chapters providing examples of how tutorials are being used by network members are also available (gcat-seek.weebly.com). Our webpage also contains information on how to join the network and information regarding future workshops.
This lesson focuses on skills related to the use of tools implemented in a Linux environment. Linux is a widely used operating system due to its ease of use for students with limited experience in computer sciences (9). This lesson provides chapters that focus on teaching students how to use tools including Trimmomatic (10), KmerGenie (11), Soap (12), MaSuRCA (13), Maker (14), Bowtie2 (15), Samtools (16), VarScan (17), SnpEff (18) and a variety of Linux commands that are useful in the context of data management and visualization. To ease access to these tools, a brief three-chapter tutorial (pre-tutorial) introduces students to Linux, molecular biology, and sequencing genomes [Supporting Materials 2]. The Linux chapter is meant to teach individuals with no Linux experience a few essentials behind navigation and basic commands. The molecular biology chapter is meant to provide an overview of the biology needed to understand the main chapters for individuals with very limited biological background. The genome-sequencing chapter is meant to provide an overview of some terms and technologies involved in modern eukaryotic genome sequencing.
INTENDED AUDIENCE
This tutorial can be effective for a variety of audiences, including undergraduates and biology faculty who are novices with respect to bioinformatic analysis. Here, we focus on the implementation of this tutorial in an undergraduate research methods laboratory course at a small liberal arts school (Juniata College). This course included sophomore, junior, and senior biology majors, all of whom had taken at least two other biology courses. In the discussion, we describe some alternative implementations. This tutorial will likely work best in an upper level genomics lab or class session with no more than 10 to 15 students per instructor (i.e. professor or well-trained TA), as questions inevitably arise that require intervention from the instructor.
Each chapter in the tutorial contains an introduction to the related genome biology and protocols for accomplishing specific bioinformatic endpoints. Specific student backgrounds can be flexible, but understanding the computational and biological content from the pre-tutorial chapters is essential for understanding the main tutorial. Introductions within the text and links to further information serve as points for in-class lectures and discussions that, at any point, may be addressed in more depth considering the primary literature backing a particular topic. In this way, the tutorial can be geared to match needs of different students. These differences may be related to student background (e.g., a 100 or 200 level biology class will need much more background than a graduate level class) or student interests (e.g., a course for computer scientists will focus more on the Linux implementations than the underlying biology that may interest a group of biologists; a course for bioinformatics students may focus more on how biological questions can be addressed computationally).
REQUIRED LEARNING TIME
Each chapter takes approximately one to four hours for students to complete (Table 2). Instructor preparation time is estimated to be double that of what it takes students to complete the lab. The three pre-tutorial chapters can either be completed in-class (if all students will require them), or outside of class (if only a few students require the remediation). We spent approximately a third of the semester working through this material. We covered roughly one chapter per class meeting, devoting one three-hour lab period to work on the tutorial, with the expectation that students read the chapter ahead of time and completed assessment questions outside of class if necessary. Students worked on and turned in answers to the assessment questions at the end of each chapter for a grade. This lab was supplemented with discussions of the primary literature (not included in this tutorial) that were related to specific projects students completed in the class.
The workflow of a standard chapter began with a brief (varying between 15 minutes and an hour, depending on whether primary literature was discussed) overview of the concepts and goals for the lesson. This introduction was generally in a discussion format without prepared slides and often centered on a primary research article including those describing the tools we were about to use (as referenced in the manual), the background material from the manual itself, and other articles relevant to the interests of the class. After the introduction, the students then worked individually and at their own pace through the tutorial. As problems and questions arose, they worked together to solve them with guidance from the instructor as needed. At the end of the class period, students compared their results, discussed any differences they noted, addressed discussion questions in groups, and wrote up answers to assessment questions individually. Some chapters require students to compare their results to those of their peers, so they are generally motivated to help make sure the work goes smoothly for everyone.
