Linking Genotype to Phenotype: The Effect of a Mutation in Gibberellic Acid Production on Plant Germination
Published online:
Abstract
Basic concepts in genetics and plant development are often taught in lecture courses without a lab component. This approach provides students with the content knowledge, but fails to make connections to research opportunities and real-world applications of the science. To help address this deficiency, we developed a hands-on activity demonstrating the effect of the plant hormone gibberellic acid (GA) on germination, and tested it within the undergraduate course lecture setting. The activity takes advantage of a mutation in a GA biosynthesis gene, demonstrating the requirement of this important hormone for normal plant development. In this exercise, students investigate the effects of a GA supplement on the germination rate of GA-producing wild-type seeds versus GA-deficient mutant seeds. Based on their observations, students are able to identify the genotype of an unknown seed sample. The simple design allows students to learn about the importance of using controls, the difference between wild type and mutant, and the phases of plant germination including the role of GA. The teachers can also use student responses to the assignments to determine and clarify common misconceptions students have developed as a result of insufficient prior knowledge. By helping students make connections between different concepts within the covered material, this lesson enables teachers to accomplish higher level learning goals, thus complementing and adding value to the lectures.
Citation
Mann, J.W., Larson, J., Pomeranz, M., Knee, E.M., Shin, D., Miller, J.A., Price, C.G., Crist, D.K., Grotewold, E., and Brkljacic, J. 2017. Linking Genotype to Phenotype: The effect of a mutation in gibberellic acid production on plant germination. CourseSource. https://doi.org/10.24918/cs.2017.18Society Learning Goals
Genetics
- Molecular biology of gene function
- How is genetic information expressed so it affects an organism's structure and function?
- Genetic variation
- How do different types of mutations affect genes and the corresponding mRNAs and proteins?
Plant Biology
- Evolution, Natural Selection and Adaptation
- How does variation among plants affect survival and reproduction?
- Plant Genetics and Variation
- Why do individuals of the same species vary in how they look and behave?
- Plant Growth and Reproduction
- How do plants grow and develop?
Lesson Learning Goals
Students will:- understand the differences between wild-type and mutant organisms.
- understand the importance of hormones for development.
- understand the effects of mutations on the phenotype of an organism.
- value the use of model organisms for elucidating biological mechanisms.
- appreciate the importance of using controls.
- understand the concepts of genotype and phenotype.
- understand how genotype affects phenotype.
Lesson Learning Objectives
Students will be able to:- identify when germination occurs.
- score germination in the presence and absence of GA to construct graphs of collated class data of wild-type and mutant specimens.
- identify the genotype of an unknown sample based on the analysis of their graphical data.
- organize data and perform quantitative data analysis.
- explain the importance of GA for plant germination.
- connect the inheritance of a mutation with the observed phenotype.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Plants play an essential, albeit often overlooked, role in our daily lives. They serve as the foundation of our food system, provide valuable environmental services, and contribute to energy production. Despite serving these roles, plants are underappreciated in much of society, a phenomenon known as "plant blindness" (1). In biology courses, the developmental processes and genetic concepts behind the growth and development of organisms, including plants, are often perceived as difficult to understand and teach. Additionally, genetics has become a controversial and sometimes sensationalized topic of discussion in our society, leading to a wide variety of misconceptions about mutations and natural variation. Hands-on exercises demonstrating a link between genotype and phenotype often require the understanding and use of molecular techniques (2,3), which can be challenging. To address these concerns simultaneously, and to complement the biology curriculum, we designed a simple plant-based hands-on exercise focused on the relationship between genes, mutations, and phenotype. This exercise was incorporated into the curriculum, taking advantage of a model plant Arabidopsis thaliana (Arabidopsis).
Arabidopsis is a weedy plant species native to Europe and Asia (4) that has become an essential tool in plant science (5,6). Its small size, short growing cycle, self-fertilization, and small genome have made Arabidopsis an ideal choice as a model organism for plant research (6). These characteristics, along with its relatively low-maintenance growing requirements, make Arabidopsis a good choice for use in K-12 and college-level classrooms. While Arabidopsis resources are easily accessible to researchers via multiple stock centers, the Arabidopsis Biological Resource Center (ABRC) has taken a lead to design hands-on teaching lessons and make this plant more readily approachable for educators as well.
