Meiosis: A Play in Three Acts, Starring DNA Sequence
Published online:
Abstract
Meiosis is well known for being a sticky topic that appears repeatedly in biology curricula. We observe that a typical undergraduate biology major cannot correctly identify haploid and diploid cells or explain how and why chromosomes pair before segregation. We published an interactive modeling lesson with socks to represent chromosomes and demonstrated that it could improve student understanding of ploidy (1). Here we present an improvement on that lesson, using DNA paper strips in place of socks to better demonstrate how and why crossing over facilitates proper segregation. During the lesson, student volunteers act out the roles of chromosomes while the whole class discusses key aspects of the steps. Strips of paper with DNA sequences are used to demonstrate the degrees of similarity between sister chromatids and homologous chromosomes and to prompt students to realize how and why homologous pairing must occur before cell division. We include an activity on Holliday Junctions that can be used during the main lesson, skipped, or taught as a stand-alone lesson.
Citation
Newman, D.L., and Wright, L.K. 2017. Meiosis: A Play in Three Acts, Starring DNA Sequence. CourseSource. https://doi.org/10.24918/cs.2017.9Society Learning Goals
Cell Biology
- Cell Cycle and Cell Division
- How do cells conduct, coordinate, and regulate nuclear and cell division?
Genetics
- Transmission - Patterns of Inheritance
- What are the mechanisms by which an organism’s genome is passed on to the next generation?
Lesson Learning Goals
- Students will understand how is DNA organized during meiotic cell division
- Students will know how genetic information is preserved and passed to the next generation
Lesson Learning Objectives
- Students will be able to identify sister chromatids and homologous chromosomes at different stages of meiosis.
- Students will be able to identify haploid and diploid cells, whether or not the chromosomes are replicated.
- Students will be able to explain why homologous chromosomes must pair during meiosis.
- Students will be able to relate DNA sequence similarity to chromosomal structures.
- Students will be able to identify crossing over as the key to proper pairing of homologous chromosomes during meiosis.
- Students will be able to predict the outcomes of meiosis for a particular individual or cell.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Meiosis is a standard part of any biology curriculum, but for at least two generations, researchers have noted that students have trouble with the concepts of meiosis (2-5). Widespread misconceptions have been documented with many different populations, including high school students, college students, and high school teachers (6-8). Students are typically introduced to the process in middle school, return to it in high school, and then biology majors repeat it again in multiple undergraduate courses such as Introductory Biology, Cell Biology, and Genetics. Unfortunately, based on our analysis of how meiosis is presented in textbooks, the lesson rarely changes much from one iteration to the next: students are typically presented with a series of pictures of discrete phases to remember and a lot of similar-sounding vocabulary to memorize. Meiosis is often taught alongside mitosis but remains unconnected to other biology topics. In addition, texts rarely reprise the topic of meiosis later in other chapters. Given this lack of novelty or context, it is not surprising that students do not tend to engage productively with the important topic of meiosis!
Many others have designed lessons about meiosis using pool noodles, pop beads, candy, or other manipulative materials (9-11). However, research showed that the concepts of chromosome structure and ploidy remained difficult for students to grasp. Therefore we designed an interactive modeling lesson to clarify these points, using socks to represent DNA (1). Although the lesson was effective in helping students understand the concept of ploidy and the outcomes of crossing over, we felt that the concept of homology was still unclear for many students and thought that the model could be extended to introduce students to the molecular mechanisms that drive crossing over. In the past, to explain the role of crossing over in homologous pairing, we would step away from the socks used in the original model to show a slide with double-stranded DNA sequences lined up and recombining. When many students expressed surprise that homologous chromosomes would be so similar at the DNA sequence level, we realized that most textbook drawings and descriptions about homologous chromosomes emphasize their differences rather than their similarity (12). For example, visual representations often show maternal chromosomes as red structures and "homologous" paternal chromosomes as blue structures without any attempt to illustrate the underlying DNA sequence on any of the chromosomes. In-class and out-of-class discussions with students consistently revealed that students don't really know the meaning of the word homology and all that it implies in biology. Students can come up with the phrase "homologous chromosomes" but cannot connect that term with conceptual understanding about the underlying history and relationship of homologous chromosomes. To address some of these problems, we redesigned the "meiosis with socks" lesson that emphasizes the nature of chromosomes at the DNA level. The new model replaces socks with long strips of paper containing DNA sequences, and removes obvious visual clues as to which pairs of chromosomes are homologous. Sister chromatids are depicted as duplicated strips of DNA sequence, and homologous chromosomes are approximately 99% identical at the DNA sequence level. Although the exact amount of similarity between homologous chromosomes will vary depending on the species, we wished to emphasize the fact that homologous chromosomes are almost identical at the sequence level. For example, the International SNP Map Working Group estimated that on average the haploid human genome contains 1 SNP per 1331 bp (13), which means that homologous chromosomes would be > 99.9% identical (not counting other types of variation, such as CNVs and rearrangements).
