A Close-Up Look at PCR
Published online:
Abstract
The Polymerase Chain Reaction (PCR) is a fundamental laboratory technique that allows for the amplification of many copies of a desired DNA target sequence. Despite its prevalence, undergraduate students often have poor comprehension about the underlying molecular mechanisms of PCR and the components necessary to carry out the reaction. We designed an interactive modeling activity to help students visualize the underlying molecular processes of denaturation, annealing, and extension, and to see how PCR parallels in vivo DNA replication. During the activity, students mimic denaturation, annealing, and extension by synthesizing DNA strands from individual nucleotides and primers in the 5' to 3' direction. Because they carry out three cycles, students construct and observe the intermediate products that lead to the exponential amplification of the target sequence. Instructors can easily assemble kits from relatively inexpensive foam nucleotide pieces, and the models can be reused indefinitely. Field testing with first and second year undergraduates suggested that students productively interacted with the models to improve their understanding of PCR.
Citation
Donahue, C.J., Adair, A.A., Wright, L.K., and Newman, D.L. 2019. A Close-Up Look at PCR. CourseSource. https://doi.org/10.24918/cs.2019.3Society Learning Goals
Genetics
- Methods & Tools in Genetics
- What experimental methods are commonly used to analyze gene structure and gene expression?
Science Process Skills
- Process of Science
- Interpret, evaluate, and draw conclusions from data
- Construct explanations and make evidence-based arguments about the natural world
- Modeling/ Developing and Using Models
- Recognize the important roles that scientific models, of many different types (conceptual, mathematical, physical, etc.), play in predicting and communicating biological phenomena
- Make inferences and solve problems using models and simulations
- Build and evaluate models of biological systems
- Quantitative Reasoning/ Using Mathematics and Computational Thinking
- Apply the tools of graphing, statistics, and data science to analyze biological data
- Communication and Collaboration
- Share ideas, data, and findings with others clearly and accurately
Lesson Learning Goals
- Students will understand the molecular basis of PCR
Lesson Learning Objectives
At the end of this lesson students will be able to...- Describe the role of a primer in PCR
- Predict sequence and length of PCR product based on primer sequences
- Recognize that primers are incorporated into the final PCR products and explain why
- Identify covalent and hydrogen bonds formed and broken during PCR
- Predict the structure of PCR products after each cycle of the reaction
- Explain why amplification proceeds exponentially
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Polymerase Chain Reaction (PCR) is an integral tool in the biology laboratory. Developed in the 1980s, PCR is a technique that allows an investigator to replicate a target region of DNA, resulting in the amplification of a specific genetic sequence (1-3). Because PCR can be used in a wide-range of applications (e.g., cloning, genotyping, phylogenetic analysis), it has become a standard part of most biology laboratory courses. Research demonstrates, however, that undergraduate students often do not understand the fundamental mechanisms of PCR (4-7).
Because PCR is often only one part of an experimental strategy, learning activities may not put enough emphasis on helping students learn specifically about PCR. For example, the activity designed by Haydel and Stout (4) combined the processes of PCR, restriction digests, and cloning techniques. While the activity overall provided a good representation of cloning, it did not provide a deep focus on the molecular interactions that occur during PCR. Many online resources about PCR exist but the viewer is usually a passive bystander during the tutorials.
PCR is a dynamic and complex process that involves repeating steps of template denaturation, primer binding, and incorporation of nucleotides into the growing strand in the 5' to 3' direction. The dynamic nature of this process may be difficult for students to conceptualize when using typical textbook resources as illustrations and diagrams are static. Additionally, the explanations and illustrations of PCR designed to help introductory students understand the process are typically simplified to diagrams representing DNA with arrows and solid lines, rather than emphasizing the molecular nature of the process and complementary binding of primers to template (unpublished data). Research suggests undergraduates struggle to conceptualize the underlying molecular/submicroscopic scale when learning topics related to genetic information (8-10). We suggest that when molecular-level details are missing from textbook and online representations of PCR (e.g. showing target DNA as a rectangular block), students are not primed to think about PCR on a molecular scale.
The DNA Triangle is a framework proposed to help educators and students to conceptualize and integrate the chromosomal, informational and molecular levels of DNA when thinking about complex molecular genetics concepts (11). Applied to PCR, the chromosomal level is akin to the macroscopic view of DNA during the process; the separation of DNA strands and the resulting expansion of the target DNA product. The informational level can be aligned with the application or underlying reason for carrying out PCR; to visualize the presence of a particular gene or, depending on the specificity of primers, the pattern of alleles present in a particular genome. The molecular level explains how the PCR process works; single stranded DNA primers bind in a sequence specific manner to two complementary regions flanking the double-stranded target sequence. We hypothesize most teaching resources focus on the informational and chromosomal levels of DNA when explaining PCR to learners at the expense of the molecular level.
