Understanding Protein Domains: A Modular Approach
Published online:
Abstract
An understanding of protein structure, function, and interaction is central to biochemistry. One angle into this topic is to engage students in considering protein domains as modules that develop form and execute function semi-independently. Here, I describe a modular guided inquiry Lesson for students with an introductory background in molecular biology and biochemistry. The Lesson enables students to further explore how we define and investigate the structure and function of protein domains in a research context. Activities focus on bioinformatic approaches and interpretation and design of experiments to investigate protein interactions. Possible extensions into wet-lab and/or research projects are also highlighted. Students from various science majors enrolled in an intermediate-level biochemistry course reported that this Lesson strengthened their ability to analyze protein sequence and structure and to understand approaches to determining protein interactions.
Citation
Hark, A.T. 2017. Understanding Protein Domains: A Modular Approach. CourseSource. https://doi.org/10.24918/cs.2017.21Society Learning Goals
Biochemistry and Molecular Biology
- Macromolecular Structure Determines Function and Regulation
- What factors contribute to the size and complexity of biological macromolecules?
- What factors determine structure?
- What is the role of noncovalent intermolecular interactions?
Science Process Skills
- Process of Science
- Locate, interpret, and evaluate scientific information and primary literature
- Plan, evaluate, and implement scientific investigations
- Interpret, evaluate, and draw conclusions from data
- Construct explanations and make evidence-based arguments about the natural world
- Address novel questions through authentic research experiences
- Modeling/ Developing and Using Models
- Build and evaluate models of biological systems
- Quantitative Reasoning/ Using Mathematics and Computational Thinking
- Apply the tools of graphing, statistics, and data science to analyze biological data
Lesson Learning Goals
- Students will understand that protein domains have structure and function that may be both semi-independent of and contribute to overall protein structure and function.
- Students will understand that protein-protein interactions can be tested experimentally both in vitro and in vivo.
Lesson Learning Objectives
- Students will be able to compare protein sequences and identify conserved regions and putative domains.
- Students will be able to obtain, examine, and compare structural models of protein domains.
- Students will be able to interpret data on protein interactions (in vitro pull-down and in vitro and in vivo functional assays)
- Students will be able to propose experiments to test protein interactions.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
When introducing students to protein structure, the modular domain aspect of proteins can be fascinating in theory, but sometimes hard for students to completely grasp. How is it that segments of a protein encoded within a single gene and translated from one mRNA can have separable functions? How might various domains of a protein contribute to overall function? How can we define domains and study how they interact with other macromolecular structures within and outside of the cell? Focusing on these questions provides students with opportunities to think about structure-function relationships, to consider the impact of molecular evolution on domain structures, and to understand how scientists can demonstrate domain interactions or interactions involving whole proteins. These aims are accepted as important outcomes in the study of biochemistry. For example, among the undergraduate learning goals identified by American Society for Biochemistry and Molecular Biology (ASBMB) are the declarations that "[s]tudents should understand that proteins are made up of domains..." and "[s]tudents should be able to discuss the interactions between a variety of biological molecules and describe how these interactions...[lead] to changes in biological function." (1).
One route to addressing these questions is through bioinformatics. The National Institutes of Health (NIH) defines bioinformatics as "research, development, or application of computational tools and approaches for expanding the use of biological, medical, behavioral, or health data, including those to acquire, store, organize, analyze, or visualize such data." (2). Bioinformatics is important for the study of biochemistry as it focuses on the analysis of molecular sequences (DNA, RNA, and proteins) and reveals the structure and function of macromolecules. I want to encourage students to use bioinformatics tools to explore relationships between primary sequence, three-dimensional structure, and biological function, both in this course and in their work in other areas. Comparison of protein sequences and structures connects to learning goals articulated within ASBMB's Concept-Driven Teaching, e.g., "Use various bioinformatics approaches to analyze macromolecular primary sequence and structure." (1). Completing these assignments reinforces concepts with which biochemistry students should be comfortable, while students are simultaneously learning bioinformatics skills that they can use across disciplines. Magana et al. among others highlight the importance of bioinformatics education for cross-disciplinary problem solving (3). They also advocate for establishment and assessment of related learning outcomes, as in this Lesson (3).