Through scaffolding by instructor questions, students are led to construct an explanation of homologous chromosomes that acknowledges the important aspect of DNA sequence. The main "eureka moment" occurs when students carrying the strips of paper realize that they can find their homologous mate by examining the sequence itself, and that sequence similarity will allow them to pair properly and segregate without a loss of information.
The lesson described here was first used with an introductory biology class for majors (100-level) and later with a cell and molecular biology class (200-level). In the intro class, we evaluated the effectiveness of the lesson with pre/post testing using selected questions from the Meiosis Concept Inventory (4).
INTENDED AUDIENCE
We taught this lesson at a large, private 4-year institution in two different courses for students with biology-related majors: freshmen in an introductory biology class and sophomores/juniors in a cell and molecular biology class. This lesson was carried out in classes of 20-50 students, and an earlier version was used successfully in a class of about 100 students. In a class with far more students (e.g., hundreds), the instructor may choose to handle this activity as a clicker-based demonstration. Alternatively, they may choose to run it in a lab or recitation section rather than in the lecture.
REQUIRED LEARNING TIME
The lesson can be completed in a 50-minute class or in a 75-minute class. In a 50-minute version, you will probably not be able to model the entire process, but will be able to complete the most important parts (chromosome structure, homology, and ploidy). In a 75-minute class, the activity on Holliday Junctions can be included. If desired, the Holliday Junction activity can also be done in a different class period.
PRE-REQUISITE STUDENT KNOWLEDGE
Students should have a basic understanding of DNA structure and base pairing as well as a familiarity with chromosome structure (e.g., be able to recognize a centromere and know that each chromatid contains a single DNA double helix) and the concept that chromosomes segregate during cell division. They should also know how sexual reproduction works at the cellular level (i.e., meiosis to form gametes, fertilization pairing male and female gamete to form a zygote).
PRE-REQUISITE TEACHER KNOWLEDGE
Instructors should have a solid understanding of the processes of meiosis and crossing over and be able to mentally move between models of chromosomes at various levels (molecular, microscopic, and genetic).
SCIENTIFIC TEACHING THEMES
ACTIVE LEARNING
A subset of students act out the parts of chromosomes during meiosis, and the entire class is engaged in group discussion and comparison of DNA sequences. Clickers may be used, especially with larger classes, but are not necessary because the instructor is consistently asking for feedback from the student "audience." We have successfully implemented this lesson with and without clickers, so instructor preference can guide the decision about clicker use.
ASSESSMENT
We employed questions from the Meiosis Concept Inventory (4) both pre- and post-lesson to measure student achievement of our learning objectives. Other related questions were included on exams.
INCLUSIVE TEACHING
Some students are involved in acting and kinesthetic learning using the large paper ("demonstration") strips of chromosomes. Others are observers who can see the big picture in a way that individualactors may not. In a large classroom, smaller versions of the chromosomes ("audience strips") can be printed on standard 8.5 x11" paper so that all students have models to manipulate. All students are encouraged to use reasoning skills and help each other make connections with material learned in a different context.
