CURE-all: Large Scale Implementation of Authentic DNA Barcoding Research into First-Year Biology Curriculum
Published online:
Abstract
Growing calls in science education reform have emphasized wide-scale engagement of first-year undergraduate students in authentic research experiences; however, large course enrollments, inadequate student experience, limited resources and departmental inertia often create obstacles to reaching this goal. To help overcome these obstacles, the Department of Biology at James Madison University (JMU) has developed a cost-effective, scalable, and transferable semester-long (14-week) course-based undergraduate research experience (CURE) designed for large enrollment introductory biology labs. In this series of labs, first-year students use DNA barcoding to engage in authentic research practices drawn from the fields of ecology, molecular biology, and bioinformatics. These labs enable students to identify local species of plants, fungi, and invertebrates using student-generated DNA barcode sequences, which are then shared through a public database. Since their implementation at JMU in 2016, students in these labs have created and shared over 1,500 unique DNA barcode sequences and documented over 300 local species of plants, fungi, and invertebrates. These data are being used in an ongoing project comparing the biodiversity of forest edge versus forest interior habitats, but the labs are adaptable to almost any habitat or taxonomic group. In this article, we provide detailed descriptions of the content, logistics, and implementation of this 14-week series of labs. To our knowledge, this is among the largest-enrollment CUREs being offered to first-year undergraduates in the United States, and we hope that it can be useful to other institutions interested in documenting biodiversity and engaging introductory biology students in authentic research.
Citation
Hyman, O.J., Doyle, E.A., Harsh, J., Mott, J., Pesce, A., Rasoul, B., Seifert, K., and Enke, R.A. 2019. CURE-all: Large Scale Implementation of Authentic DNA Barcoding Research into First-Year Biology Curriculum. CourseSource. https://doi.org/10.24918/cs.2019.10Lesson Learning Goals
- Understand and apply the process of science
- Understand and apply the conventions of experimental design
- Understand the relationships between taxonomy, evolution, and cladistics
- Understand the importance of biodiversity
- Understand and apply basic techniques and concepts from ecology, evolutionary biology, molecular biology, and bioinformatics
- Understand and apply the conventions of displaying scientific data and communicating research findings
Lesson Learning Objectives
Students will be able to: Week 1-4: Fundamentals of Science and Biology- List the major processes involved in scientific discovery
- List the different types of scientific studies and which types can establish causation
- Design experiments with appropriate controls
- Create and evaluate phylogenetic trees
- Define taxonomy and phylogeny and explain their relationship to each other
- Explain DNA sequence divergence and how it applies to evolutionary relationships and DNA barcoding
- Define and measure biodiversity and explain its importance
- Catalog organisms using the morphospecies concept
- Geographically map organisms using smartphones and an online mapping program
- Calculate metrics of species diversity using spreadsheet software
- Use spreadsheet software to quantify and graph biodiversity at forest edges vs. interiors
- Write a formal lab report
- Extract, amplify, visualize and sequence DNA using standard molecular techniques (PCR, gel electrophoresis, Sanger sequencing)
- Explain how DNA extraction, PCR, gel electrophoresis, and Sanger sequencing work at the molecular level
- Trim and assemble raw DNA sequence data
- Taxonomically identify DNA sequences isolated from unknown organisms using BLAST
- Visualize sequence data relationships using sequence alignments and gene-based phylogenetic trees
- Map and report data in a publicly available online database
- Share data in a formal scientific poster
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Engagement in undergraduate research (UR) is widely held by the scientific community as one of the most impactful learning practices for students in preparation for the science careers and challenges of the 21st century. Reflecting this view, national calls in reform documents over the prior three decades have advocated the wide-scale adoption of authentic research as a feature of the college science experience for all students (e.g., 1-4). As a scalable and inclusive means to engage students in research, Course-based Undergraduate Research Experiences (CUREs) are becoming increasingly popular as a standard part of the curriculum at a growing number of institutions. Prior studies have documented a wide range of outcomes as potential benefits of CUREs for students, including the enrichment of research-related skills, increased understanding of the process of scientific discovery, and enhanced interest in STEM careers (5-8). Students who participate in CUREs are also more likely to graduate and complete STEM degrees - especially students from groups traditionally underrepresented in these fields (e.g. 9-10).