Another set of skills that we seek to develop in undergraduate science students is their ability to propose experiments and to interpret data. This "ability to apply the process of science" is reflected as one of the Vision and Change Core Competencies for undergraduate biology education (4). The Modules described here can help build skills identified as learning goals for undergraduate biochemistry majors by the ASBMB (Students should be able to: "interpret data pertaining to tertiary and quaternary structure of molecules;" "propose a purification scheme for a particular molecule;"and "propose appropriate...chemical biology approaches to explore the...interactions of biological macromolecules.") (1).
To help students accomplish the aims described above, I developed this series of guided inquiry exercises. This approach resembles problem-based learning (PBL) in that, for at least some students, the problems introduce new content and/or require research (5). The problems may also reinforce concepts introduced earlier in the course and curriculum. This Lesson supports the aims identified by Eberlein et al. for PBL and related approaches: "To promote higher-order thinking skills; to help students learn to reason though problems, instead of using algorithmic approaches; to build conceptual understanding through active engagement with the material; to foster growth in teamwork and collaborative problem-solving skill" (5). This Lesson also offers an opportunity for student-centered learning advocated for by the Vision and Change report, as the projects are inquiry-driven, require active participation, and benefit from student collaboration (4). Please see the Scientific Teaching Themes section for further discussion.
The exercises I developed focus on a specific protein, the transcriptional coactivator ADA2. This protein is known to work with GCN5, a histone acetyltransferase, in various eukaryotic species. The choice of ADA2 provides a connection to a research project in my lab, which uses Arabidopsis thaliana as a model system. In Arabidopsis, gene duplication has created two versions of ADA2, referred to as ADA2a and ADA2b. ADA2b seems to have more significant biological function (6), and so I used Arabidopsis ADA2b as our primary starting point for some of the activities. As discussed later in this article, these Lesson activities could be adapted for any protein for which there is information about amino acid sequence, structural data, and in vitro or in vivo interactions. Alternatively, these inquiries could lay the foundation for novel research investigations, thereby building new scientific investigations on what is already known.
INTENDED AUDIENCE
The Lesson was used in an intermediate-level biochemistry course at a liberal arts college. Students enrolled in the course were sophomore through senior undergraduate science majors.
REQUIRED LEARNING TIME
The four Modules that comprise this Lesson were deployed throughout an academic semester. For each module, background information (if needed) and the assignment were provided a week in advance of a lab section during which time was allotted for students to ask questions about the Module. Following that meeting, students typically had two additional weeks to submit their work (Table 1). Scheduling of the Modules throughout the semester allowed for assessment and feedback on each assignment before the next was due. However, since the skills needed for each module are distinct, this sequential timing is unlikely to be essential. Following submission of each complete assignment, students in the Spring 2017 cohort were asked to (voluntarily) indicate how much total time they had spent preparing the assignment. In all cases, the majority of respondents indicated that they had spent either less than three hours or three to five hours engaged in these problems. That time including talking with me and/or their peers and preparing their final answers.
PRE-REQUISITE STUDENT KNOWLEDGE
Students should be familiar with concepts underlying the Central Dogma of molecular biology and with basic protein structure including amino acid functionality. If students are not familiar with BLAST and/or multiple sequence alignment or protein structural viewers, the instructor will need to provide background tutorial materials or other approaches to help students develop skill in using these programs. Similarly, if students have no or limited knowledge of SDS-PAGE, protein purification (GST pull down), and/or transgenic organisms, review of these approaches could be built into Modules III and IV.
PRE-REQUISITE TEACHER KNOWLEDGE
Instructors who teach molecular and cell biology at the introductory college level or above should have the prerequisite knowledge. Skill development would be similar as that outlined for students above.
SCIENTIFIC TEACHING THEMES
ACTIVE LEARNING
As noted above, this deployment of the Modules represents guided inquiry or problem-based learning. Students have to collect and interpret data and design and explain experimental approaches to answer scientific questions. Students will need to leverage prior knowledge and acquire new knowledge and skills to prepare Module answers. Collaboration is strongly encouraged in engaging with these problems. Students in my course are accustomed to working in problem-solving groups in class and so many engage in this high impact practice [as defined by the American Association for Colleges and Universities (7)] for this project. These active learning approaches in STEM fields have been highlighted as important for enhancing student engagement, performance, and retention by Project Kaleidoscope (8), the Vision and Change report commissioned by the National Science Foundation and the American Association for the Advancement of Science (4), and by numerous individual science education studies [a sampling summarized in (9)].