LESSON PLAN
OVERVIEW
In this lesson, students are recruited to play the parts of chromosomes in a cell undergoing meiosis in three acts: 1) Identifying Homology; 2) Crossing Over; and 3) Cell Division. Instead of focusing on visually recognizable cellular structures or the phase names, this activity focuses on the behavior of chromosomes during the process. Table 1 lists specific concepts and explains how they are addressed in the activity. Supporting Table 1 provides a timeline for the instructor. Nuclear and cell membranes are not part of the model; it is sufficient to indicate areas of the room as different "cells." If students mention the dissolution of the nuclear membrane, the instructor should agree that it does occur but redirect the focus to chromosomes. The instructor can also say that nuclear breakdown is not essential. For example, the nuclear envelope remains intact in fungi. Experienced students may also talk about the names of phases. If so, the instructor can confirm/correct/provide information, but should point out that the phase names are artificial constructs, i.e. names placed on somewhat arbitrary segments of a continuous process.
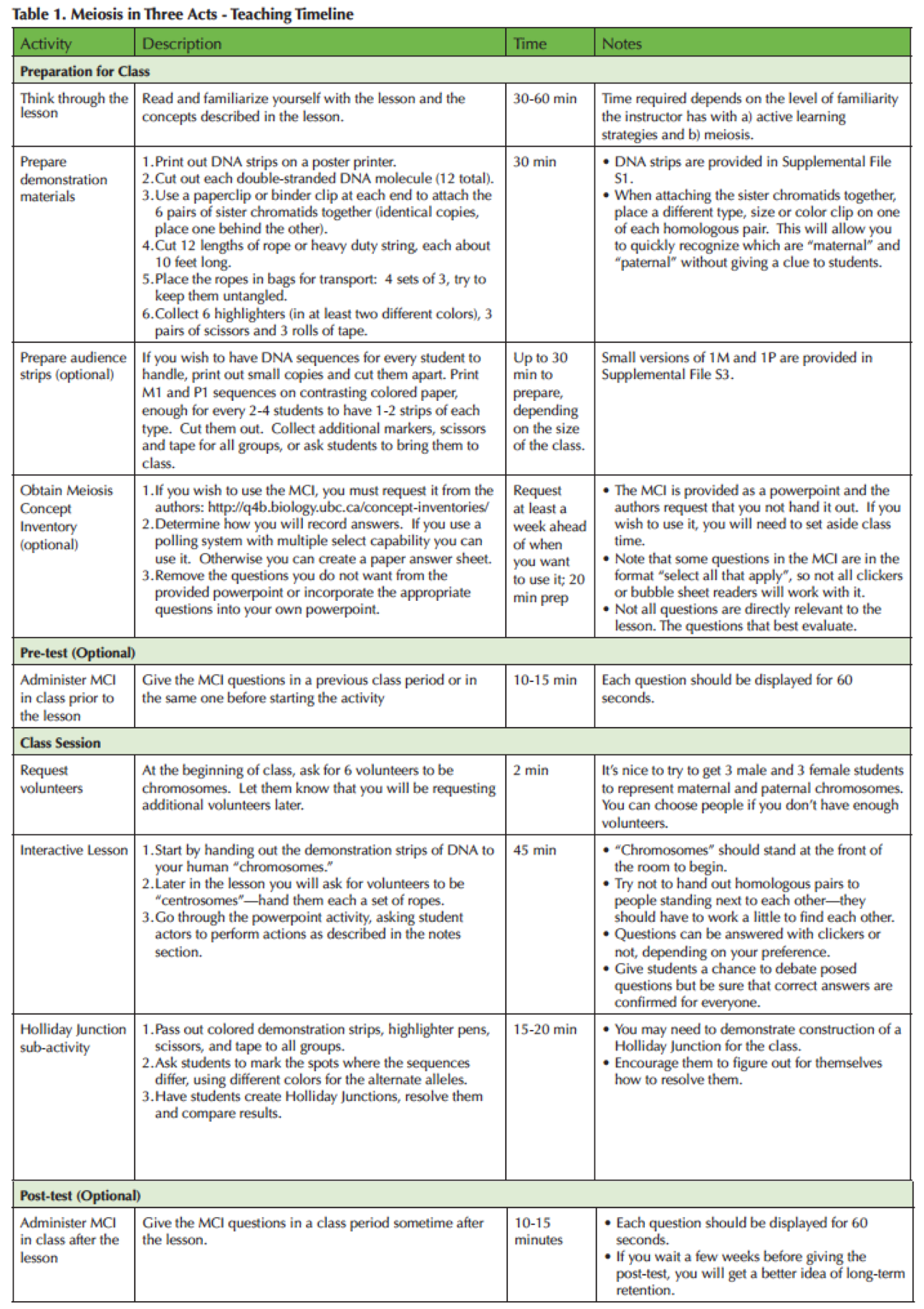
Table 1. Meiosis in Three Act-Teaching Timeline
The starting cell contains six chromosomes (three pairs of homologues). This number was chosen deliberately; many textbook illustrations show cells with only two or four chromosomes, which allows students to arrive at four daughter cells by simply dividing chromosome structures in half twice. In our education research, we find that many incorrect ideas about the process will be revealed when students are faced with a diploid cell containing six chromosomes instead of two or four (1,8). They can't simply divide six chromosomes twice, since that division results in 1.5 chromosomes. If students start with four chromosomes, they can end up with a haploid cell that looks right, even though they incorrectly understand the process. The "divide by two" strategy is entrenched: when faced with six chromosomes, students who misunderstand the process sometimes drop whole chromosomes or fuse them to make the product match their expectations.