In light of such potentially positive impacts, STEM educators and administrators have been encouraged to include discovery-based CUREs within the first two years of college; a critical time in the recruitment and retention of STEM majors (4). While an emerging catalogue of CURE models have been reported (see https://serc.carleton.edu/curenet/index.html), the ability to scale these types of experiences to meet the needs of large-enrollment first-year courses can be difficult (1). Furthermore, the generalizability and transferability of some CUREs that have been successfully developed for large first-year courses may be limited if they depend heavily on extramural funding, rely on tenure-track faculty/primary investigator investments, or require access to a very specific geographic location or organism. To offset such limitations and better align with national reform efforts, we developed a transferable, scalable, and low-cost semester-long (14-week) research experience based on a DNA barcoding experimental workflow (Figure 1).
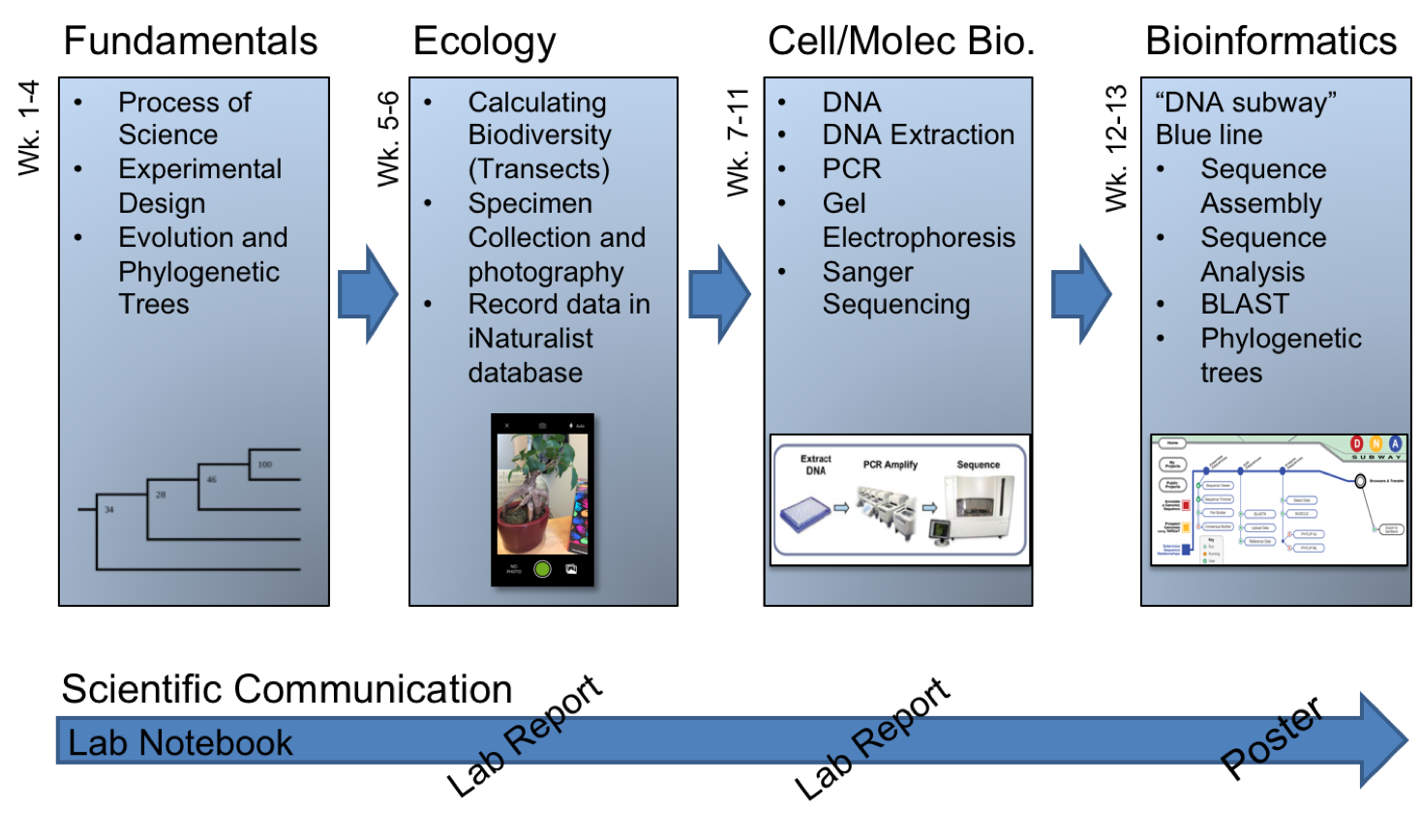
Figure 1. Overview of the topics covered during this semester-long DNA barcoding lab experience
DNA barcoding is a molecular technique that can help non-experts determine the taxonomic identity of living specimens using short (~500 base pair) DNA sequences, or barcodes (11). Just like the barcode on a box of cereal tells the grocer what brand it is, DNA barcodes act as unique nucleotide sequences that can often reveal the genus or species identity of a biological specimen. Though not entirely foolproof, over the last decade, DNA barcodes have been developed and verified for a wide variety of taxa, making them useful in the identification of a wide variety of organisms (11). In some cases, DNA barcoding can provide an alternative to traditional taxonomic identification, which often relies on expert morphological analysis of whole specimens and the ability distinguish subtle differences between closely related species. DNA barcodes, on the other hand, can be identified using tissue from damaged or incomplete specimens, and enable non-expert users, such as college undergraduates, to reliably identify organisms using a simple and repeatable workflow.
The DNA barcoding workflow requires extracting DNA from fresh, frozen, damaged, or processed biological samples. After DNA extraction, barcoding regions are amplified by polymerase chain reaction (PCR), visualized via gel electrophoresis, and subsequently sequenced using taxa specific primers (12). DNA sequences can then be analyzed through freely available, user-friendly software that uses curated databases of known organisms to determine the taxonomic identity of the barcoded sequence. Because DNA barcoding can be used in tandem with classical morphological analysis to identify taxa, it has a wide variety of research applications including biodiversity assessments (13), disentangling cryptic species complexes (14), or uncovering product mislabeling (15). This has resulted in several large DNA barcoding initiatives (http://www.ibol.org/phase1/about-us/campaigns/) .