Like any other plant, Arabidopsis seeds require a combination of the right conditions to germinate. Temperature, light, and water are the most important environmental stimuli influencing germination. Additionally, complex chemical processes within the plant interact with these external factors to control germination. Gibberellic acid (GA) is a plant hormone required for seed germination. GA is released by the seed embryo and diffuses into the aleurone layer (the outermost cell layer of the endosperm) signaling the metabolism of lipids, which are utilized by the embryo for growth and germination (7). GA also triggers a vacuolation process, weakening the aleurone cells around the radicle (embryo root) tip allowing it to more easily break through the seed coat (8). If an Arabidopsis plant is unable to produce its own GA, for example because of a mutation in a GA biosynthetic gene, then GA must be added to initiate germination (9).
To demonstrate the requirement of GA for germination, we selected two different seed strains, a Landsberg erecta (Ler-0) (referred to here as the wild type) and a ga1-2 mutant. The mutant strain was generated from wild type Ler-0 plants by mutagenesis and contains a mutation that disrupts the function of the GA1 gene (10). Since the GA1 gene product is one of the essential enzymes in GA biosynthesis (Supporting Material S1), this mutation eliminates the plant's ability to synthesize GA (11,12). This lesson only explores the effect of the ga1-2 mutation on germination, although notably the mutation causes other developmental consequences such as dwarfism (8) (Figure 1). The effects of GA on plant height have been described extensively by others (13,14).
Figure 1. Wild type (Ler-0) and mutant (ga1-2) Arabidopsis thaliana plants shown 27 days after planting. Mutant seeds were soaked in 200 µM GA solution for 3 days to initiate germination. No GA was added during subsequent growth. The plant containing the mutation exhibits a severe dwarf phenotype.
We tested this hands-on exercise and established that it worked remarkably well at the introductory biology undergraduate course level. At the beginning of the activity, students planted wild type, mutant, and an unknown strain of Arabidopsis on petri dishes containing either water or GA. Before recording germination data, they predicted the outcome of the experiment based on their current understanding of the topics. They followed germination rates over a period of five days and recorded their observations, after which they graphed and analyzed class average results. Based on this analysis, the class was able to contrast the germination rates of the wild-type and mutant seeds, to deduce the genotype of the unknown seed sample, and to compare their germination predictions with the obtained results. At the end of the exercise, we assigned homework to help students make connections between genotype and phenotype, as well as synthesize knowledge related to concepts such as photosynthesis, Mendelian genetics, gene expression, and gene regulation.
A number of recent research studies demonstrate that the basic molecular machineries controlling a plant and an animal organism are much more similar than previously thought. Therefore, plants represent excellent models to study both genetic and epigenetic control of gene expression which in turn drives the differentiation and development of both types of organisms. The examples of successful application of the knowledge obtained on plants and applied to animals are numerous and range from the transduction of and response to chemical signals (including hormones), to gene silencing and chromatin modifications-directed stem cell programs (15,16,17). Consequently, the conclusions students make studying a mutant in a plant hormone biosynthesis can be easily extrapolated to mutations affecting processes regulated by hormones in animals, including humans. After completing this exercise, students are equipped with enhanced understanding necessary tomake the connection between genetics and human health and disease.
INTENDED AUDIENCE
This lesson is intended for introductory undergraduate biology and plant biology courses. Although originally designed for high school biology, feedback from college instructors who used the lesson demonstrated it is a valuable resource for the college classroom as well. Regardless of student level, the activity remains the same, but the other material in this lesson can be easily adjusted to include more complex treatment of the subject matter. Included in the supplements are the assignments used in two separate introductory biology courses at a large land-grant research university. One of the courses, "Biological Sciences: Form, Function, Diversity, and Ecology," focused on evolution and diversity (Evolution), and the other, "Biological Sciences: Energy Transfer and Development," focused on energy transformation and genetics (Genetics). Both courses consisted primarily of undergraduate science majors with students ranging from freshmen to seniors. Due to the large class size, the student population varied widely in their science background and level of understanding, giving us the opportunity to apply the activity successfully to varying student populations. We designed this activity with the flexibility to be adapted to both upper- and lower-level courses.
REQUIRED LEARNING TIME
The experiment requires time in four different class periods. The experimental setup takes place during the first class period and requires 40-45 minutes. The other three periods require approximately 10 minutes each to collect, report, and record data as a class. The remainder of the project requires students to complete homework assignments, which we collect in a later class period.