The class (Biological Science Research Methods) started with a focus on students' semester-long research project in evolutionary genomics of fishes, and moved into bioinformatics training once project-related wet-lab work was completed. A list of possible project directions derived from the instructor's research program was provided to students. Students selected projects during the first three weeks of the semester. All project options involved whole-genome shotgun sequencing of a single male rockfish from two closely related species (Sebastes chrysomelas and S. carnatus). Project goals were either to characterize functional variants at the individual SNP level between species in order to better understand the processes leading to their divergence (3), or to characterize polymorphisms between X and Y sex chromosomes (5). A reference genome was available from a closely related member of the genus (S. rubrivinctus) for alignment of short reads from which variants from the species of interest could be identified and analyzed. Individual students chose different bioinformatic approaches to pursue the question they chose. We began with three weeks of combined literature review related to the projects, and DNA isolation of rockfish samples. Once sufficient quality and quantity of DNA was confirmed, samples were shipped to the Center for Genomics and Bioinformatics at Indiana University for whole-genome sequencing. The sequencing center barcoded the two samples during sequencing library construction, and performed 75bp single-end read sequencing using a Illumina NextSeq High Output run (~30 billion bases providing about 20X nucleotide coverage per species), costing $2.6k. The tutorial was completed over the next four weeks, during which time data was returned from the sequencing center. After completion of the tutorial, students had developed sufficient skills to engage in a student-led independent research project during the remaining seven weeks of the semester. The students then worked on these projects in small groups for the remainder of the semester. The results of the research projects were reported in the form of a scientific manuscript. Making explicit from the beginning of the class the need to use data analysis tools for the project helped motivate the students to learn the data analysis skills. Students weren't just doing the lessons because they were told to, but rather to develop the skills they would need to address an authentic research project.
The NCBI short read archive (SRA) contains raw sequencing data from every published experiment that used high-throughput platforms, and could be used to identify other projects of appropriate scope and difficulty for just about any class. The first two steps of genome analysis for species lacking a closely related reference involve de novo genome assembly and gene annotation. Given the high rate of publication of new tools for assembly and annotation, instructors could have students attempt to re-assemble and/or re-annotate previously assembled genomes using new tools, and determine effects on published downstream analyses. Instructors with their own data could focus on assembly and annotation with their students, using varying methodologies. For species with very large genomes (>1Gbp), instructors may opt to complete assembly on their own, given that this task may take weeks of computational time to complete. The students would then participate in downstream applications like characterization and comparison of gene sequences to related species, additional sequencing of variants with interesting phenotypes, comparison of gene expression levels (RNAseq; 8), or epigenetic state among samples (ChIPseq).
PRE-REQUISITE STUDENT KNOWLEDGE
This tutorial focuses on both an introduction to the biology of genomic analysis, and teaching the computer competencies necessary to conduct appropriate analysis. A basic working knowledge of computers is required (e.g., how to install programs on your own computer). Since most of the actual computation is performed on super computers, only basic computers are needed to complete the tutorial. Three computer programs need to be installed on the computers used by the students: a File Transfer Protocol (FTP) application to move and view files (e.g. Cyberduck), the Integrated Genome Viewer, and a program to connect to other computers via ssh (secure shell) connections. Linux and Mac computers should already have an SSH connection program in default installations, and windows machines can connect using PuTTY (http://www.chiark.greenend.org.uk/~sgtatham/putty/download.html). No background in Linux or computer programming is assumed, since these skills are taught to the extent that they are required. It is important to note that this lesson is no substitute for an actual computer programming class.
Together with a basic working knowledge of computers, students should have a basic background knowledge of statistics and biology, such as would be gained in an introductory sequence in each discipline. The specific biology and statistics concepts necessary for the learning goals of a particular course could be taught alongside this tutorial without impeding student progress on the computer work. Students with more extensive backgrounds in biology, statistics, or computer science will find much of the content easier to understand, though a lack of that knowledge will not prevent them from completing the lessons.
PRE-REQUISITE TEACHER KNOWLEDGE
This tutorial also assumes that the instructor has some background in these fields. It is strongly recommended that the instructor using these materials be comfortable with the general concepts being taught, as well as with the requisite skills and tools (e.g., Linux and general principles of computer programming). Working through the tutorial completely may be sufficient, but reading the cited primary literature is recommended. Practical Computing for Biologists (19) is an excellent background text for instructors wishing to bolster informatics skills. The pre-tutorial includes additional "getting up to speed" suggestions. In addition, there are a number of options to develop those skills, including workshops offered by GCAT-SEEK each summer where this tutorial is used to teach faculty (http://gcat-seek.weebly.com/). Instructors may request accounts on Juniata's cluster to run through the tutorial themselves and/or with groups of students from the corresponding author.
SCIENTIFIC TEACHING THEMES
This tutorial was implemented as part of a Course-Based Undergraduate Research Experience (CURE). CURE's are emerging as an effective way to directly engage large numbers of students in the research process and networks are developing to share the resources necessary to conduct them (20). As such, this tutorial is primarily a hands-on tool, pushing students to directly interact with the data and including all members of the class in the analysis. The end goal for this tutorial was to ensure that students learn the concepts, tools, and skills needed to successfully conduct an independent research project related to genome analysis.