A growing body of evidence shows the incorporation of physical, interactive models into course activities helps students understand foundational concepts in molecular biology and chemistry (12-14). To help students understand the molecular basis of PCR, we designed a model-based activity that allows students to "see" the underlying molecular interactions that are essential for PCR. To create an activity to help our students understand the molecular nature of PCR, we first created a paper-based PCR modeling activity (which included DNA sequence) to an honors section of Introductory Biology students. The activity involved cutting out DNA sequences from strips of paper, binding primers via taping paper oligonucleotides, and writing out DNA sequences. After we administered the activity and corresponding worksheet to our class, we discovered students still had a lot of confusion about PCR. Additionally, having students cut strips of paper during the activity may have led some students to conclude that restriction enzymes were part of PCR and that a shorter target product was obtained by simply cutting the longer template being cut.
Using feedback and observations from the early iteration of this activity, we revised our activity extensively and created an improved physical model-based activity. Using the foam nucleotide pieces from 3-D Molecular Design's Flow of Genetic Information Kit(C) , we designed an interactive model that helps students understand denaturation, primer annealing and primer extension. Students use pre-designed templates and primers with free nucleotide pieces to complete three rounds of PCR. Pre-post testing and field observations during the activity showed improvement in understanding the mechanism of PCR, identifying the components of the process, understanding primer binding specificity and the exponential nature of PCR.
Intended Audience
The intended audience for this activity is novice undergraduate students majoring in biology or a related field; however, the activity could be used with advanced-level high school biology courses or upper level undergraduate courses that require a thorough understanding of PCR. The activity was developed for a class of 24 students in an honors-level Introduction to Biology course and has also been used in a Molecular Biology course. Whatever the course size, we suggest that students work in groups no larger than six for this activity.
Students typically come to our classes with a varied background of PCR experience both at the practical level and conceptual level. Few students seem to have a very complete understanding of PCR, even if they have used this technique in the past (e.g., through coursework or a research project). Because different students bring different experiences and perspectives, they may be able to help each other achieve deeper understanding during the activity.
Required Learning Time
The time needed to complete the hands-on model-based activity and worksheet is approximately 60 minutes.
Prerequisite Student Knowledge
Students should have prior knowledge of nucleotide and DNA structure, as well as the cellular process of DNA replication, including a basic understanding of the function of DNA polymerase, helicase, primers, deoxyribonucleotides (dNTPs) and templates. If students have used the Flow of Genetic Information Kit(C) (3D Molecular Designs) in the past, they will be familiar with the foam nucleotide pieces and may remember how to identify the nitrogenous base, sugar group, phosphate group and 3' and 5' ends of the molecule. If students are unfamiliar with the kit, the instructor should help students compare the foam nucleotide pieces to the chemical structure of dNTPs and DNA.
Prerequisite Teacher Knowledge
Instructors must have working knowledge of the process of DNA replication and PCR. If instructors have not worked with the Flow of Genetic Information Kit(C), they should take the time to familiarize themselves with the foam puzzle-like pieces that represent nucleotides and how they form a strand of DNA.
SCIENTIFIC TEACHING THEMES
Active Learning
This highly visual and kinesthetic learning activity encourages students to model three rounds of PCR by interacting with physical models. During the activity students pull apart a model of double stranded DNA to mimic denaturation, identify 3' and 5' ends of primers and DNA strands by examining model pieces, identify primer binding sites in the template DNA by comparing nucleotide sequences, anneal complementary primers to the correct locations on the template strands, and mimic DNA synthesis by incorporating complementary nucleotide pieces on the 3' ends of annealed primers. The activity worksheet provides directions and scaffolding support for learners and allows students to reflective about what they experienced and draw conclusions from the activity.
Assessment
Instructors measured learning through both formative and summative assessment. During the model-based activity, students complete a worksheet that requires them to record primer binding sequences, draw/illustrate the comparative lengths of DNA sequences at various points throughout the PCR process, recognize PCR as a DNA synthesis reaction, and recognize the various steps and mechanisms within the PCR process. Summative assessment is facilitated by an eight-question quiz composed of open response, forced choice, and multiple select questions. This assessment was designed to probe students' knowledge about the mechanisms and products of PCR. In the assessment, students are expected to relate primer binding to the specificity of the reaction, relate PCR to the synthesis of macromolecules, explain the exponential nature of PCR, explain at each step how the different chemical bonds found in DNA are impacted, and predict the products of PCR.