Previous studies have also demonstrated the effectiveness of DNA barcoding as a tool in biology education (16-19). This is in part because the aforementioned barcoding workflow is engaging, open-ended, easy to implement, and flexible in the questions it can address. It also incorporates important tools, technologies, and concepts from a broad range of biological disciplines. Perhaps most importantly, DNA barcoding allows relatively inexperienced users to create reliable data for larger scientific endeavors (16-19). For these reasons, several DNA barcoding teaching and outreach programs already exist.
For example, at the secondary school level, the Cold Spring Harbor Laboratory DNA Learning Center has created several after-school programs that select and train small teams of high schoolers and educators to document the biodiversity of Long Island and the five boroughs of New York City using DNA barcoding (https://www.dnabarcoding101.org/programs/). Similarly, Harris and Bellino (16) developed a year-long DNA barcoding curriculum used by high school aged students to document biodiversity in New York City and Belize. In Canada, Barcoding Life's Matrix (http://www.studentdnabarcoding.org) provides resources and workshops to train 11th and 12th grade teachers to guide their students through a three-week DNA barcoding pipeline culminating in contributions to an international DNA barcode database (17). Even younger students (ages 9+) are participating in a national scale, two-week program in which students use Malaise traps to capture insects which are barcoded by the Centre for Biodiversity Genomics (CBG) at the University of Guelph, Ontario (18).
At the post-secondary level, upper division biology students at University of California San Diego are barcoding local organisms as part of a two-week long module (http://sdbiodiversity.ucsd.edu) aimed at documenting local biodiversity (19). Similarly, students in their last semester of the biotechnology program at Tulsa Community College in Oklahoma, participate in a three-week DNA barcoding module in which students identify local organisms and present their findings to the community (19).
Despite this great wealth of resources and programs currently available for incorporating DNA barcoding into the classroom, few of these programs are specifically targeted at large enrollment, first-year introductory undergraduate courses. Nor do these courses typically include lesson plans that explicitly address some of the core concepts underlying DNA barcoding research, such as experimental design, evolution, reading phylogenetic trees, and the general process of scientific discovery. In this article, we build on currently available DNA barcoding education resources by outlining the design, content, and logistics of a scalable, semester-long (14-week) sequence of lab modules specifically designed for implementation in a large enrollment, first-year undergraduate biology course.