PRE-REQUISITE STUDENT KNOWLEDGE
The procedure involved in setting up the experiment is simple and does not require students to have any background knowledge. All instructions for completing the activity are provided to the students in PowerPoint slides shown during the class period (Supporting Material S2, S3), as well as in a handout posted online or distributed prior to the start of class (Supporting Material S4). There are two versions of the post-experiment homework assignment, one for each course (Supporting Material S5). Although no pre-requisite knowledge is required for completing the experiment, some prior knowledge is important for completing the homework assignments. In the Genetics Course, students should be given an introduction to cell communication (including hormone signaling) prior to setting up the experiment to make the use of GA more meaningful. Before completion of Part 1 of the assignment, students should have learned about photosynthesis, Mendelian Inheritance including inheritance of dominant and recessive traits, the definition of a wild-type strain, the difference between genotype and phenotype, how to calculate the probability of a specific genotype or phenotype from a cross, and the use of a test cross to determine the genotype of an unknown. Before completing Part 2 of the assignment (Genetics Course), students should additionally have learned about the levels of regulation of gene expression in eukaryotes and how gene expression affects phenotypes. In order to complete the homework assignment in the Evolution Course, students should know basic plant biology including the differences between vascular and nonvascular plants, be able to differentiate between an angiosperm and a gymnosperm, the basics of plant germination, and where plant biomass comes from. Both versions of the assignment require the students to construct and interpret a graph of the collated class data.
PRE-REQUISITE TEACHER KNOWLEDGE
All instructions for this activity are included and do not require any pre-requisite knowledge from the teacher to implement in the classroom. A basic understanding of GA and its importance for plant germination is required in order to explain the experiment to the students. To use the homework assignments as presented the teacher would need to know the same concepts described as pre-requisite knowledge for the students. Campbell Biology (10thEdition) was utilized as the textbook in the courses as a resource for the students and the teacher to gain the pre-requisite knowledge of a majority of the concepts (18). For additional information on GA in plants see (19).
SCIENTIFIC TEACHING THEMES
ACTIVE LEARNING
This activity engages all students in a hands-on experiment within a lecture setting. Students work collaboratively in groups of two or three to complete all aspects of the project. During the first class period, students set up an experiment and make a prediction of their results. In the following class periods they analyze and report their data. Data collection can be done using clickers (recommended) or by manually recording results. This method requires participation by all students and allows them experience in the collection of real data (20). The data are collected in class so students can graph and analyze the data as part of homework assignments (Supporting Material S5).
ASSESSMENT
Students were assessed via graded homework assignments and with a question on their final exam (Supporting Material S6). The assignments were designed to help students make connections between the lecture material and the experiment, and to give students practice in constructing and interpreting graphs. The assignments differed between the two courses. The homework for the Evolution Course was assigned after students learned about the germination process. The homework for the Genetics Course was divided into two parts. Part 1 was assigned after students learned about photosynthesis, and Part 2 was assigned after students learned about regulation of gene expression and Mendelian genetics. The assignments were adjusted to match the learning goals for each course: in the Evolution Course, we focused on directly addressing the connections between the activity and plant structure, function and development (Supporting Material S6, questions 2-4, 7-9), while the Genetics Course covered topics of photosynthesis, cell signaling (Supporting Material S6, Part 1 questions 2 and 3, respectively), Mendelian genetics, gene regulation, and development (Supporting Material S6, Part 2 questions 5, 1-4 and 1, respectively). These assignments are formative and give the instructor the opportunity to identify common misconceptions on a variety of topics covered during the semester. Some common misconceptions targeted include the difference between differentiation and determination, the differences in gene regulation between prokaryotes and eukaryotes, where photosynthesis occurs in plants, and from where plants get a majority of their dry mass. Common mistakes found during the grading of the homework assignments were addressed in class in a following lecture. These assignments can be modified for use as summative assessments depending on the overall grading structure of the course. The assignments can be easily modified to assess student understanding of different concepts depending on the learning objectives of the course being taught. To assess whether students understood the experiment and the graphing exercise, we included a question in a final exam asking students to draw a conclusion from data presented in a graph (Supporting Material S6).
INCLUSIVE TEACHING
This activity was brought into a lecture setting instead of a laboratory setting specifically for the purpose of engaging students with course material in an inclusive format as active learning strategies increase student learning and success in STEM courses, including underrepresented minority students (21,22). This lesson works very well in an introductory biology course for students with varying backgrounds in biological sciences. Because this activity does not require students to have prerequisite knowledge from other courses or specific laboratory skills, all students participate in the experimental setup and procedures. To address the diversity of learning types within the classroom and to help make sure all students have the opportunity to understand the activity, instructions are provided both in print-form on a handout (Supporting Material S4) and presented on PowerPoint slides (Supporting Material S2) as the students set up the experiment. Students perform the entire activity with a partner, giving them the opportunity to discuss their results and the assignment with at least one other person.