Inclusive Teaching
The model-based activity allows students to "see" a dynamic molecular process that is not directly visible to them. As groups of students work through this activity, they create a physical model that becomes the shared mental model for how PCR works. Because the modeling activity is administered to small groups and requires three rounds of "PCR" (template denaturation, primer annealing and synthesis), it encourages all students to get involved and contribute to the activity. The nature of the foam pieces (i.e., all the same shape, but different colors) encourages students to work together to identify primer binding sites and hunt for the next complementary nucleotide during extension. All students in the group, regardless of experience level or English Language status, will have something to point to, pick up and manipulate which allows for non-verbal communication to occur during the activity.
LESSON PLAN
Overview
In this lesson, students are exposed to an interactive modeling activity to demonstrate the process of PCR. The activity focuses on hands-on learning using a physical model. This way, students avoid dealing with cognitive overload or failure to notice salient details, and instructors can more easily assess and appropriately correct individual comprehension of PCR. Table 1 provides a recommended timeline for instructors. Table 2 provides a list of specific responses for instructors to employ if they notice or hear a student behavior while engaging with the model.
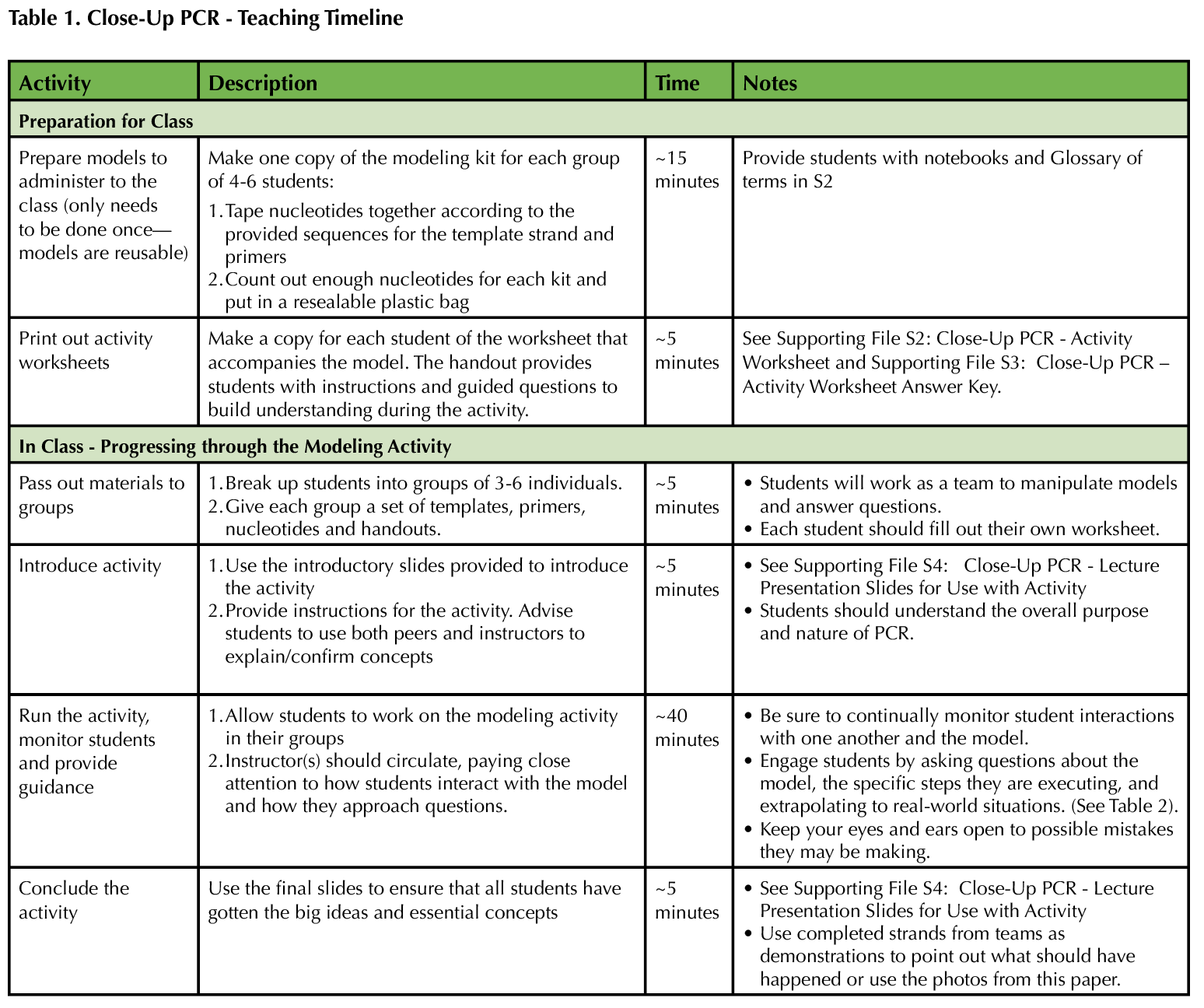
Table 1. Close Up PCR - teaching Timeline
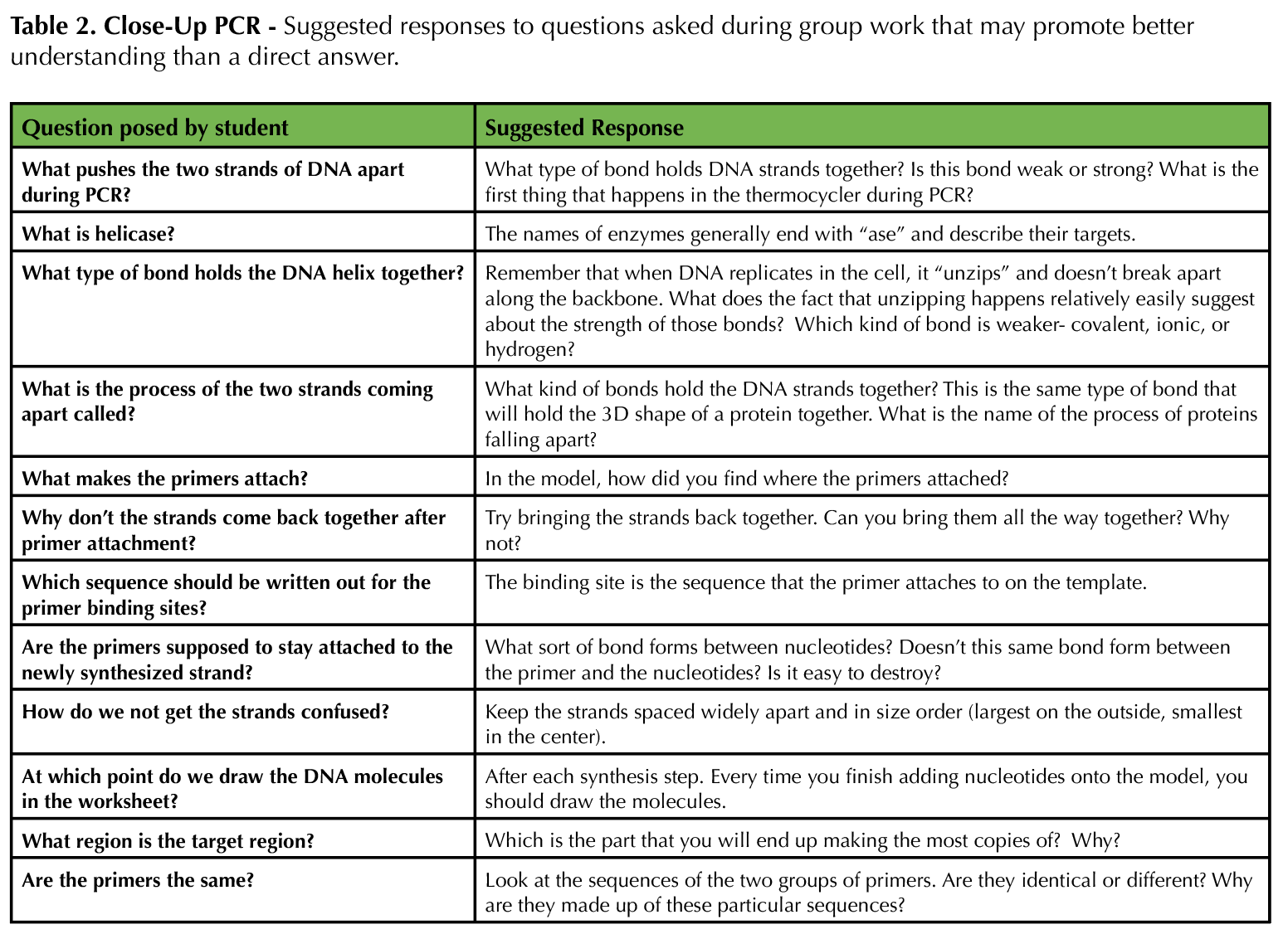
Table 2. Close-Up PCR - Suggested responses to questions asked during group work that may promote better understanding than a direct answer.