The DNA sequences are designed so that reading the English letters left to right is the same as moving 5' to 3' along the DNA molecule. The two strands are positioned in antiparallel fashion (one "upside down" compared to the other), with short lines representing the hydrogen bonds between bases. This lack of a clear "top" and "bottom" may at times cause students to conclude that they have no matching sequence when in fact they are just looking at the wrong strand, an important lesson about the nature of DNA.
To simulate the random nature of molecular movement, students are encouraged to mill around unless held in place during the exercise.
We have also added an activity on Holliday Junctions. This activity can be skipped if you do not have time or do not want to go into this level of detail with your class.
PRE-CLASS PREPARATION
PREPARE THE DEMONSTRATION MATERIALS
1. Large ("Demonstration") Strips of DNA Sequences representing the six chromosomes: As described in Table 1, DNA sequences must be printed out, cut apart, and sorted, as explained below. Supporting File S1 contains twelve double-stranded DNA sequences, intended to be printed on poster paper (42"x30"). If you do not have access to a large format printer, you can use smaller printouts. Although audience members will not be able to read the individual sequences on either poster-sized or smaller paper from their seats, the larger version of the sequences will be more obvious to the audience and thus will better reinforce the idea of the underlying structure of a chromosome as a chain of base pairs. Cut each sequence along the dotted lines. Labels are for your use only; do not include them on the strips of paper because the labels can give away information to the students that will circumvent the lesson. Attach each pair of identical sequence (e.g., the strips designated 1M) with paperclips at each end, so that one strip is hidden behind the other. These strips are the props given to students at the start of the lesson. Each chromosome also has a nearly identical homologue. Identify the three homologous pairs (1M goes with 1P, etc.). All three chromosomes are about the same length so that students do not try to use visual differences to identify pairs. Note that one pair differs slightly in length due to a deletion of two base pairs on one chromosome compared to its homologue. Identify one chromosome from each homologous pair in a subtle way, such as using a different kind of paperclip. This strategy will allow you to differentiate between maternal and paternal chromosomes during the lesson without giving a clue about pairing to the students (e.g., we have successfully used plain steel paperclips on all P chromosomes and colored plastic-coated ones on all M chromosomes). Shuffle the strips so that you do not hand them out in order during class, which can result in all people standing next to their homologue from the beginning.
2. Spindle Fibers: Prepare the spindle fibers by cutting 12 lengths of rope or twine to 10 feet long. Coil four sets of three ropes together and place each in a large baggie. It will slow down the lesson if the ropes need to be untangled during class.
3. Tools for Showing Crossing Over: Prepare three sets of the following: 1. Two highlighters of different colors, so that maternal and paternal chromosome differences can be marked; 2. A pair of scissors to cut the DNA sequences, simulating recombination; 3. A roll of clear tape, to attach the recombinant chromosomes together. Place each set of materials in a baggie for ease of distribution. The audience in a large class will also need these crossing over materials (see below.)