ASSESSMENT
Individual written responses to short answer questions were assessed for a grade. As students are allowed to collaborate as they work through the questions, there is also a great deal of self-assessment that goes on, as student reconcile their thinking with that of their peers. As elaborated on in the Discussion, student understanding of the concepts and skills connected to the Modules was accessed through short answer questions on the final exam (e.g. interpreting a sequence alignment or data from a pull-down assay).
INCLUSIVE TEACHING
These Modules represent learner-center instruction. Students are provided with background information and/or an assignment prompt to review outside of class. This gives each student time and space to connect this new work to previously developed knowledge and skills and frame questions. Individual students/small groups have opportunities to ask questions during a lab period and/or during office hours after this initial read-through. Students may also use any resources they like (with the exception of reading another student's written responses) in developing their answers. Encouraging this variety of ways to engage with the material aligns with inclusive teaching, described by Cornell's Center for Teaching Innovation as incorporating "any number of teaching approaches that address the needs of students with a variety of backgrounds, learning styles, and abilities." (10).
Aspects of this work can also prompt thought and discussion around issues of access and equity in science. For example, the bioinformatics utilities chosen are freely available and data excerpts can be chosen from research papers that are Open Access. The studies used here include authors of various nationalities working as collaborators; in general, looking at the authors of the published works chosen provides an entry point to discussing the (lack of) diversity in scientists. This latter point is reflected in a reflection prompt on inclusive teaching strategies provided by the University of Michigan's Center for Research on Learning and Teaching (11).
LESSON PLAN
Overview
These Modules were used as part of a 15-week course scheduled for three hour-long lecture meetings and one three-hour lab meeting per week. For each module, an assignment sheet and in some cases, specific background reference materials were provided a week in advance of the lab meeting in which time was allotted for students to ask questions about the assignment. Following that meeting, students typically had two additional weeks to submit their work (Table 1).
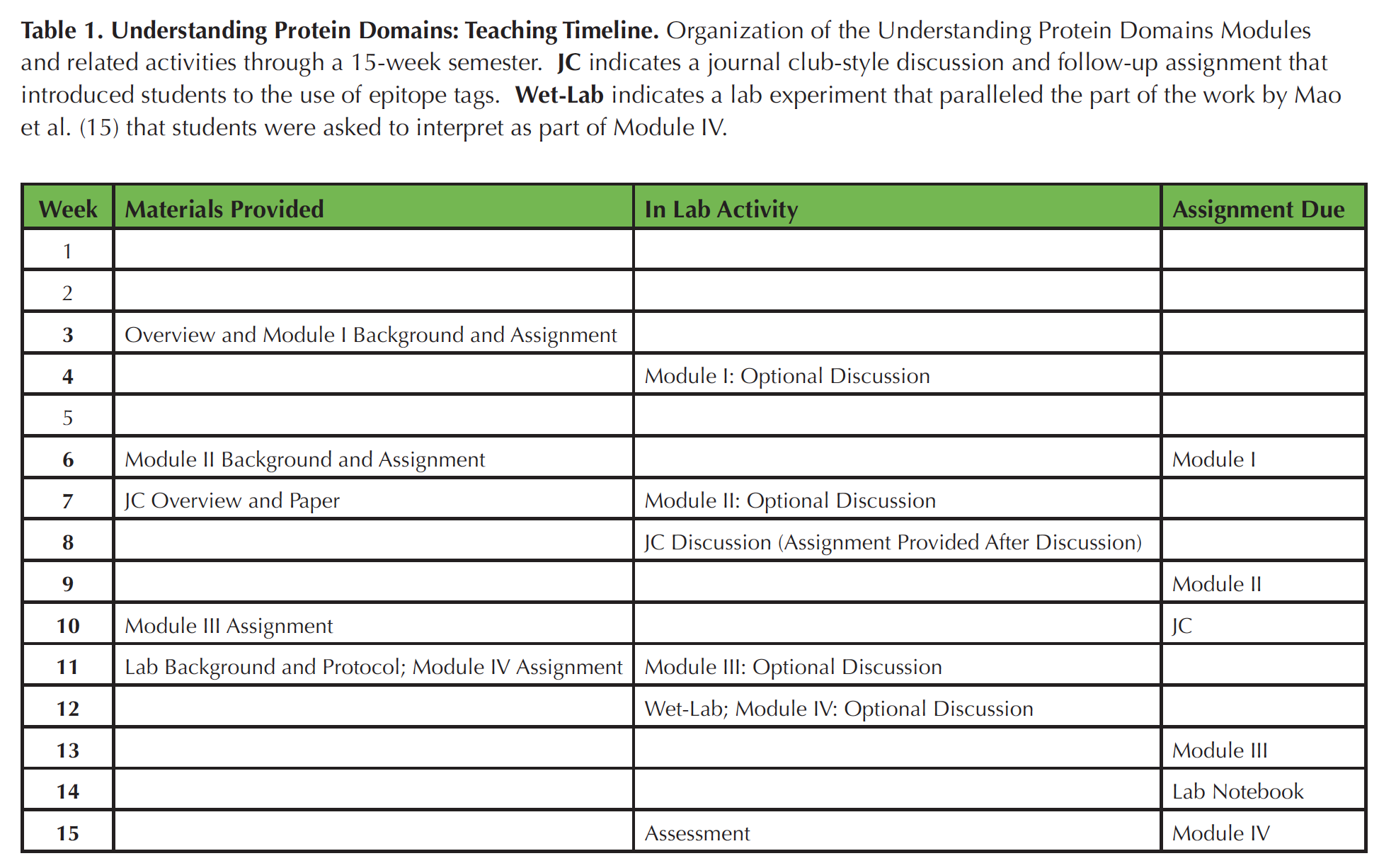
Table 1. Organization of the Understanding Protein Domains Modules and related activities through a 15-week semester. JC indicates a journal club-style discussion and follow-up assignment that introduced students to the use of epitope tags. Wet-Lab indicates a lab experiment that paralleled the part of the work by Mao et al. (15) that students were asked to interpret as part of Module IV.