In these lessons, students use DNA barcoding to assess the biodiversity of different habitats from a campus arboretum. Students also learn core biological concepts such as experimental design and reading phylogenetic trees. These modules should serve as an adoptable model for faculty interested in adding authentic research experiences into their introductory biology courses or developing CUREs to help first-year students collect and interpret DNA barcode data for biodiversity assessments, taxonomic identifications, or even product mislabeling.
Intended Audience
These lessons are intended for introductory biology students at a large research university, small college, community college, or high school seniors. In our labs, students are divided into laboratory sections of 24 students that meet once a week for two hours and 50 minutes for 14 weeks. Each laboratory section has an instructor and an optional undergraduate teaching assistant. Depending on the activity, students work individually, in pairs, or in teams of four, but each student is assigned an individual organism to identify via DNA barcoding.
Required Learning Time
This course is designed as a semester-long laboratory series with ~14, two to three-hour, weekly laboratory meetings (Supporting File S0.1: DNA Barcoding - Sample Syllabus.docx).
Pre-requisite Student Knowledge
No pre-requisite student knowledge is required, but very basic background knowledge about DNA, cells, and taxonomy is certainly advantageous.
Pre-requisite Teacher Knowledge
No pre-requisite teacher knowledge is required, though previous background in molecular biology, natural history, and ecology are certainly beneficial. The lab coordinator or lead organizer should have prior experience with basic techniques in molecular biology (pipetting, PCR, gel electrophoresis, and preparation of associated reagents: primers, agarose, TE buffer, etc.)
SCIENTIFIC TEACHING THEMES
Active Learning
To inform the development of the learning activities and broader course design, we integrated the five essential dimensions for a CURE as identified by Auchincloss and others (20). This framework involves the meaningful engagement of students in: 1: Multiple scientific practices that reflect activities comparable to the community of practice, such as asking questions, using various tools of science, gathering and analyzing data, navigating the messiness of real-world data, and communication. 2: Discovery through a novel research question or problem that may advance our understanding of the natural world. 3: Research that is relevant beyond the scope of the course with the potential of furthering scientific knowledge. 4: Collaborative in terms of student-student and student-instructor interactions for problem solving. 5: Iterative by providing opportunities for trying, failing, and trying again, which is central to the learning process as well as personal development (21). In addition, drawing from literature on apprentice-like undergraduate research (e.g., 22-23), emphasis was placed on providing multiple opportunities for 6: scientific communication in oral and written formats to contribute to the progression of research-related and life-long skills.
Given the nature of the design framework and associated course activities, students are inherently engaged as active learners. The described lessons encourage student autonomy as they navigate a wide variety of hands- and minds-on activities in the laboratory and in the field. Students work both individually and collaboratively over the term as they gain exposure to the disciplinary knowledge and research-skills necessary to successfully complete the DNA barcoding project as well as the broader nature of the process of science. For this, the lessons are designed with minimal lecturing and scaffolded to help students progressively advance their understanding and skills with the instructor taking the role of a lab mentor, providing enough guidance to keep students on track, while maintaining distance that promotes autonomy and project ownership.
Assessment
Formative assessments: demonstration of research-related skills (e.g., molecular techniques, communication), pre- and post-lesson worksheets, individual and group writing assignments, in-class exercises, in-class discussions, participation, and maintenance of a laboratory notebook.
Summative assessments: post-lesson quizzes, scientific posters, lab reports, and a cumulative final examination.
Self-reflection: evaluation of each group member's contribution to projects and activities including their own contribution.
Detailed information on these assessments is included in the outlines of each weekly lesson below. In addition, we've included a comprehensive list of assessments aligned with broad course objectives (Supporting File S0.2: DNA Barcoding - Alignment of objectives and assessments.docx).