LESSON PLAN
Materials
Seeds can be obtained as a set (catalog #CS27971) from the Arabidopsis Biological Resource Center (ABRC) through The Arabidopsis Information Resource (TAIR, arabidopsis.org). The set contains 300 seeds of each of two samples, wild type (Ler-0) and mutant (ga1-2), which is enough to facilitate this lab with a class of 30 students. Additional sets of seeds can be ordered to accommodate larger classes. Supplementary resources, such as instructional videos and background information can be obtained through the ABRC Education and Outreach website under the Life in Bloom module (abrcoutreach.osu.edu). Additional lessons using Arabidopsis to teach a variety of science concepts are also available for download from this site.
Designate Ler-0 and ga1-2 seeds as either "Unknown A" or "Unknown B". In advance of the first class period, seed samples should be divided into separate Eppendorf tubes, each containing at least 20 seeds of a single genotype. For each genotype, label two-thirds of the tubes so that the genotype is known (WT [Ler-0] or mutant [ga1-2]), and the remaining one-third of the tubes for each genotype with its designated unknown label (A or B). At the end of this step, teachers should have enough tubes so that each group will receive one tube each of WT and mutant and one tube containing an unknown genotype. Sealing film (e.g., Parafilm), wax paper, and aluminum foil can be pre-cut before class to decrease the amount of in-class time required for the activity (Supporting Material S7). A 200 umol GA solution should be prepared before class according to the following steps:
- Preparation of 1 M GA stock solution: Dissolve 0.346 g of GA powder in 1 mL of either water or ethanol. (Some types of GA powder are water-soluble, but others will only dissolve in 70% ethanol.) Warm the solution in a hot water bath (50°C) for 5-10 minutes to help the GA powder dissolve better in either water or ethanol.
- Preparation of 200 uM working GA solution: Mix 6 uL of 1 M GA stock solution with 30 mL of distilled water.
- Storage: Place both GA solutions in a sealed container (eppendorf tube or a glass bottle) completely covered with aluminum foil and store in a standard refrigerator (4°C). GA degrades in the light, and must be stored in dark conditions for best results.
- Immediately preceding the first class, aliquot 10 mL of the 200 umol GA solution into a 15 mL tubes. Each group will need one tube of GA solution.
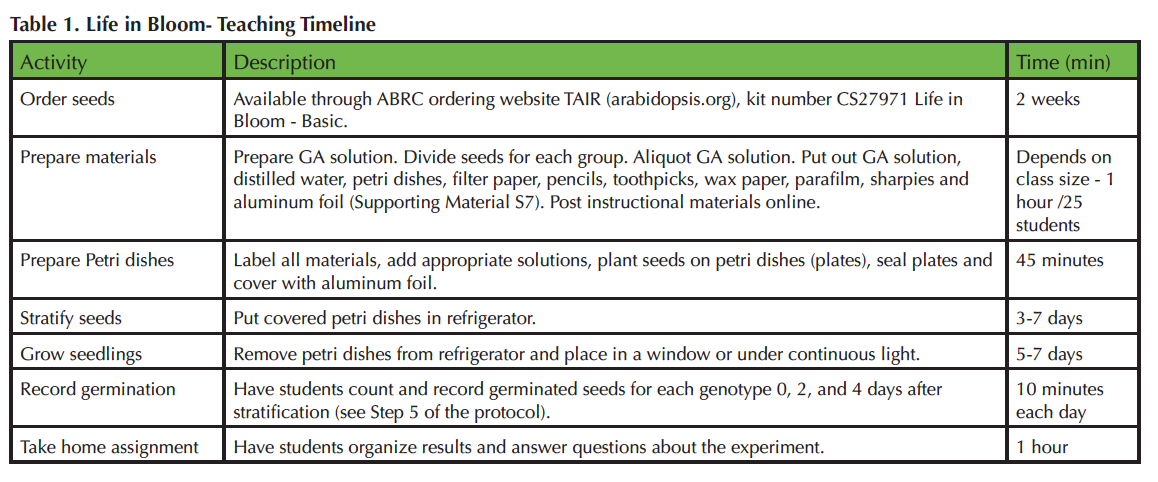
Table 1. Life in Bloom-Teaching Timeline
In-class Facilitation
Prior to class, step-by-step instructions were provided to the students in a handout posted online (Supporting Material S4). During class, the instructions were displayed on PowerPoint slides (Supporting Material S2). This activity was carried out in a college-level class with approximately 50 students. To facilitate this activity in a similar setting, set up two stations within a lecture hall with all needed supplies (see Timeline and Supporting Material S7). After students are arranged into groups, instruct one person from each group to pick up the necessary supplies from one of the stations. As a precaution, students should wear gloves. The following step-by-step guide is provided as an overview of the protocol:
- Separate students into groups of two or three. One student in the group should prepare the plate with the water treatment and the other should prepare the plate with the GA treatment. Have students use a #2 pencil to label a piece of circular filter paper for each of the GA and water treatments (Figure 2). We recommend using a pencil rather than a pen as some inks blur when wet.