Pre-Class Preparation
This activity assumes that students have already been introduced to the process of DNA replication, and we believe it is helpful to pair the activity with a laboratory exercise where students perform PCR for real. Instructors will need to dedicate time to the construction of the materials, but once created, they are reusable for future classes. The kits are assembled from foam pieces purchased from 3D Molecular Designs(R) (FGIK replacement nucleotides: each set contains 40 deoxyribonucleotides of each type) and foam tape. Note that the nucleotides are not permanently altered and only held together with removable tape, so they can be repurposed for another use if desired. Multiple kits will be required per class; we recommend groups of three to six students per kit to maximize learning comprehension. Step-by-step instructions and visual representations on how to build the kits can be found in Supporting File S1: Close-Up PCR - Instructions for Assembling Kits. Creating the appropriate number of primers for each kit is essential for ensuring that students do not attempt to "reuse" primers and thus break them off from synthesized PCR products. Similarly, it is important to have enough nucleotides that students will not need to break apart their strands to make new ones.
Before class, individual activity worksheets must be printed for each student. To cut down on grading load and promote interaction among students, we recommend having each group turn in a single worksheet. Instructors who chose to do that should print a separate copy of the activity for each group on colored paper.
Instructors should have a full understanding of how the model works and the concepts covered before implementing the activity. Looking over the pre-and post-test questions, referring to the learning goals, and reviewing Supporting Files S2-S4 will help instructors comprehend the full extent of the model's purpose so that they get the most out of the activity. The activity runs more smoothly with multiple instructors/TAs facilitating, as students will often try to take shortcuts that counteract the lesson or fail to make important connections if not monitored.
If desired, the pre-test can be given at any time before the activity is implemented (Supporting File S5: Close-Up PCR - Pre/Post Assessment with Answer Key).
During class
Instructors
Although we provide a fact sheet informing students about the components of the modeling activity, it is beneficial to explain how the foam pieces of the kit represent the nucleotides that are assembled into DNA during PCR to students prior to the activity's start. In our class, students had used these pieces previously for learning about DNA replication and transcription, but if they are new to the students, ensuring that they know which end of each piece represents the 3' and 5' ends and the nitrogenous base will help prevent confusion or delay getting started. In addition, showing students which pieces represent the template strand of DNA, which position the template strands should be in, and which pieces represent the primers also help students get started. Though we suggest not telling students which primer represents the forward primer and which represents the reverse, organizing the primers into two separate groups (forward and reverse) on the table or lab bench helps shorten the activity's running time.
Instructors should explain that groups will receive a modeling kit to work with, along with an accompanying packet of instructions and questions to answer. It is important not to jump ahead of the worksheet, to let the worksheet guide them along the activity, and to work efficiently and effectively. Lastly, instructors should inform participants that everyone in the group must interact with the model as the activity progresses. Encourage students to ask clarifying questions along the way and pay close attention to the directions.
The instructor (and TAs if possible) should facilitate discussion if a group is struggling to collaborate, offer advice when students ask questions about the process of PCR and the modeling activity, ensure that students are synthesizing new strands of DNA properly, and keep students on track when their discussions start to veer towards common misconceptions. For a class of 24 (six groups), we had three facilitators. Table 2 provides suggestions of how to address student questions. Note that most questions are not answered directly; the Socratic method of teaching helps guide students down the path to building their own knowledge, rather than trying to provide answers that they are then more likely to forget. After the students complete the lesson, review the big ideas that students should take away. The final slides in Supporting File S4: Close-Up PCR - Lecture Presentation Slides for Use with Activity are useful for this purpose. If desired, the instructor can give the post-test (Supporting File S5: Close-Up PCR - Pre/Post Assessment with Answer Key) at any time to determine how well students achieved the learning objectives.
Students
Students should complete the activity in conjunction with the activity's accompanying worksheet. Each student should have an individual worksheet on which they record their own thoughts and answers to each question. Students should then come together to discuss potential question answers as a group, and then record a group answer on the group worksheet, which is identical to each individual student worksheet except printed on colored paper. This worksheet will be turned in and graded at the end of the activity.
The worksheet functions to guide the students through the model. In addition to asking questions about PCR, the activity instructs students when to complete each component of each PCR cycle (denaturation, annealing, extension) with the model. How each of these steps should be specifically performed with the model is explained below
Complete the first PCR Cycle (1 DNA molecule --> 2 DNA molecules; Figure 1)
- Denature: The two strands of foam nucleotides should be pulled apart from each other, resulting in two single strands. The strands come apart very easily, simulating breaking the relatively weak hydrogen bonds between bases.