Inclusive Teaching
The situation and design of the course facilitates an inclusive learning environment in multiple ways. First, as the gateway introductory biology course required for all biology majors and some other programs, all our biology students have equal access to an authentic research experience early in their academic career. Second, the term-long barcoding project is centered on exploring the natural diversity of the JMU Arboretum. These sorts of natural habitats can often be unfamiliar to students, so we make great efforts to make them accessible to all students regardless of their familiarity or comfort with nature. Thus, these labs provide an opportunity for all students to learn about the organisms and habitats in their own proverbial backyard. Furthermore, students are free to select the organism they investigate, which supports student comfort, creative autonomy, and project ownership. Likewise, the data students generate are added to an open-access species database that other researchers, arboretum staff, and the general public can use for research, outreach, learning natural history, or conservation purposes. Open access to these student-generated data not only enhances the inclusivity of these labs, but also has the potential to increase the meaningfulness of these labs for both students and non-student users. For example, data generated by students at JMU have been used in faculty research to identify unique or hard to identify species of beetles and beetle larvae. Students have also contributed data for fungal surveys conducted by the Mycological Society of America. Furthermore, students are free to select an organism of interest to investigate, which supports creative autonomy and project ownership. Third, as referenced above as a design framework feature, the field and lab activities are collaborative and require effective teamwork inside and outside of the classroom for the successful completion of tasks. Fourth, varying modalities of instruction incorporated into the lessons such as hands-on tasks, online modules, mini-lectures, discussions, and demonstrations, accommodate student preferences in learning. Similarly, as described above, varying forms of formative and summative assessments are used to measure student learning trajectories and guide instruction. As possible, assignments are criterion-scored using standardized rubrics made available in advance to make student expectations explicit (e.g. Supporting File S6.5: Week 06 - Biodiversity Lab Report - Guidelines, Rubric, and Template.docx, but see lesson plan descriptions below for detailed supporting files). Lastly, in that the lab activities are situated in three differing areas of study, ecology, molecular biology, bioinformatics, it helps speak to students with varying interests or proclivities.
LESSON PLAN
Broad Outline and Logistics
At JMU these labs typically enroll ~500-600 students per semester. Students are divided into laboratory sections of 24 students that meet once a week for two hours and 50 minutes in each of 14 weeks. Each laboratory section has an instructor and an optional undergraduate teaching assistant. Each laboratory section uses an online course management page (we use Canvas) where weekly assignments are posted, though labs could easily function without this software. Lab sections are linked to lecture, but the lab content is independent of the lecture content. In lab, students work individually, in pairs, and in groups of four, but each student gets to barcode their own organism that they select. Some activities ask students to use a shared digital notebook, in which case we use google docs, though other types of notebooks could be used.
Working together, students learn the underlying theory, concepts, and techniques needed to understand DNA barcoding, then apply this knowledge to catalogue the biodiversity of areas surrounding campus (Figure 1 & 2; Supporting File S0.1: DNA Barcoding - Sample Syllabus.docx). These labs are designed to have students compare the biodiversity of edge and interior habitats of a nearby forest, but they could easily be adapted to have students quantify biodiversity of any habitat of interest, even urban landscapes (https://www.dnabarcoding101.org/programs/). Each student collects a small piece (~1 cm diameter) of fresh tissue from an unknown plant, invertebrate, or fungus that they select, identifies the organism using DNA barcoding, then adds it to a publicly available database. Detailed sample collection protocols are included (see Supporting File S5.2: DNA Barcoding - Week 05 - Biodiversity 1 Handout INSTRUCTOR KEY.docx; and Supporting File S5.3: DNA Barcoding - Week 05 - Biodiversity 1 Prep Notes.docx), but typically, any sample of fresh, soft, fleshy, non-desiccated leaf, invertebrate, or fungal tissue works well.