PREPARE THE AUDIENCE MATERIALS
1. Smaller DNA Sequence Strips: If the class is larger than 24 students, prepare small DNA strips, printed on 8 x 11" paper, so that everyone has a chance to examine DNA sequence. As you did for the poster-sized "demonstration strips," print out, cut, and organize strips from Supporting file S2 so that every two to four students will have their own set. For these "audience strips," use different colored paper for the maternal and paternal versions. It is important to use two different colors (even if one is white) because it helps students visualize the crossover products. The file contains numerous copies of the DNA sequence for maternal chromosome 1 (1M) and paternal chromosome 1 (1P); audience members will only work with that set.
2. Tools for Showing Crossing Over: Prepare a baggie with the crossing over tools (two highlighters, scissors, tape) for every two to four students in the audience. They can share the materials as needed. Alternatively, students can be asked to bring these materials to class.
INSTRUCTOR PREPARATION
Review the Presentation File (Supporting File 3) and adapt if necessary to your own specifications. You may also choose to run the lesson without projected slides, if that is your style. The presentation is provided to guide what you should be talking about at each point in time.
Test out the Holliday Junction activity and make sample recombinant chromosomes as models, in case they are needed. Photos showing the strips before, during, and after recombination are shown in Supporting File 4.
DETAILED ORDER OF EVENTS
Supporting Table 3 provides a resource for instructors to help them align the actions in the play and the slides in the presentation (Supporting File 3), with the phases of meiosis in a series of traditional images.
ACT 1: IDENTIFYING HOMOLOGY
The instructor calls for six actors to act as chromosomes in the play. Try to choose three men and three women without being obvious about it. (Part of the lesson is the surprise in discovering that they will pair as male and female.) Each student volunteer is given a strip of DNA sequence. If possible, give all maternal "M" chromosomes to women and all paternal "P" chromosomes to men. The instructor must be explicit about the students representing chromosomes. Inform the class that this group of students represents a diploid cell and ask for "n," the number of chromosomes that contains one complete set of genetic information. This is a good time to bring up the idea that n is a characteristic of a species, and therefore should not change at all during the exercise; the cell may go from 2n to n (or other multiples of n in some species), but the value of n remains constant for that species.
The class is asked to debate what should happen first if this cell will undergo meiosis. Once the class decides that DNA replication must occur, the student "chromosomes" are asked to replicate their DNA. They will probably have already noticed that they actually have two strips clipped together and will naturally pull the second strip of DNA from behind the first (Figure 1A). Ask the "chromosomes" to compare their two sequences. They should notice that the two sequences of each chromosome are 100% identical. (If a student "chromosome" says that their sequences are different, ask the student to try turning one strip upside down; the student may be comparing the wrong strands.)

Figure 1. Students engaged in the interactive meiosis activity. A. At the beginning of the lesson, six volunteer “chromosomes” go through “DNA replication” so that each “chromosome” now contains two copies of identical DNA sequences.
Next, the class is asked to identify the number of chromosomes now present in the cell, either by clicker or by shouting it out or by a show of hands. Many students will say twelve, a common mistake. The instructor points out that the students, not the strips of paper are the chromosomes. The instructor asks what the strips represent. Someone will likely come up with the correct answer: the DNA in sister chromatids. This distinction can be discussed further and connected to the concept of ploidy: although the amount of DNA doubled, DNA replication did not add any new information content to the cell. When asked again how many chromosomes are present, students will revise their answer to six. The instructor can point out now that we can differentiate types of chromosome: unreplicated and replicated. Both types are considered chromosomes and contain the same information, but one has two copies of the DNA helix instead of one.