- Anneal: Students should attempt to find each primer-binding site by picking up each primer and scanning along the single stranded template until a match is found. Each foam nucleotide fits into its complementary partner with puzzle-like precision, and each primer binds to one specific location on either the A or B DNA strand. Each primer should be bound to the template strand by fitting the piece into its complementary nitrogenous bases.
- Extend: Nucleotides should be sequentially inserted, one at a time, until the end of each template strand is reached. The first new nucleotide is added to the 3' (arrow) end of the primer, according to the complementary nitrogenous base of the template strand. It is important that students do not add the complimentary nucleotides non-sequentially, as this does not mimic actual DNA synthesis and could create an incorrect mental model of the PCR process. After complete extension, each group should have two DNA molecules that are partially double-stranded and partially single-stranded.

Figure 1. First Cycle of PCR. The top panel shows the model after the first denaturation step, the middle panel shows the model after primer annealing and the bottom panel shows the model after the first extension has been completed.
Complete the second PCR Cycle (2 DNA molecules --> 4 DNA molecules; Figure 2)
- Denature: Each DNA molecule generated in the previous cycle should be broken apart, resulting in four single-strands of DNA.
- Anneal: For each single strand of DNA, one primer, either forward or reverse, should be attached to its binding site. Two forward and two reverse primers will be needed to bind to the four single-strands of DNA.
- Extend: Nucleotides should be sequentially added to the 3' end of the primer, one at a time, until the end of each new template strand is reached. Upon completion of extension, each group should have four partially double-stranded DNA molecules.
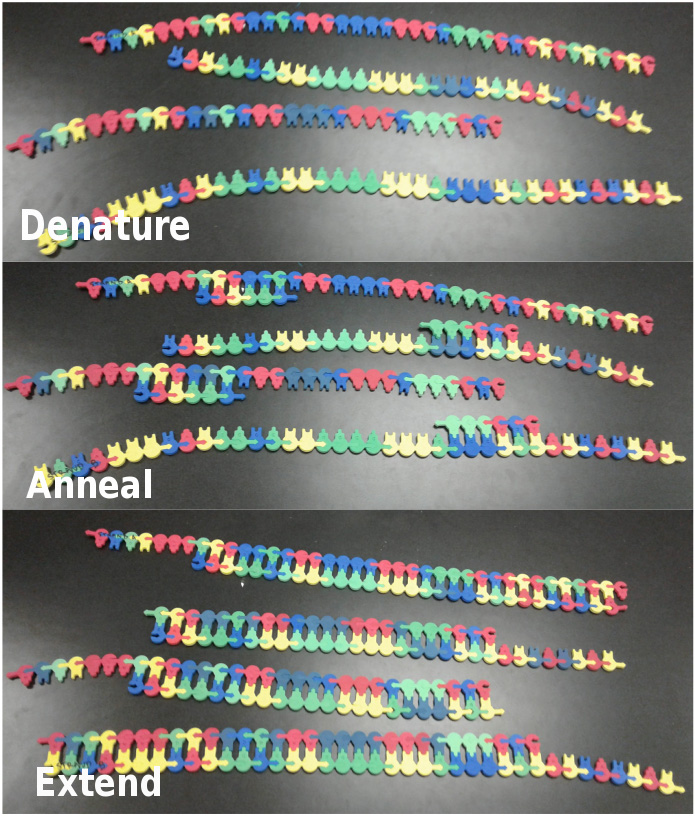
Figure 2. Second Cycle of PCR. The top panel shows the model after the second denaturation step, the middle panel shows the model after primer annealing and the bottom panel shows the model after extension has been completed a second time.
Complete the third PCR Cycle (4 DNA Molecules --> 8 DNA molecules; Figure 3)
- Denature: Each DNA molecule generated in the previous cycle should be broken apart, resulting in eight single-strands of DNA, with several different lengths.
- Anneal: For each single strand of DNA, one primer, either forward or reverse, should be attached to its binding site. Four forward and four reverse primers will be needed at this stage.
- Extend: Nucleotides should be sequentially added to the 3' end of the primer, one at a time, until the end of each new template strand is reached. At the end of this step, each group should have eight molecules of DNA, six of which are partially single-stranded and two that are completely double-stranded. The molecules without single-stranded overhangs represent the amplified target region of the DNA. Students should now be able to visualize what happens in later steps--this shortest molecule is amplified exponentially, the middle-sized ones increase in a linear fashion, and no new full length strands are ever made.