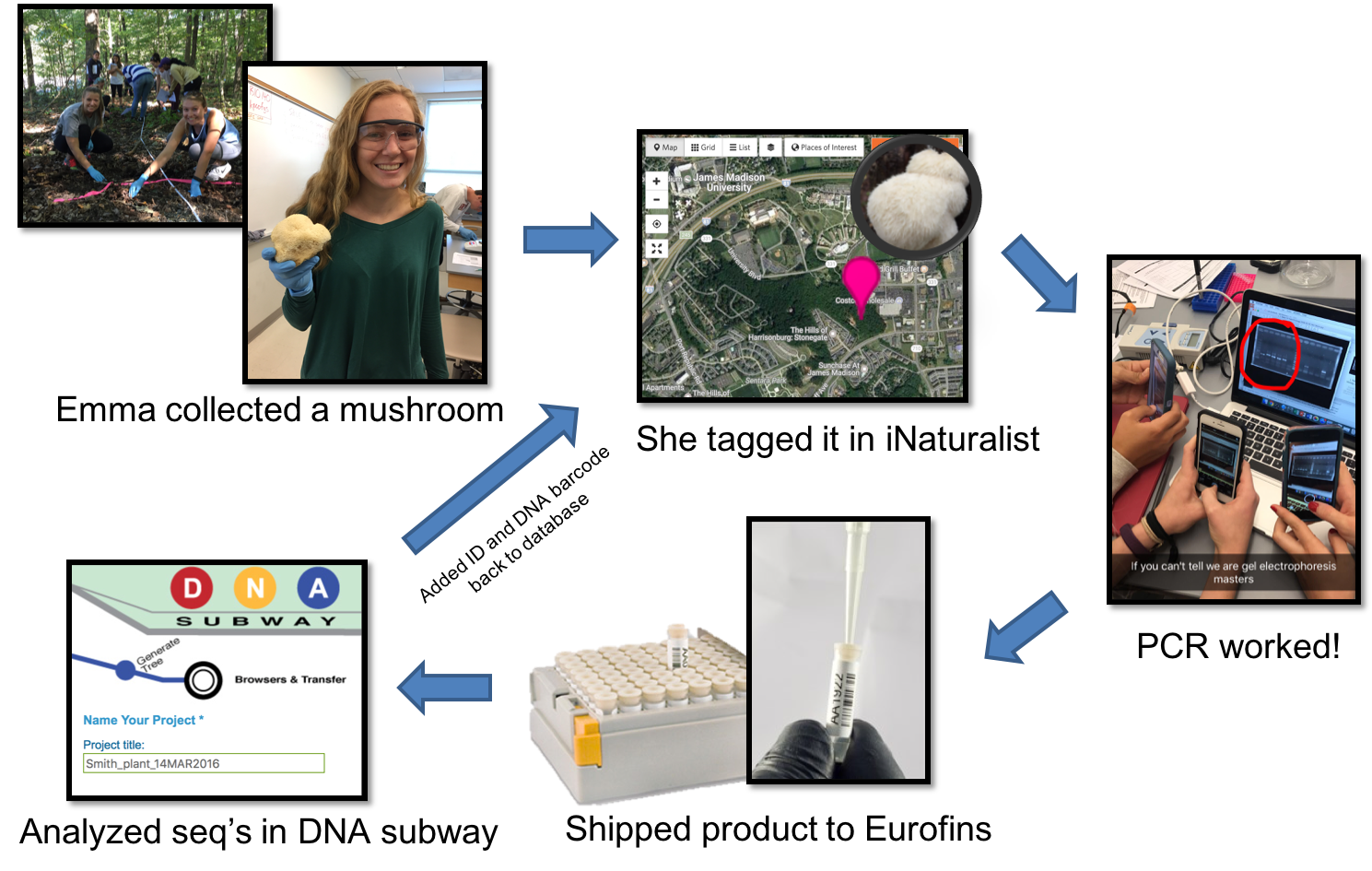
Figure 2: Lab workflow from the perspective of an individual student
Broadly speaking, these are an inquiry-based series of labs designed to teach students the basic skills, content knowledge, and theory needed to successfully identify an unknown organism using DNA barcoding and to share these findings (Figure 1). This lab sequence is designed to integrate concepts from a variety of scientific disciplines including ecology, evolutionary biology, systematics, taxonomy, molecular biology, and bioinformatics. These labs are also designed to help promote project ownership, enhance student confidence and self-identification as a scientist, and build a foundation in scientific writing and communication skills. The more specific goal of these labs is to use DNA barcoding to catalog and compare species diversity in forest edges versus interior habitats to address the broader question of how habitat type and degradation can influence biodiversity. Students sample organisms from forest edges and interior habitats in our campus arboretum and compile their data in a publicly available database to compare these habitats. In addition, as semesters pass and the database grows, students and instructors will have ability to add additional sites or develop labs to address additional questions such as temporal trends, succession, species accumulation curves, or introductions of new or non-native species. Alternatively, if these topics are not feasible or of interest to your institution, these labs can be adapted to identify specific taxonomic groups (ex. mosquitos), habitats (ex. urban parks), or questions relevant to your geographic location or faculty research interests. For example, honors sections of our labs have been involved in identifying species of dung beetles on farms, identifying fungi collected by the Mycological Society of America, and identifying cryptic species of tadpoles from Borneo. A more thorough discussion of alternative implementations of these labs is included in the teaching discussion section of this manuscript.