Now, by clicker or soliciting suggestions, the instructor asks for the next step. Often students will suggest "lining up" in the center of the cell, and "chromosomes" may move to the center without pairing. The instructor should stop them and ask how the chromosomes "know" where to go and how they "know" their pairing partner. With some Socratic dialogue, students will eventually realize that chromosomes must find their homologous mate first and that their mate is identifiable because of shared DNA sequence. Allow students to compare sequences to find out which chromosomes are most similar (Figure 1B). Give them highlighter pens to highlight any differences between homologous chromosomes, marking the sequences that differ on both maternal and paternal chromosome, with the maternal highlighted in one color and the paternal chromosome in another. Make sure students mark both strands of the DNA, not just one (Figure 1C). They will find very few differences, leading to the realization that homologous chromosomes are extremely similar at the DNA sequence level (see Supporting File 1 and Supporting File 4, Figure S1). The chromosome that contains a deletion compared to its homologue often causes trouble at this point. Students may think that the chromosomes are completely different after the insertion/deletion, so be sure to lead them to discover that the chromosomes are the same from both ends with only a small change in the middle. In a class with more than 24 students, the instructor can distribute smaller "audience strips" of M1 and P1 sequence to groups of students (Supporting File 2) at this time so that all students can examine the DNA sequences.

Figure 1. Students engaged in the interactive meiosis activity. B. Students try to find their “homologues” by comparing DNA sequences.

Figure 1. Students engaged in the interactive meiosis activity. C. Once the “homologous pairs” have formed, students examine their DNA sequences more closely for sequence differences.
ACT 2: CROSSING OVER
The students are now asked how this comparison of DNA sequences would happen molecularly. In other words, how do strands of DNA find other strands that share sequences with them? This question will lead to a discussion of the mechanism of crossing over. Depending on the class and the instructor's learning goals, crossing over can be explained in very general terms as DNA molecules hybridizing their sequences with each other, or in detail including strand invasion and repair.
If you want to use the Holliday Junction exercise, explain or demonstrate to students how to form the four-strand structure. If the class is small, all students can gather around the three large pairs of chromosomes to help. If the class is large, groups of students can work with the audience strips (Supporting File 2) to allow everyone to model the process. Students will cut through the hydrogen bonds in the centers of two homologous chromosome strips, and tape them so that the opposite chromosomes are now paired (see Supporting File 4, Figure S2). It is important to remind students that DNA does not begin unwinding from the ends of actual linear chromosomes, so opening the structure in the middle of the model is more realistic. Make sure students cut the hydrogen bonds between the areas of sequences differences between the two homologues. Students will then resolve the structures by cutting through the backbone of one strand on each molecule and "ligating" (taping) it closed (see Supporting File 4, Figure S3). Depending on how the students resolve the junctions, they will create either recombinant or non-recombinant outcomes (see Supporting File 4, Figure S4). Once the recombination process is complete, ask the students to hold up their two final chromosomes. They should notice that some students have chromosomes with the same original alleles and some will have chromosomes with swapped alleles. (If not, you can show the demonstration models that you made before class.) Ask them how this happened; the answer is, "it depends on where you made your cuts." It is important for the instructor to point out that although, in both cases, portions of the maternal and paternal strands do change places, the genetic information does not necessarily have to change. The instructor may also want to ask students about how the choice of location of the event affected their outcomes. If the crossing over had not occurred in a region between two mutations, there would not be any new combinations of alleles. This realization may help students overcome the common misunderstanding that crossing over is only for the purpose of producing genetic diversity.
Once homologues have found each other and completed crossing over, the students (in the front of the room) are asked to link arms to show that they are synapsed (Figure 1D). They are encouraged to continue to mill around as a pair. If the instructor has handed out DNA strips according to gender, the pairs should each consist of a male and a female. The analogy to maternal and paternal chromosomes is obvious and should be discussed.

Figure 1. Students engaged in the interactive meiosis activity. D. Each synapsed homologous pair consists of a “maternal” and a “paternal” chromosome, visualized as female and male students with arms linked. The allelic differences have already been marked on the DNA strips at this point.
ACT 3: CELL DIVISION
Two new volunteers are recruited to act as spindle organizing centers, or centrosomes. They are each given three lengths of rope ("spindle fibers") and instructed to attempt to "lasso" each pair of synapsed homologous chromosomes. "Chromosomes" will help out by grabbing the rope as it is tossed at them, but each individual must remain linked to their homologue and can only hold one rope at a time. Once an attachment is made, the "centrosome" should pull the "chromosome" toward them. Since the pairs are linked, tugging from both sides will result in the pairs "lining up" in the center. Pairs are instructed to unlink arms at a signal from the instructor (e.g., a clap), which represents a molecular cell cycle signal that causes the cohesins to release. Since the "centrosomes" are continuing to pull on the "spindles," the homologous pairs will separate, with one moving to each side of the cell. An imaginary line is drawn between the two groups of students to suggest cytokinesis and the creation of two cells.