Figure 3. Third Cycle of PCR. The top panel shows the model after the third denaturation step, the middle panel shows the model after primer annealing and the bottom panel shows the model after extension has been completed a third time.
TEACHING DISCUSSION
Activity's effectiveness at achieving the stated learning goals and objectives
We field tested the activity on students enrolled in a Cell & Molecular biology class (n=24), who were given the pre/post assessment (Supporting File S5). Based on verbal and written feedback, after participating in the activity, students had a better understanding about the molecular basis of PCR. Even students that claimed to be familiar with and have experience doing PCR found the activity beneficial. The self-correcting nature of the activity (i.e. the single stranded primer is complementary to only one stretch of sequence in the template) allowed students to observe that primer binding occurs through sequence complementarity and why the location of primers delineates the final products of PCR. By incorporating new nucleotides (one at a time) at the 3' end of the annealed primers, students realized why primers are incorporated into final PCR products. Because the foam nucleotide pieces from the Flow of Genetic Information Kit (3D Molecular Designs(R)) were designed to help students differentiate covalent bonds (stronger connections) and Hydrogen bonds (weaker connections) students are able to identify the covalent and Hydrogen bonds created during PCR. Table 3 illustrates how the activity and the pre/post assessment questions align with the stated Learning Objectives.
Pre/post assessment indicated that students improved in their ability to identify primers and templates, explain their roles in PCR, predict the structure of PCR products, and describe the exponential amplification of product (Figure 4). Questions were a mixture of open-ended and multiple select formats, so students could receive partial credit for each of the seven questions. The only question that did not show significant improvement was #6, which has been reworded for improved clarity in Supporting File S5: Close-Up PCR - Pre/Post Assessment with Answer Key. We did not test this revised version of the question.
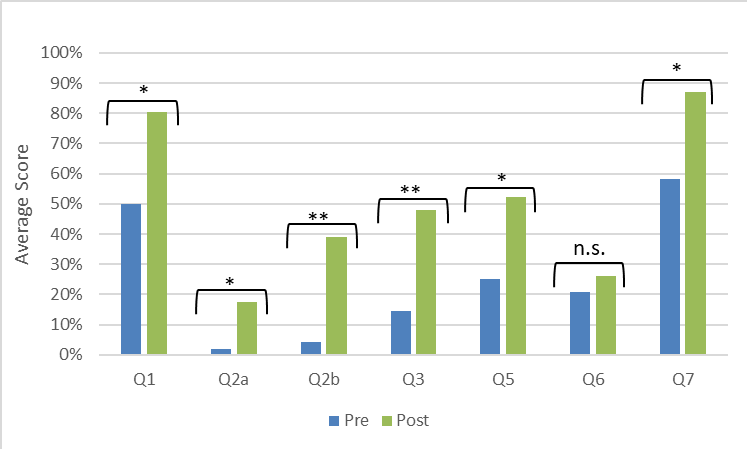
Figure 4. Students improved their understanding of the concepts after participating in the activity. The assessment questions (see Supporting File S5: Close-Up PCR - Pre/Post Assessment with Answer Key) were designed to address common misconceptions related to PCR. Question 4 was excluded from analysis due to a confusing typo in the original assessment. Significance of the differences were determined by t-tests: * p < 0.05, ** p<0.01.
Reactions to the lesson
In general, students liked the activity and had fun assembling the DNA strands. Although the activity was carefully designed to help students learn how PCR "works" at the molecular level, we emphasize that an observant instructor, TA or learning assistant must also be involved in the activity or students may try to skip steps or devise a strategy that lets them finish the activity very quickly that short-circuits the learning process. During our trial runs, we have observed students synthesizing new DNA strands by what we called "chunk synthesis"; students will build short stretches of complementary DNA (by looking at the sequence) without building them off of the template. These chunks may be randomly inserted until the stretch between the primers are filled in. Students may also try to insert all of the As at once, then all of the Gs, etc. This tactic may lead to poor comprehension because it does not demonstrate the correct 5' to 3' directionality of synthesis, does not emphasizes the role of the primer (to provide a free 3'OH end on which to extend) or reinforce the idea that primers are part of the final PCR product. Students may also try to skip building the intermediate DNA structures produced in rounds 1 and 2 of PCR. This short-cut should also be discouraged because students won't visualize the various intermediate DNA structures that form during the reaction.