The four Understanding Protein Domains (UPD) modules were worth 10% of the overall course grade, with the first two assignments being weighted slightly more due to the higher estimate of amount of work required. Based on the peer learning environment I seek to foster in my courses and the collaborative nature of science, I encouraged students to discuss use of the web-based utilities, experimental approaches, and even the UPD questions with their colleagues. However, each student was responsible for constructing their own answers prior to submission.
Module I: Comparing Sequences
The first Module (S1) involves comparing protein sequences. Students were encouraged to review written background [adapted from Young (12)] and instructor-generated screencasts reviewing BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and Clustal (https://www.ebi.ac.uk/Tools/msa/clustalo/) as needed. This Module benefits from this opportunity for learner-specific instruction, as some students are experienced with these programs while others may have only been introduced to them. In addition, students were free to complete the assignment with comparable programs if they preferred. Students were directed to align eight sequences and identify domains primarily on the basis of primary sequence conservation (S1). In this assignment we studied the ADA2 transcriptional coactivator, for which paralogs exist in several species. To minimize complexity, after consultation of the literature (13), I provided students with an accession number for the Arabidopsis thaliana ADA2b protein as a starting point and also specified the species in which students should search for orthologs.
Module II: Comparing Structures
In this Module (S2), students use bioinformatics approaches to examine secondary, tertiary, and quaternary protein structure. Students were again encouraged to review written background information [adapted from (12)] and/or instructor-generated screencasts reviewing NCBI Structure/Cn3D (https://www.ncbi.nlm.nih.gov/structure), although other structural viewers could be used as well. For ADA2, the structures of only some specific domains have been solved, which reinforced domains as protein sections that can fold and function semi-independently. This module also reinforced conservation at the structural level (S2, question 4) and offered one way to look at structural interactions (S2, question 3).
Module III: Test of Interaction
The third Module provides an opportunity for students to consider which domains of ADA2 and GCN5 might mediate chemical interactions between the two proteins. Students are asked to interpret a Glutathione S-transferase (GST) pull down experiment (14) and then propose an experiment to extend their understanding of the protein interactions involved (S3). In this example, the latter portion builds on findings from Module I, as students are asked to outline a way to test if GCN5 interacts with the plant-specific domains of ADA2b they had previously identified.