ACTIVE LEARNING
All students move through the tutorial at their own pace, with peer and/or instructor guidance available when needed. Additional instruction, such as lectures or discussions on the underlying biology, may be necessary. Students use the tutorial to guide their active exploration and support their learning of the primary learning goals of each chapter. In addition, students troubleshoot problems as they arise, particularly when a second student encounters similar errors. These troubleshooting steps are not intentionally inserted into the lesson, but students tend to make small errors when running the commands that cause errors until they are identified.
Also contributing to active learning, students help each other during the troubleshooting process, which appears to help both the helper and the struggling student to learn in more depth. The entire analysis, from raw sequence data to downstream interpretation, is carried out directly by the students, creating substantial buy in and individual investment. This buy-in is further increased by utilizing data for a research question that reflects the student's interest; many data sets, representing a broad variety of data, are freely available online for the students to use to answer their own questions. For example, a segment of genomic DNA pre-screened by the instructor to contain genes of interest can be used to learn annotation.
ASSESSMENT
During the tutorial phase of the class, we assessed students primarily via successful completion of the required tasks, followed by either graded end-of-chapter assessment questions or quizzes to assess student knowledge and ability. Quizzes used in the class are available (Supporting Materials S3). During the assessment, students were allowed as much time as they needed to obtain the result, and were provided with assistance as needed, since the goal was to ensure they developed sufficient skill to conduct their research project. Additional course assessments, including preparation of a formal manuscript, were structured around the students' projects. Skills students developed in the tutorial (See Table 2) were used to complete their manuscripts.
INCLUSIVE TEACHING
The successful completion of this tutorial requires every student to complete their assigned portion of the analysis, enforcing inclusion and encouraging cooperation in overcoming hurdles. The active learning required at each step of the manual, and teamwork that instructors should encourage to complete tutorials questions, may help students from varying backgrounds engage in this challenging area. The GCAT-SEEK network has recently completed faculty-development workshops at several minority-serving institutions (MSIs). We will monitor implementation and efficacy of the lesson at those institutions in comparison to non-MSIs to determine and adjust protocols to relevant challenges.
LESSON PLAN
The best preparation for teaching this tutorial is to work through it yourself on Juniata College's Linux computer cluster described in the first pre-tutorial, after obtaining an account from the primary author. Many of the common pitfalls will become apparent and the additional readings listed at the end of each tutorial chapter will prepare you for many of the questions the students are likely to raise during the class. Before working on the tutorial, contact the author to obtain access to join the GCAT-SEEK network and access Juniata's Linux computer cluster. Apart from resources that may be available at one's home institution, free public access to Linux supercomputer resources are available for education purposes, including the National Center for Genome Analysis Support and the iPlant collaborative. Contact one of those groups for more information on how to meet the specific needs of your classroom. If not using Juniata's cluster, one would need to: i) obtain access to a Linux cluster; ii) download the data and install all required software; iii) confirm that all of the steps work at your location; and iv) change any necessary steps in the tutorial to reflect your particular Linux system. Installation of software on the Linux OS for teachers not using Juniata's server is covered in general in some texts (19), and each program comes with specific instructions. Because installation on a new system and requisite changes to the manual may take days to weeks, we recommend using the Juniata's cluster for instructors just getting started, since all software is already installed and available for teaching purposes.
If not using the Juniata's computer, once you have access to a Linux computer cluster, transfer the data (Supporting File S4) to the system. Unpack the data (e.g., tar -zxvf fileName.tar.gz) and you are ready to get started. Because each Linux system is a bit different, you will need to work through the tutorial on your particular system to ensure that all of the steps work properly, particularly the installation steps and scripts detailing file locations. A sample of a completed tutorial is included in the Supporting materials (Supporting File S5). If any of these steps cause problems, please contact the GCAT-SEEK network for help. For any steps that do not work smoothly on your system, the information in the manual pages linked within the tutorial, combined with online searches, should solve the issues you may be having. It will be important for you to edit the manual to reflect the differences that you discover, because novice users (like the intended student audiences) rely heavily on very specific directions.
It is important to consider that the preparation for teaching the course will take several days or even weeks, as setting up accounts on new systems is not always an easy process (and often each student will need an account), so give yourself plenty of time. If you are planning to work through this tutorial in class, we recommend assigning the background reading from the manual and selected primary literature to students before class, and to expect both pre-tutorial and tutorial chapters to take about a lab or two to complete (in a two or three-hour lab). That should provide sufficient time for the tutorial with additional instruction in the lab and lecture periods.