- Stack the labeled piece of filter paper on top of an unlabeled piece of filter paper with the label facing up. Place the resulting filter paper stack in the bottom of the plate, with the label still facing upwards. Use a finger or the blunt end of a pen to smooth the edges of the filter paper along the inside edges to flatten the filter paper on the bottom of the petri dish. Add water or 10 mL GA solution, depending on the treatment, to the filter paper. The filter paper should be wet while avoiding having standing liquid at the bottom of the dish. Pour off any excess liquid into a sink.
- Sprinkle half of the seeds of a single genotype or unknown onto a piece of wax paper for the water treatment. Sprinkle the remaining half of the seeds onto a separate piece of wax paper for the GA treatment. Wet the tip of a toothpick by touching it gently to the wet filter paper. Use the wet tip of the toothpick to place nine seeds, one at a time, from each sample type onto the appropriately labeled section of the filter paper (Figure 2). Placing nine seeds of each type will allow for data collection using a clicker response system limited to responses of 0-9. Place the seeds in an evenly spaced row to make data collection easier. Use a new toothpick and a new piece of wax paper for each treatment and seed type to prevent cross-contamination.
Figure 2. Template for labeling filter paper
- Cover the prepared plates with lids and wrap the edges with parafilm to prevent excess evaporation. Stack the two dishes from each group together, and wrap the stack with aluminum foil. Ask students to submit a prediction of their results as part of the in-class portion of the assignment (Supporting Material S2, S5).
- Place the plates in a horizontal position at 4°C (any standard refrigerator) for 3-7 days. This process, called stratification, simulates winter and breaks the seeds out of a state of dormancy. It also synchronizes the germination of the different seed samples. Three days is sufficient to break dormancy and synchronize seed germination for most Arabidopsis accessions (23).
- Bring the plates back to the class after stratification and remove the aluminum foil. Students should count the number of seeds that have germinated in their assigned plate (Figure 3). Provide each group with a small magnifying glass to assist with data collection. If necessary, additional liquid may be added to the dishes at this time so the seeds remain hydrated until the next observation. Be careful not to add so much liquid that the seeds become displaced on the labeled filter paper.
Figure 3. Seed germination. (A) Seed with no signs of germination (B) Seed in the early stages of germination as evidenced by the presence of a radicle that has just broken seed coat. (C, D) Root elongation, (E) Hypocotyl emergence, and (F) Greening of the emerging plantlet.
- Have students record germination data for each plate (Day 0, Supporting Material S5). Class germination data can be recorded on an excel spreadsheet shared with the class or by using the Turning Technologies clicker system if it is available. Supporting Material S3 and S8 can be used with the clicker system to store responses, automatically calculate averages, and display the results to the class. When clickers are not available, or if a clicker system not capable of collecting numerical responses is used in the classroom, students can enter the totals into Supporting Material S9, which automatically calculates a class average without the use of a clicker system.
- Re-wrap the plates with parafilm (but not aluminum foil) and place in a horizontal position in a window or under a continuous light source. Arabidopsis grows best at 120-150 µmol/m2s continuous light and a temperature of 22-23°C (23). The suggested light source is a 40W daylight fluorescent lamp positioned 14.5"- 15" away from the plates. The number of germinated seeds should be counted and recorded in class on Day 2 and Day 4 using the same procedure discussed in steps 6 and 7.
- Since the seed samples do not contain any recombinant DNA, you may discard the plates with regular waste at the conclusion of the experiment (or students may take them home to observe further if they would like).
- Have students use the germination class averages to graph the data as part of the homework assignment (Supporting Material S5). Example germination results are provided in Supporting Material S10.