Students had been introduced to the related idea of co-immunoprecipitation through lecture and had encountered the use of epitope tags through a journal club-style review of an unrelated article from the primary literature (Table 1). However, student questions and answers revealed that more specific background about pull-down assays would be beneficial. In the Spring 2016 offering of the course, an instructor-generated screencast outlining the method was posted along with the answer key, to help increase student understanding of this approach. Please see the Teaching Discussion section for further commentary on revisions to Module III.
Module IV: Test of Function
In this last Module, I wanted to provide students with an opportunity to think about how one might test the function of a given protein domain (S4). The example here is specific to published literature on ADA2, which identifies a region of the plant-specific domain as a target for acetylation by GCN5 and then tests the requirement for a specific lysine residue through site-directed mutagenesis and an in vivo test (15). This example also afforded connection to a similar test of the function of the entire plant-specific domain that has been carried out in my research lab (S4; also see Extensions section below). No specific background was provided for this assignment, since students were completing this module in the last weeks of the semester and were asked to draw on prior course knowledge and problem solving skills. For example, students did not have specific experience with acetylation assays as shown in panel B in S4, but they had run SDS-PAGE in lab. Students were familiar with sequence alignment (from Module I) and histone acetylation (from lecture), which were likely helpful in interpreting panel C mentioned in S4. The experiment referenced in the second part of S4 is similar to an approach students were introduced to through a wet-lab exercise (Table 1).
TEACHING DISCUSSION
Student Outcomes
Student understanding of the concepts and skills connected to the UPD Lesson was accessed on the final exam. For example, in Spring 2016, a question that required interpretation of a sequence alignment was answered correctly by two-thirds of class (77% in Spring 2017) and a question relating to a pull down assay was answered correctly by 60% of students (64% in Spring 2017). In addition, students were asked to complete an indirect assessment of learning gains related to the goals of the UPD modules (Table 2). In general, students agreed that engaging in the UPD modules strengthened their understanding of protein domains, facilitated their ability to use sequence and structural data, and helped them interpret protein interaction.

Table 2. Self-reported confidence gains in understanding of and ability to study protein domains and interactions. Students in the Fall 2015 and Spring 2016 cohorts completed anonymous evaluations in the last week of class. For evaluation purposes, numerical values were assigned to confidence levels with “strongly agree” =5 and “strongly disagree”= 1.
Students were least confident in their ability to design experiments to study protein interactions. This challenge was reflected in direct assessment as well. The assignment for Module 3 asks students to propose an experiment to test which domains may support protein-protein interactions. In the Spring 2016 cohort (N=35), the mean grade on this assignment was a 73%, while the combined average for the other three assignments was an 86%. This lower score may have resulted from lack of specific background on pull down assays (the focus of this assignment). I provided more explicit instructions and a review of a pull down assay via screencast in Spring 2017, and the average on the Module 3 assignment increased to 87%, although averages across the board were slightly higher for this cohort (90%).
The Modules Themselves
Data suggested that students spent the most time on Module I: 14 out of 23 respondents spent three to five hours and six students spent six or more hours working on this assignment. The higher time spent may be due in part to this being the first assignment of this type in the semester. One specific area of challenge for students was trying to determine putative domains based on sequence conservation. In my view there is no single "right" answer here. When students discuss this challenge with me, I remind them to consider general estimates of minimum domain length (and to think about what drives this minimum). Another challenge comes from the fact that, while my students have reviewed amino acid chemistry and functionality, they are still practicing weighing identity vs. similarity when looking at sequence comparisons. Again, when asked for advice, I indicate that they should be consistent with the stringency used to choose putative domains throughout the primary sequence.
When evaluating BLAST search results, students may have difficulty recognizing multiple entries of the same protein or paralogs within a species. I encourage them to look beyond BLAST score and consider length of match, identity and similarity, etc. Module II, in which students compare structures, seems to be a little more straightforward, perhaps because there are a more limited number of structures in the databases. I encourage students to recognize this issue and consider why this might be the case.
As mentioned above, my students find proposing an experiment to test protein-protein interactions (in Module III) to be one of the more difficult items in this Lesson. I deliberately left the question fairly open, so that students could reiterate a GST pull down experiment similar to the one they had just reviewed or consider other approaches (e.g. yeast two hybrid, structural methods). In Spring 2017, I altered the instructions slightly, indicating a point value along with items that should be included in their answer: how to obtain/prepare potential interacting partners, how to assay interaction, and what controls should be used. I think this additional information helped highlight the importance of these experimental aspects. Depending on their level of expertise, students may need to research these experimental approaches before completing the assignment. Alternatively, an instructor could choose to provide more detailed background in this area prior to assigning Module III or modify his/her/their assessment.