TEACHING DISCUSSION
Students appeared to learn sufficient genome analysis strategies to be well prepared for summer research or graduate projects working with bioinformatics data. Students who read assigned background material from the manual and assigned primary literature before class found the material more approachable and were more productive and comfortable within class. Students whose projects were more tightly linked to the skills and content taught in the manual were better able to see the utility of the acquired skills. In spite of the ultimate positive outcomes, initial misgivings and push back are likely to be common when this tutorial is used at any level. The transition from graphical user interfaces to command line can be intimidating for many novice computer users. However, by helping students to build those skills slowly, and by reminding them why they are learning the skills, the instructor can help students gain confidence. Students in the referenced class completed the Grinnell College CURE pre- and post-reflections. Grinnell College CURE report results indicated highest learning gains of my students (N=9) compared to all students (N=8581) in the areas of science writing (+0.49 SD) and understanding that scientific assertions require supporting evidence (+ 0.4 SD). Other notable positives compared to all students included understanding how scientists work on real problems (+0.33 SD), ability to analyze data and other information (+0.24 SD), understanding the research process (+0.20 SD), and tolerance for obstacles faced in the research process (+0.1 SD). The most negative learning gain was in skill in giving oral presentations (-1.4 SD), which my students did not do. Spring 2016 was the first time the tutorials were used as the basis for a semester-long course.
Alternative Implementations
In addition to the undergraduate course, this tutorial has been used in other contexts and could be readily extended to more. One of the primary motivations of the GCAT-SEEK group, for which this tutorial was developed, is to train faculty to bring Next Generation Sequencing tools to undergraduate classrooms. To this end, this tutorial is also being used as part of the GCAT-SEEK faculty workshops. Of the faculty who attended one of the two workshops in which this set of tutorials was taught, many were amazed at how quickly they picked up the computer programming portions of the tutorial. All of the faculty have, to the best of our knowledge, continued to analyze sequence data from their own research projects. The faculty workshops are much shorter than the course, with only 18 hours of instruction time over three days, and thus necessarily provided much less information beyond the tutorial itself. As such, the focus was on learning the general process and identifying the way to analyze your own data. Faculty evaluation comments along the lines of, "I can't believe how much we've learned this week," are representative. The manual has also been used as a self-paced tutorial to train summer undergraduate researchers, followed by development of skills and concepts needed for the research project at hand.
It is also possible to work through any single chapter of this tutorial for an individual class. Each chapter can also stand on its own if an instructor wants to use just a small portion, but understanding the pre-tutorial chapters is essential background. Input files necessary to complete each chapter are provided, so it is not necessary to complete prior chapters. We also include the final output files from each chapter (Supporting File S6) for reference. The instructor could use the entire manual with existing raw datasets from the SRA archive, or just the last chapter on variant calling, if re-sequencing new individuals from a reference genome.
Changes to the tutorial: Several things about this tutorial have changed since it was initially developed. A number of chapters were replaced as we learned the challenges students faced in learning the material and the topics of highest interest to the students. Software was chosen to be approachable by the target students, while providing research-grade analysis for publication. Flowcharts of methods have been developed to help keep the reader oriented since many programs and protocols are used. If you encounter any similar hurdles, either for your own work of that of your students, please contact us so that we may update this instrument to serve as wide an audience as possible.
SUPPORTING MATERIALS
In addition to the versions archived with this publication, we will continue to update these tutorials at the GCAT-SEEK webpage (gcat-seek.weebly.com).
- S1. Eukaryotic Genome Analysis-Seven tutorial chapters, covering assembly, annotation, and variant discovery
- S2. Eukaryotic Genome Analysis-Three pre-tutorial chapters, covering topics in basic Linux, relevant molecular biology, and genome biology
- S3. Eukaryotic Genome Analysis-Quizzes accompanying pre-tutorial and tutorial chapters
- S4. Eukaryotic Genome Analysis-Data directories accompanying tutorial chapters needed to complete each tutorial chapter (this file can be accessed at http://tinyurl.com/EukaryoticGenomeAnalysis)
- S5a. Eukaryotic Genome Analysis-Answers to pre-tutorial and tutorial chapter questions
- S5b. Eukaryotic Genome Analysis-Answers to pre-tutorial and tutorial chapter questions
- S6. Eukaryotic Genome Analysis-Completed Linux directories for all tutorials (this file can be accessed at http://tinyurl.com/EukaryoticGenomeAnalysis)
ACKNOWLEDGMENTS
This work was funded by the National Science Foundation (DBI-1248096) and a grant to Juniata College from the Howard Hughes Medical Institute through the Precollege and Undergraduate Science Education Program. There are no potential conflicts of interest. We would also like to thank the other instructors at the GCAT-SEEK workshops, Mark Peterson, Arthur Hunt, Nancy Trun, Jeff Newman, Tammy Tobin, Andres Aguilar, and Regina Lamendella, for their support in development of the program and this tutorial. We extend a special thanks to Chris Walls for cheerful cluster maintenance and operations.