Expected Results
The GA-producing wild-type seeds are expected to germinate on both the GA and water plates, while GA-deficient mutant seeds are expected to germinate only on the GA plates (Figure 4). The mutant seeds should not germinate on plates lacking GA solution because of a mutation in the GA1 gene. This mutation disrupts the function of ent-copalyl diphosphate synthase (CPS), an enzyme responsible for converting geranylgeranyl pyrophosphate (GGPP) into ent-copalyl diphosphate (CDP), the essential first step of the GA biosynthetic pathway (Supporting Material S1) (11,12). The seeds of unknown genotype should have germination results corresponding to either the wild type or the mutant (Figure 4). Students are expected to be able to determine the genotype of the unknown by comparing the germination results with those observed for the mutant and wild-type samples.
Figure 4. Wild type (Ler-0), mutant (ga1-2), and unknown seeds/seedlings on plates containing (A) water or (B) GA solution. The picture was taken 4 days after the plates have been exposed to light.
TEACHING DISCUSSION
INTRODUCTORY BIOLOGY COURSES
We used this lesson to reinforce the learning goals and objectives of two introductory biology courses. To emphasize the value of quantitative data analysis, students were given a take-home assignment requiring them to graph and analyze the collated data from the in-class experiment. The graphing exercise served as an effective tool to demonstrate the importance of using controls, replicates, and larger sample sizes for accurate data analysis in a format much like other similar exercises for which class average data were used to graph the results (13,14). In addition to asking students to organize data in graphical form, the homework assignments also were designed to help students make connections between various components of the course and to assess their mastery of course-related biological concepts specific to each course (Evolution and Genetics). The homework assignments contained both lower-level (definitions, remembering understanding) and higher-level learning questions (application and analysis). While it was expected that a majority of students would be able to correctly answer the lower-level questions, the majority of students were able to answer homework questions requiring higher-level thinking. For example, most of the students came up with reasonable interpretations when asked to explain the difference in phenotypes seen between the wild-type and mutant seed strains. One particular question served as an excellent formative assessment. Question 2 in Part 2 of the Genetics Course homework assignment (Supporting Material S6) is a higher-level question and asks students to make connections between several concepts covered during the semester (e.g., gene expression regulation, the effect of a mutation on phenotype) by describing two possible locations within the genome where a mutation could have occurred to result in the phenotype exhibited by the GA mutant strain. Based on their responses, about 67% of the students made the expected connections between the activity and the lecture material as they included descriptions of epigenetic effects, mutations in a site affecting the splicing of the pre-mRNA, mutations within the promoter or enhancer regions of a gene involved in GA biosynthesis, and mutations in the gene for a transcription factor required for activating a gene involved in GA biosynthesis.
We found this lesson to be helpful in identifying and clarifying common challenge students previously had with relating the genotype of an individual to its phenotype by observing firsthand how a mutation can affect plant growth. The responses we obtained from the take-home assignments suggested that, after this activity, students almost always successfully connected the phenotype to mechanisms of gene expression (Supporting Material S6, Genetics Course homework assignment Part 2, question 2). For the Evolution Course, two questions in the homework assignment, directly related to this activity, forced students to think about plant mass and seed resources that need to be utilized by the seedling until it becomes capable of performing photosynthesis (Supporting Material S6, questions 8 and 9).
Overall, the simple design of the activity enabled most students to complete it fully, including setting up the experiment, constructing the graph, and answering the questions, without further assistance. Nearly all students were able to correctly interpret a graph of data on the final exam (Supporting Material S6; 93% of students in the Evolution Course, 98% of students in the Genetics Course) demonstrating that they understood the experiment. Besides being simple, this lesson helps students understand both basic scientific principles (such as the experimental process and how to use data and graphing to understand it) as well as higher level scientific principles (e.g., relating findings on plants to human biology; understanding a correlation between genotype and phenotype). This lesson can be used as-is or as a template with the teacher molding discussions and homework assignments for their own goals. Some ideas for alterations are presented below. Taken together, the incorporation of this module into class represents a small investment of class time that returns as a significant impact on student learning.
ADAPTING MATERIAL FOR MORE ADVANCED COURSES
The concepts covered in this lesson can be easily related to the biology of humans and other animals. The idea that hormonal regulation of various functions/processes is similar between plants and animals is illustrated in Supporting Material S11. Demonstrating to students how understanding systems in plant biology relates to similar systems in animal physiology may help to pique student interest in other plant biology topics.