Students are now asked to count chromosomes again. They should identify three chromosomes per cell and should note that each chromosome is still composed of two sister chromatids. The connection with ploidy is again made. Comparing sequences will show that each chromosome in the new cell holds completely different information, and this is what makes the cell haploid.
Another pair of "centrosomes" is recruited and given a set of ropes to simulate the next, mitosis-like division. This time, each "chromosome" holds two ropes, one from each "centrosome." Spindle fiber tension will again line up the "chromosomes" in the center. The instructor now points out that the model would require tearing students apart to make one replicated chromosome into two unreplicated chromosomes. Since that is not possible, additional students are recruited to stand behind the original "chromosomes" and act as new chromosomes when the sister chromatids separate. Chromosomes are counted in the final configuration (three chromosomes in each of the four cells). The genetic content is also compared: while all cells contain a copy of all information, no two cells are exactly the same due to crossing over.
ASSESSMENT
To gauge the student learning, we tested the students before and after the activity with nine questions from the Meiosis Concept Inventory (MCI) (4). With no other instruction on meiosis or mitosis in the setting that was tested, students improved from an average of 26% to 42% correct (n = 20, p = 0.003). Note that the MCI does not address DNA sequence directly. We also included questions about meiosis on the final exam (see Supporting File 5). Some of the questions go beyond the lesson itself, but they can also provide inspiration for further discussion in class or as homework assignments. Another idea for assessment is having the students create a short video to explain the process of meiosis or how meiosis can go wrong (e.g., nondisjunction leading to Down syndrome). One student from an introductory biology class used this exercise as inspiration for a video that fulfilled a class requirement that could have focused on a variety of different topics (see Supporting file 6).
TEACHING DISCUSSION
In our experience, students enjoy participating in the "skit" and getting to work with physical models. We find that many students are making important conceptual connections throughout the activity. The main lesson is especially useful for helping students understand the relationships between sister chromatids and homologues and the basic idea of homologous pairing. Most of our students have never considered before how homologous pairing occurs; they just know that it happens and lively discussion usually accompanies this activity. It also helps them remember the order in which events must occur. The Holliday Junction activity is good for a more in-depth understanding of homologous pairing, and clarifying how genetic recombination occurs. Again, this process is usually familiar to students, but only as a black box; they have never known how it happens.
In our experience, students made strong gains on the MCI questions that relate to chromosome structure, homology, and the mechanism of meiosis. They were also better able to answer novel questions about recombination, such as reasoning out how non-meiotic recombination events might take place to form translocated chromosomes.
SUPPORTING MATERIALS
- S1. Meiosis in Three Acts-Large version of DNA Sequences. This file contains 12 dsDNA sequences to be printed as model chromosomes on a poster printer (42" x 30" paper).
- S2. Meiosis in Three Acts-Small version of DNA sequences. This file contains multiple copies of the 1M and 1P dsDNA sequences to be printed on 8.5" x 11" paper. Use of this material is optional but will allow all students in a larger class to participate in comparing DNA sequences and performing the Holliday Junction activity.
- S3. Meiosis in Three Acts-PowerPoint file that can be used during the interactive lesson. Instructions for the instructor are provided in the notes box below each slide.
- S4. Meiosis in Three Acts-PowerPoint file containing Supporting Figures 1-4 that detail the Holliday Junction sub-activity.
- S5. Meiosis in Three Acts-Suggested assessment questions.
- S6. Meiosis in Three Acts-Student-created video to explain meiosis after experiencing the lesson.
ACKNOWLEDGMENTS
We thank Pam Kalas for providing the Meiosis Concept Inventory; Jordan Cardenas, Katie Palmer, Brianna Santangelo, Zack Santos, Megan Stefkovich, and Nick Young for posing as student participants in the photos; Hanna Berga and Ryan Hertzell for creating and providing their video.
References
- Wright LK, Newman DL. 2011. An interactive modeling lesson increases students' understanding of ploidy during meiosis. Biochem Mol Biol Educ 39:344-351.