References
- Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. 2013. The next-generation sequencing revolution and its impact on genomics. Cell. 155:27-38
- Hoskins SG, Lopatto D, Stevens LM. 2011. The C.R.E.A.T.E. approach to primary literature shifts undergraduates' self-assessed ability to read and analyze journal articles, attitudes about science, and epistemological beliefs. CBE Life Sci Educ 10:368-378.
- Buonaccorsi VP, Narum SR, Karkoska KA, Gregory S, Deptola T, Weimer AB. 2011. Characterization of a genomic divergence island between black-and-yellow and gopher Sebastes rockfishes. Mol Ecol 20:2603-2618.
- Via S. 2009. Natural selection in action during speciation. Proc Natl Acad Sci U S A 106 Suppl :9939-9946.
- Fowler BLS, Buonaccorsi VP. 2016. Genomic characterization of sex-identification markers in Sebastes carnatus and S. chrysomelas rockfishes. Mol Ecol.
- Buonaccorsi VP, Boyle MD, Grove D, Praul C, Sakk E, Stuart A, Tobin T, Hosler J, Carney SL, Engle MJ, Overton BE, Newman JD, Pizzorno M, Powell JR, Trun N. 2011. GCAT-SEEKquence: Genome consortium for active teaching of undergraduates through increased faculty access to next-generation sequencing data. CBE Life Sci Educ 10:342-345.
- Buonaccorsi VP, Peterson M, Lamendella G, Newman J, Trun N, Tobin T, Aguilar A, Hunt A, Praul C, Grove D, Roney J, Roberts W. 2014. Vision and Change through the Genome Consortium on Active Teaching using Next-Generation Sequencing (GCAT-SEEK). CBE-Life Sciences Education. 13: 1-2.
- Peterson, M., Malloy, J., Buonaccorsi, V. and Marden, J., 2015. Teaching RNAseq at undergraduate institutions: a tutorial and R package from the Genome Consortium for Active Teaching. Couresource 2:1-8.
- Rajaravivarma R. 2005. A games-based approach for teaching the introductory programming course. ACM SIGCSE Bull 37:98-102.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30:2114-2120.
- Chikhi R, Medvedev P. 2014. Informed and automated k-mer size selection for genome assembly. Bioinformatics 30:31-37.
- Miller JR, Koren S, Sutton G. 2010. Assembly algorithms for next-generation sequencing data. Genomics. 95:315-327.
- Zimin A V., Mar?ais G, Puiu D, Roberts M, Salzberg SL, Yorke JA. 2013. The MaSuRCA genome assembler. Bioinformatics 29:2669-2677.
- Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Alvarado AS, Yandell M. 2008. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 18:188-196.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357-359.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078-2079.
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. 2012. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22:568-576.
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly. 6:80-92.
- Haddock SHD, Dunn CW. 2011. Practical computing for biologists. Sinauer Associates, Sunderland, MA.
- Auchincloss LC, Laursen SL, Branchaw JL, Eagan K, Graham M, Hanauer DI, Lawrie G, McLinn CM, Pelaez N, Rowland S, Towns M. 2014. Assessment of course-based undergraduate research experiences: a meeting report. CBE-Life Sciences Education. 13:29-40.
Article Files
Login to access supporting documents
An Introduction to Eukaryotic Genome Analysis in Non-model Species for Undergraduates: A tutorial from the Genome Consortium for(PDF | 135 KB)
S1. Eukaryotic Genome Analysis-Seven tutorial chapters covering assembly annotation and variant discovery_0.docx(DOCX | 1 MB)
S2. Eukaryotic Genome Analysis-Three pre-tutorial chapters covering topics in basic Linux relevant molecular biology and genome biology_0.docx(DOCX | 387 KB)
S3. Eukaryotic Genome Analysis-Quizzes accompanying pre-tutorial and tutorial chapters.docx(DOCX | 405 KB)
S5a. Eukaryotic Genome Analysis-Answers to pre-tutorial and tutorial chapter questions_0.docx(DOCX | 32 KB)
S5b. Eukaryotic Genome Analysis-Answers to pre-tutorial and tutorial chapter questions.pdf(PDF | 393 KB)
- License terms

Comments
Comments
There are no comments on this resource.