Module IV covers both in vitro and in vivo tests of function. Necessary background will likely depend on the course and curriculum structure as well as the general student ability. For example, I use analysis of the second figure from Mao et al. (15) as a way to introduce conventional site directed mutagenesis leading to a single amino acid change (e.g. K215R). Since my students are very familiar with the amino acids by this point, many can make the leap or they can check with me or classmates. Also, my students have been introduced to the idea of transgenes and genetic rescue in another lab module; in other cases, this approach would likely require some explanation.
The Project Overall
Others have reported pedagogical strategies to help students develop bioinformatic skills including aligning sequences and investigating protein structure-function relationships, both within individual courses (e.g., 16-18) or in larger curricular structures (19,20). The Modules described here bring together these bioinformatics approaches with data analysis of wet-lab experiments designed to interrogate protein domain structure and protein-protein interactions that drive function. Because this approach can be based on existing data, it is cost effective. These Modules could be embedded within a biochemistry, molecular biology, or cell biology course.
Extensions and Adaptations
I see this course structure as adaptable in several significant ways. One could choose a protein of research or curricular interest and develop exercises using this scaffold. Focusing the exercises on a protein of interest would facilitate coupling Modules III and/or IV to wet-lab experiments, if feasible and desirable. In addition, one could adapt any of the modules to be novel investigations into the structure or function of protein domains of interest or simply rely on students' analysis of database information and published literature as I have done here. Additional extensions could involve students constructing phylogenetic trees (21) or exploring domains through other common utilities (e.g. PROSITE, https://prosite.expasy.org/).
From a pedagogical perspective, one may wish to adapt how the assignments are given depending on one's own teaching and/or course design preferences and based on the student population. Suggestions about places in which additional content background or skill development may be helpful are included in the reflection on "The Modules Themselves" in the Teaching Discussion. In addition, I employed a learner-center approach, which places responsibility on each student to review the assignment and any necessary background information prior to an optional class discussion. Students could be offered incentives to read the material in advance and/or be required to participate in preliminary class discussion as ways to enhance engagement and collaborative learning.
Aims
This modular approach can be used as an entire series or as individual problem-based learning experiences. It could be adapted to integrate with lab exercises or an area of research driven by the instructor or curriculum. While the Learning Goals and Learning Objectives listed earlier were developed for this specific Lesson, they connect directly to ASBMB's core concepts, specifically the idea that macromolecular structure determines function and regulation (1). The bioinformatics skills are also referenced under the Foundational Concept of Discovery (1).
SUPPORTING MATERIALS
- S1. Understanding Protein Domains-Module I assignment
- S2. Understanding Protein Domains-Module II assignment
- S3. Understanding Protein Domains-Module III assignment
- S4. Understanding Protein Domains-Module IV assignment
ACKNOWLEDGMENTS
I would like to thank Muhlenberg College students who were enrolled in BIO 220 classes and who engaged in these exercises. I also thank past research students who conducted wet-lab experiments that motivated this work, in particular Nora Hudis, Hillary Gordon, Evan Sheppard, and Dana Tedesco. I appreciate suggestions from both Editors and Reviewers that allowed me to strengthen these materials.
References
- American Society for Biochemistry and Molecular Biology. Foundational Concepts and Skills. http://www.asbmb.org/education/teachingstrategies/foundationalconcepts/. Accessed May 26, 2017.
- NIH Bioinformatics Definition Committee. NIH Working Definition of Bioinformatics and Computational Biology. http://www.binf.gmu.edu/jafri/math6390-bioinformatics/workingdef.pdf. Accessed May 26, 2017.
- Magana AJ, Taleyarkhan M, Alvarado DR, Kane M, Spring J, Clase K. 2014. A Survey of Scholarly Literature Describing the Field of Bioinformatics Education and Bioinformatics Educational Research. CBE Life Sci Ed 13: 607-623.
- American Association for the Advancement of Science. Vision and Change in Undergraduate Biology Education: A Call to Action. http://visionandchange.org/files/2013/11/aaas-VISchange-web1113.pdf. Accessed June 1, 2017.