- Kindfield ACH. 1994. Understanding a basic biological process: Expert and novice models of meiosis. Sci Educ 78:255-283.
- Kindfield ACH. 1991. Confusing Chromosome Number and Structure: A Common Student Error. J Biol Educ 25:193-200.
- Kalas P, O'Neill A, Pollack C, Birol G. 2013. Development of a Meiosis Concept Inventory. CBE Life Sci Educ 12:655-664.
- Brown CR. 1990. Some Misconceptions in Meiosis Shown by Students Responding to an Advanced Level Practical Examination Question in Biology. J Biol Educ 24:182-86.
- Stewart J, Dale M. 1989. High school students' understanding of chromosome/gene behavior during meiosis. Sci Educ 73:501-521.
- Dikmenli M. 2010. Misconceptions of cell division held by student teachers in biology: A drawing analysis. Sci Res Essay 5:235-247.
- Newman DL, Catavero C, Wright LK. 2012. Students Fail to Transfer Knowledge of Chromosome Structure to Topics Pertaining to Cell Division. CBE-Life Sci Educ 11:425-436.
- Locke J, McDermid HE. 2005. Using Pool Noodles to Teach Mitosis and Meiosis. Genetics 170:5-6.
- Wells RF. 1982. Simple Chromosome Models. Am Biol Teach 44:311-312.
- Mary F. Durham. 2015. Demonstrating Meiosis using Manipulatable Chromosomes and Cells. Genet Soc Am Peer-Rev Educ Portal.
- Wright LK, Catavero C, Newman DL. 2017. The DNA Triangle and Its Application to Meiosis.
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, Hunt SE, Cole CG, Coggill PC, Rice CM, Ning Z, Rogers J, Bentley DR, Kwok P-Y, Mardis ER, Yeh RT, Schultz B, Cook L, Davenport R, Dante M, Fulton L, Hillier L, Waterston RH, McPherson JD, Gilman B, Schaffner S, Etten WJV, Reich D, Higgins J, Daly MJ, Blumenstiel B, Baldwin J, Stange-Thomann N, Zody MC, Linton L, Lander ES, Altshuler D. 2001. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409:928-933.
Article Files
Login to access supporting documents
Meiosis: A Play in Three Acts, Starring DNA Sequence(PDF | 982 KB)
S1. Meiosis in Three Acts-Large version of DNA Sequences.pdf(PDF | 202 KB)
S2. Meiosis in Three Acts-Small version of DNA sequences.pdf(PDF | 205 KB)
S3. Meiosis in Three Acts-PowerPoint file that can be used during the interactive lesson.pptx(PPTX | 306 KB)
S4. Meiosis in Three Acts-PowerPoint file containing Supporting Figures 1-4 that detail the Holliday Junction sub-activity.pptx(PPTX | 69 MB)
S5. Meiosis in Three Acts-Suggested assessment questions.docx(DOCX | 268 KB)
S6. Meiosis in Three Acts-Student-created video to explain meiosis after experiencing the lesson.mp4(MP4 | 34 MB)
Supporting Table 1. Meiosis in Three Acts-Conceptual Design of the Lesson .pdf(PDF | 32 KB)
Supporting Table 3. Meiosis in Three Acts-Alignment of meiotic phases with chromosome number ploidy and slide number in the presentation for the lesson.pdf(PDF | 101 KB)
- License terms

Comments
Comments
1 Like
Michael Gilchrist @ on (Edited: @ on )
1. My TAs just tried this in 6 sections of a class of 140 general genetics students and it went very well.
2. I modified the materials a bit to better fit in the material I am covering.
3. We didn't assign the students any material related to it before hand as I had just lectured on the topic. Instead, TAs showed this short video: https://www.youtube.com/watch?v=kQu6Yfrr6j0
Copy link Report abuse
0 Like
Kristina Marie Whittle @ on
The files cannot be downloaded. There is no link. :(
Copy link Report abuse
0 Like
Drew LaMar @ on
Hi, Kristina. If you become a member of CourseSource (by clicking this link) you will be able to download the supporting files.
Copy link Report abuse