Regardless of the type of implementation, this 14-week, semester-long experience is broken into four basic units: (1) Fundamentals of Science and Biology, (2) Ecology, (3) Cell & Molecular Biology, and (4) Bioinformatics (Table 1; Figure 1&2). This parent document and the paragraphs that follow provide a general overview of what occurs during each week of these units. The details needed to teach each weekly lesson are provided as supporting documents, grouped by week. These supporting documents include lesson plans, lab preparation notes for the lab coordinator(s) and instructor(s), lecture slides, handouts, worksheets, and grading rubrics. Each of which is described in detail below.
Unit 1: Fundamentals of Science and Biology
Overview
Students spend the first four weeks of the semester in the Fundamentals unit conducting activities that teach about the process of scientific discovery, hypothesis testing, study design, taxonomy, phylogeny, and DNA sequence divergence. These activities lay the groundwork for students to understand the fundamental ideas underlying their upcoming semester-long project that involves quantifying biodiversity in forest edge vs. interior habitats, DNA barcoding, and bioinformatic analyses.
Week 1 - The Process of Scientific Discovery
Adapted with permission from https://undsci.berkeley.edu/lessons/introducing_flow_hs.html
(Detailed outline - Supporting File S1.1: DNA Barcoding - Week 01 - Process of Science Lesson Plan.docx)
This lesson adapted from https://undsci.berkeley.edu/lessons/introducing_flow_hs.html is an icebreaker activity that introduces students to the process of scientific discovery. Students begin with a simple warm up activity where they use mystery boxes (https://undsci.berkeley.edu/search/lessonsummary.php?&thisaudience=9-12&resource_id=204) to test hypotheses, conduct simple experiments, and collaborate with each other (Supporting File S1.2: DNA Barcoding - Week 01 - Process of Science Handout INSTRUCTOR KEY.docx) to decipher what's inside of a sealed box. Following this activity, the class discusses whether they were doing science and identify aspects of the activity that make it scientific. Next, students are introduced to a flowchart developed by educators at UC Berkeley (https://undsci.berkeley.edu/lessons/pdfs/complex_flow_handout.pdf) that depicts the process of science (Supporting File S1.3a: & Supporting File S1.3b: DNA Barcoding - Week 01 - Process of Science FLOWCHART.pdf). Students then read a story that describes how the geoscientist, Walter Alvarez, compiled over 20 years of evidence to support the hypothesis that an asteroid was responsible for the extinction of dinosaurs (https://undsci.berkeley.edu/lessons/pdfs/alvarez_hs.pdf). Students identify phrases within the story that indicate that Alvarez was doing science and use these to plot the pathway of Walter Alvarez through the process of science flowchart (Supporting File S1.4: DNA Barcoding - Week 01 - Process of Science Handout STUDENT.docx). Students are asked to identify important, and often misunderstood aspects of the process of science (Ex. pathway to discovery is often nonlinear, science involves creativity and collaboration, etc.). The process of science flow chart also acts a touchstone for students to reflect on their own activities throughout their semester-long DNA barcoding project. This lesson requires ~1.5 hours to complete and acts as a nice ice breaker that can easily be combined with a general introduction to the course (syllabus, laboratory safety, etc.) and any other first-lab-of-the-semester activities. Likewise, it can easily be made up by those students who don't enroll until the second week of class.
A detailed lesson plan (Supporting File S1.1: DNA Barcoding - Week 01 - Process of Science Lesson Plan.docx), instructor key (Supporting File S1.2: DNA Barcoding - Week 01 - Process of Science Handout INSTRUCTOR KEY.docx), student handout (Supporting File S1.4: DNA Barcoding - Week 01 - Process of Science Handout STUDENT.docx), teaching and preparation tips (Supporting File S1.5: DNA Barcoding - Week 01 - Process of Science Prep Notes.docx) and lecture slides (Supporting File S1.6: DNA Barcoding - Week 01 - Process of Science Lecture Slides.pptx) are available in the supporting materials.