By adding different mutants into the experimental protocols, instructors can also cover more advanced concepts in genetics and plant physiology. By examining seeds with mutations in different parts of the same gene, students can be taught how different gene regions are important or how different protein domains function within a GA biosynthetic enzyme. Seeds with mutations affecting different stages in the GA biosynthetic pathway can be analyzed to further explore the biochemistry involved when a plant produces GA (Supporting Material S1). Likewise, a mutant affecting germination rate, but which has little effect on germination percentage, can be used to demonstrate how some mutations in the GA pathway only partially inhibit germination. A GA-insensitive mutant (gai) can be used to explore the genetics involved in gene regulation and signaling. Some of these ideas have already been incorporated into a different education kit (Life in Bloom, ABRC catalog# CS19988), along with the accompanying protocols and handouts, which are available at https://abrcoutreach.osu.edu/educational-kits.
Taking advantage of Arabidopsis as safe and easy to grow in the classroom (shared among all plant models) and its other advantages, such as small size and short growing cycle (shared with Wisconsin Fast Plants), several other teaching modules have been designed using this weedy plant as a model. ABRC has put a significant effort in collecting and distributing undergraduate genetics teaching modules utilizing this plant. "Genetics of Arabidopsis thaliana" by Scott Poethig (CS19998), "Molecular Genotyping of Arabidopsis" by the Dolan DNA Learning Center (CS19996), "Transposing from the Laboratory to the Classroom" by Susan Wessler (CS19995), "Expression analysis of light-regulated genes in the det1-1 mutant" by Kim Loney (CS19997) (https://abrcoutreach.osu.edu/trained-educational-kits), are just a few examples of teaching units used to complement undergraduate genetics courses and aid in clarifying common conceptual difficulties students have with genetics.
SUPPORTING MATERIALS
- S1. Life in Bloom-GA Biosynthetic Pathway: A scheme showing the GA biosynthetic pathway modified from references 9 and 10
- S2. Life in Bloom-Activity Part 1: PowerPoint lecture presentation slides
- S3. Life in Bloom-Activity Part 2: PowerPoint lecture presentation slides. The last four slides are programmed to work with the TurningPoint Technologies clicker system
- S4. Life in Bloom-Activity Handout: Student handout
- S5. Life in Bloom-Homework Assignments: Homework questions from two courses
- S6. Life in Bloom-Homework Assignments with Key and Final Exam
- S7. Life in Bloom-List of Materials
- S8. Life in Bloom Data Collection Table Clickers: Excel sheet formatted for clicker data entry and, programmed to automatically calculate the average number of germinated seeds
- S9. Life in Bloom Data Collection Table no Clickers: Excel data entry sheet programmed to automatically calculate average number of germinated seeds
- S10. Life in Bloom-Example Results: Excel sheet with example germination results and graphs
- S11. Life in Bloom-Hormone signaling: Hormonal regulation is similar between plants and animals
ACKNOWLEDGMENTS
We greatly appreciate NSF for their continued support of ABRC, as well as the initial support received from the American Society of Plant Biologists (ASPB) to generate educational resources at ABRC. Thank you to Maarten Koornneef and Jan Zeevaart for donating the Ler-0 and ga1-2 seeds, respectively. We recognize and appreciate all the hard work Sebastian Matt put into the development of the experiments and original protocols this lesson is based on. We are also very grateful to Molly Marcus and Benson Lindsey for their assistance with the "Life in Bloom" video and to Chris Bartos for the design of the ABRC outreach website.
References
- Wandersee J, Schussler E. 2001. Toward a theory of plant blindness. Plant Science Bulletin, 47, 2-9.
- Phelps-Durr TL. 2016. Using computational molecular modeling software to demonstrate how DNA mutations cause phenotypes. CourseSource. https://doi.org/10.24918/cs.2016.16.
- Briju BJ, Wyatt SE. 2015. Grocery Store Genetics: A PCR-Based Genetics Lab that Links Genotype to Phenotype. The American Biology Teacher, 77(3):211-214.
- Mitchell-Olds T. 2001. Arabidopsis thaliana and its wild relatives: a model system of ecology and evolution. Trends in Ecology & Evolution. 16(12):693-700.
- Koornneef M, Meinke D. 2010. The development of Arabidopsis as a model plant. The Plant Journal. 61:909-921.
- Provart N, Alonso J, Assman S, Bergmann D, Brady S, Brkljacic J, Browse J, Chapple C, Colot V, Cutler S, Dangl J, Erhardt D, Friesner J, Frommer W, Grotewold E, Meyerowitz E, Nemhauser J, Nordborg M, Pikaard C, Shanklin J, Somerville C, Stitt M, Torii K, Waese J, Wagner D, McCourt P. 2015. 50 years of Arabidopsis research: highlights and future directions. New Phytologist. 209(3): 921-944.