Adaptations
We also implemented this activity during a laboratory portion of an Introductory Biology course to help prepare students for an upcoming wet-bench PCR experiment. Though the activity can be completed without a laboratory component, the model-based activity may help students achieve a better understanding of actual PCR results. It could easily be extended to support other activities, such as reading a primary literature paper, working through a case study or solving a forensics problem set. While we tested this activity with first and second year students, this model has also been used with a more advanced upper-division laboratory course. In our experience, even the upper-level students, who have experience running PCR in the lab, seemed to enjoy and learn from the activity. This activity could also be used with non-majors biology students, a forensics-type laboratory/course or even an Advanced Placement (AP) or International Baccalaureate (IB) High school biology students. While we believe group size should be kept to six or fewer students, an instructor could scale this activity up for a large class size if s/he had enough nucleotide pieces and Teaching Assistants to help. We only recommend doing this activity in a classroom setting if students have access to large desks or tables because the completed models take up so much physical space. Instructors of very large courses (>100 students) may consider using this model-based activity during recitation or review sessions when a smaller number of students would be present.
SUPPORTING MATERIALS
- S1: Close-Up PCR - Instructions for Assembling Kits. This file gives instructions with visual aids to help the instructor put together the materials needed for the lesson.
- S2: Close-Up PCR - Activity Worksheet. This is the handout for students with instructions for carrying out the simulated PCR and questions for them to answer.
- S3: Close-Up PCR - Activity Worksheet Answer Key.
- S4: Close-Up PCR - Lecture Presentation Slides for Use with Activity.
- S5: Close-Up PCR - Pre/Post Assessment with Answer Key.
ACKNOWLEDGMENTS
We thank the Thomas H. Gosnell School of Life Sciences at RIT for purchasing materials to construct the models and the Learning Assistant Program at RIT for providing support for CJD and AAA.
References
- Innis MA, Gelfand DH, Sninsky JJ, White TJ. 2012. PCR Protocols: A Guide to Methods and Applications. Academic Press.
- Mullis KB. 1990. The Unusual Origin of the Polymerase Chain Reaction. Sci Am 262:56-65.
- Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. 1986. Specific Enzymatic Amplification of DNA In Vitro: The Polymerase Chain Reaction. Cold Spring Harb Symp Quant Biol 51:263-273.
- Haydel SE, Stout V. 2015. A Kinesthetic Modeling Activity to Teach PCR Fundamentals. CourseSource 2.
- Phillips AR, Robertson AL, Batzli J, Harris M, Miller S. 2008. Aligning Goals, Assessments, and Activities: An Approach to Teaching PCR and Gel Electrophoresis. CBE-Life Sci Educ 7:96-106.
- Robertson AL, Phillips AR. 2008. Integrating PCR Theory and Bioinformatics into a Research-oriented Primer Design Exercise. CBE-Life Sci Educ 7:89-95.
- Wright LK, Newman DL. 2013. Using PCR to Target Misconceptions about Gene Expression. J Microbiol Biol Educ 14:93-100.
- Johnstone AH. 1991. Why Is Science Difficult to Learn? Things Are Seldom What They Seem. J Comput Assist Learn 7:75-83.
- Newman DL, Catavero C, Wright LK. 2012. Students Fail to Transfer Knowledge of Chromosome Structure to Topics Pertaining to Cell Division. CBE-Life Sci Educ 11:425-436.
- Newman DL, Wright LK. 2017. Meiosis: A Play in Three Acts, Starring DNA Sequence. CourseSource https://doi.org/10.24918/cs.2017.9.
- Wright LK, Catavero CM, Newman DL. 2017. The DNA Triangle and Its Application to Learning Meiosis. Cell Biol Educ 16.
- Newman DL, Stefkovich M, Clasen C, Franzen M, Wright LK. 2018. Physical Models can provide superior learning opportunities beyond the benefits of active engagement. In review.
- Copolo CE, Hounshell PB. 1995. Using three-dimensional models to teach molecular structures in high school chemistry. J Sci Educ Technol 4:295-305.
- Linn MC, Petersen AC. 1985. Emergence and characterization of sex differences in spatial ability: a meta-analysis. Child Dev 56:1479-1498.
Article Files
Login to access supporting documents
A Close-Up Look at PCR(PDF | 1 MB)
S1. Close-Up PCR - Instructions for assembling kits.docx(DOCX | 3 MB)
S2. Close-Up PCR - Activity Worksheet.docx(DOCX | 47 KB)
S3. Close-Up PCR - Activity worksheet answer key.docx(DOCX | 110 KB)
S4. Close-Up PCR - Lecture presentation slides.pdf(PDF | 354 KB)
S5. Close-Up PCR - PrePost assessment with key.docx(DOCX | 197 KB)
- License terms

Comments
Comments
There are no comments on this resource.