- Eberlein T, Kampmeier J, Minderhout V, Moog RS, Platt T, Varma-Nelson P, White HB. 2008. Pedagogies of engagement in science. BAMBEd 36: 262-273.
- Hark AT, Vlachonasios KE, Pavangadkar, KA, Rao S, Gordon H, Adamakis I-D, Kaldis A, Thomashow MF, Triezenberg SJ. 2009. Two Arabidopsis orthologs of the transcriptional coactivator ADA2 have distinct biological functions. BBA - Gene Regulatory Mechanisms 1789: 177-124.
- American Association of College and Universities. High-Impact Educational Practices. https://www.aacu.org/leap/hips. Accessed November 21, 2017
- American Association of College and Universities. Sign the Pledge: Improve Undergraduate Stem Education on Active Learning Day. https://www.aacu.org/whats-new/sign-pledge-improve-undergraduate-stem-education-active-learning-day. Accessed November 26, 2017.
- Freeman S, Eddy SL, McDonough M, Smith MK, Okoroafor N, et al. 2014. Active learning increases student performance in science, engineering, and mathematics. Proc Natl Acad Sci USA 111: 8410-8415.
- Cornell University Center for Teaching innovation. Inclusive Teaching Strategies. https://www.cte.cornell.edu/teaching-ideas/building-inclusive-classrooms/inclusive-teaching-strategies.html. Accessed November 22, 2017.
- University of Michigan Center for Research on Learning and Teaching. Inclusive Teaching Resources and Strategies. http://www.crlt.umich.edu/multicultural-teaching/inclusive-teaching-strategies. Accessed November 22, 2017.
- Young PG. 2008. Web-based Bioinformatics Tutorials: Exploring Genomes. New York, NY: W.H. Freeman.
- Srivastava R, Rai KM, Pandey B, Singh SP, Sawant SV. 2015. Spt-Ada-Gcn5-Acetyltransferase (SAGA) Complex in Plants: Genome Wide Identification, Evolutionary Conservation and Functional Determination. PLoS One 10: e0134709.
- Stockinger EJ, Mao Y, Reiger MK, Triezenberg SJ, Thomashow MF. 2001. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res 29: 1524-1533.
- Mao Y, Pavangadkar KA, Thomashow MF, Triezenberg SJ. 2006. Physical and functional interactions of Arabidopsis ADA2 transcriptional coactivator proteins with the acetyltransferase GCN5 and with the cold-induced transcription factor CBF1. Biochim Biophys Acta 1759: 69-79
- Terrell CR, Listenberger LL. 2017. Using Molecular Visualization to Explore Protein Structure and Enhance Student Facility with Computational Tools. BAMBEd 45: 318-328.
- Hati S, Bhattacharyya S. 2016. Incorporating modeling and simulations in an undergraduate biophysical chemistry course to promote understanding of structure-dynamics-function relationships in proteins. BAMBEd 44: 140-159.
- Jaswal SS, O'Hara PB, Williamson PL, Springer AL. 2013. Teaching structure: student use of software tools for understanding molecular structure in an undergraduate biochemistry course. BAMBEd 41: 351-359.
- Wightman B, Hark AT. 2012. Integration of bioinformatics into an undergraduate biology curriculum and the impact on development of mathematical skills. BAMBEd 40: 310-319.
- Banta LM, Crespi EJ, Nehm RH, Schwarz JA, Singer S, et al. 2012. Integrating Genomics Research Throughout the Undergraduate Curriculum: A Collection of Inquiry-Based Genomics Lab Modules. CBE-Life Sciences Ed 11: 203-208.
- Hall, BG. 2011. Phylogenetic Trees Made Easy, 4(th) edition. Sunderland, MA: Sinauer Associates, Inc.
Article Files
Login to access supporting documents
Understanding Protein Domains: A Modular Approach(PDF | 140 KB)
S1. Understanding Protein Domains-Module I assignment.doc(DOC | 52 KB)
S2. Understanding Protein Domains-Module II assignment.doc(DOC | 51 KB)
S3. Understanding Protein Domains-Module III assignment.doc(DOC | 54 KB)
S4. Understanding Protein Domains-Module IV assignment.doc(DOC | 52 KB)
- License terms

Comments
Comments
There are no comments on this resource.