- Penfield S, Rylott E, Gilday A, Graham S, Larson T, Graham I. 2004. Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires phosphoenolpyruvate carboxykinase1. Plant Cell. 16(10): 2705-2718 .
- Bethke P, Libourel I, Aoyama N, Chung Y, Still D, Jones R. 2007. The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiology. 143(3): 1173-1188.
- Debeaujon I, Koornneef M. 2000. Gibberellin requirement for Arabidopsis seed germination is determined both by Testa Characteristics and Embryonic Abscisic Acid. Plant Physiology. 122(2):415-424.
- Koornneef M, van der Veen J. 1980. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theor Appl Genet. 58(6):257-263.
- Taiz L, Zeiger E, Moller I, Murphy A. 2015. Hormones synthesized via Isoprenoid pathways. Plant physiology and development sixth edition. Appendix 3.
- Hedden P, Phillips A. 2000. Gibberellin metabolism: new insights revealed by the genes. Trends in Plant Science. 5(12):523-530.
- Farmer J, Gotwals B, DeVenny A. Gibberellic Acid's Effect on Plant Growth. http://www.ncsec.org/models.cfm.
- Baxter LH. Investigating Plant Physiology with Wisconsin Fast Plants(TM). https://fastplants.org/wp-content/uploads/2017/02/WFPphysiology-06web.pdf
- Morehead Planetarium and Science Center. 2013. Same Genes, Different Fates. https://arabidopsis.osu.edu/static/docs/SameGenesDifferentFates.pdf.
- Ghildiyal M, Zamore PD. 2009. Small silencing RNAs: an expanding universe. Nature Reviews Genetics 10:94-108.
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo Q-M, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 462:315-322.
- Reece JB, Urry LA, Cain ML, Wasserman SA, Minorsky PV, Jackson RB. 2014. Campbell Biology (Tenth edition). Boston: Pearson
- Gupta R, Chakrabarty SK. 2013. Gibberellic acid in plant: Still a mystery unresolved. Plant Signal. Behav. 8:e25504
- Pelletreau KN, Andrews T, Armstrong N, Bedell MA, Dastoor F, Dean N, Erster S, Fata-Hartly C, Guild N, Greig H, Hall D, Knight JK, Koslowsky D, Lemons PP, Martin J, McCourt J, Merrill J, Moscarella R, Nehm R, Northington R, Olsen B, Prevost L, Stoltzfus J, Urban-Lurain M, Smith MK. 2016. A clicker-based study that untangles student thinking about the processes in the central dogma. CourseSource. https://doi.org/10.24918/cs.2016.15
- Freeman S, Eddy SL, McDonough M, Smith MK, Okoroafor N, Jordt H, Wenderoth MP. 2014. Active learning increases student performance in science, engineering, and mathematics. PNAS. 111:8410-8415
- Eddy SL, Hogan KA. 2014. Getting under the hood: How and for whom does increasing course structure work? CBE Life Sci. Educ. 13:453-468
- Rivero L, Scholl R, Holomuzki N, Crist D, Grotewold E, Brkljacic J. 2014. Handling Arabidopsis plants: growth, preservation of seeds, transformation, and genetic crosses. Methods Mol Biol. 1062:3-25.
Article Files
Login to access supporting documents
Linking Genotype to Phenotype: The Effect of a Mutation in Gibberellic Acid Production on Plant Germination(PDF | 1 MB)
S1. Life in Bloom-GA Biosynthetic Pathway.pdf(PDF | 158 KB)
S2. Life in Bloom-Activity Part 1- PowerPoint lecture presentation slides.pptx(PPTX | 894 KB)
S3. Life in Bloom-Activity Part 2-PowerPoint lecture presentation slides.pptx(PPTX | 874 KB)
S4. Life in Bloom-Activity Handout.docx(DOCX | 775 KB)
S5. Life in Bloom-Homework Assignments.docx(DOCX | 21 KB)
S6. Life in Bloom-Homework Assignments with Key and Final Exam.docx(DOCX | 247 KB)
S7. Life in Bloom-List of Materials.docx(DOCX | 17 KB)
S8. Life in Bloom Data Collection Table Clickers.xlsx(XLSX | 21 KB)
S9. Life in Bloom Data Collection Table no Clickers.xlsx(XLSX | 14 KB)
S10. Life in Bloom-Example Results.xlsx(XLSX | 34 KB)
S11. Life in Bloom-Hormone signaling.jpg(JPG | 57 KB)
- License terms

Comments
Comments
There are no comments on this resource.