Week 2 - Hypothesis Testing
(Detailed outline - Supporting File S2.1: DNA Barcoding - Week 02 - Hypothesis Testing Lesson Plan.docx)
This laboratory is designed to introduce students to experimental design. The lab manual handout (Supporting File S2.2: DNA Barcoding - Week 02 - Hypothesis Testing Handout INSTRUCTOR KEY.docx) introduces students to key terms and concepts related to study design and hypothesis testing. The lab starts with an interactive lecture PowerPoint (Supporting File S2.3: DNA Barcoding - Week 02 - Hypothesis Testing Lecture Slides.pptx) that reviews key concepts related to experimental design. Following the introductory PowerPoint slides, students spend five to ten minutes observing and learning about live pillbugs. Details on materials and pillbug care are included in supporting prep notes (Supporting File S2.4: DNA Barcoding - Week 02 - Hypothesis Testing Prep Notes.docx). Following this five to ten-minute period of initial research, the whole class is provided with a single observation: pillbugs are consistently found under logs. They then generate hypotheses to explain this observation and share them with the class. Next students narrow the proposed hypotheses to those that are testable with the limited resources in the lab room (we provide lamps, foil, heat pads, food sources, soil, etc.) and identify three hypotheses to test. Each hypothesis is tested by two groups to see if outcomes are reproducible.
Once each group is assigned a hypothesis, they design and execute an experiment to test it. During this activity students identify independent/dependent variables, diagram their experimental design, create a data table, and graph their prediction(s). After completing data collection, each group creates a poster to present their findings to the class. During presentations, students in the audience must fill in a table provided in the lab manual (Supporting File S2.5: DNA Barcoding - Week 02 - Hypothesis Testing Handout STUDENT.docx), that asks them to determine whether each group's hypothesis was supported by their findings. Lastly, students must use their conclusions from their first round of experiments to generate a new hypothesis and propose a second experiment to test it, assuming unlimited resources, time, etc. This last activity is designed to demonstrate the iterative nature of scientific research.
A detailed lesson plan (Supporting File S2.1: DNA Barcoding - Week 02 - Hypothesis Testing Lesson Plan.docx), student handout (Supporting File S2.5: DNA Barcoding - Week 02 - Hypothesis Testing Handout STUDENT.docx), instructor key (Supporting File S2.2: DNA Barcoding - Week 02 - Hypothesis Testing Handout INSTRUCTOR KEY.docx), teaching and preparation tips (Supporting File S2.4: DNA Barcoding - Week 02 - Hypothesis Testing Prep Notes.docx) and lecture slides (Supporting File S2.3: DNA Barcoding - Week 02 - Hypothesis Testing Lecture Slides.pptx) are available in the supporting materials.
Week 3 - Phylogeny 1
(Detailed outline - Supporting File S3.1: DNA Barcoding - Week 03 - Phylogeny 1 Lesson Plan.docx)
This laboratory is designed to be a first encounter with taxonomy and phylogeny. This background will be vital to help students understand and interpret analyses of their DNA barcodes in subsequent labs. Before coming to lab students read a user-friendly primer on taxonomy, phylogeny, and systematics in the lab handout (Supporting File S3.2: DNA Barcoding - Week 03 - Phylogeny Handout INSTRUCTOR KEY.docx, Supporting File S3.3: DNA Barcoding - Week 03 - Phylogeny Handout STUDENT.docx) and then receive an interactive lecture PowerPoint (Supporting File S3.4: DNA Barcoding - Week 03 - Phylogeny Lecture Slides.pptx) at the beginning of the lab that reinforces these concepts. Following this introduction, students are asked to build a simple phylogeny with five playing cards (the in-group) and an outgroup. They also fill in a character table associated with their phylogeny, applying the terms ancestral and derived. To emphasize that phylogenies are hypotheses, they are next asked to build a tree with a different topology, using the same six taxa, but new characteristics. Students then complete a series of activities that illustrate the relationships between taxonomy and phylogeny by matching given characters to the appropriate taxonomic groups and testing whether their phylogeny is consistent with the current taxonomy. These activities help students interpret phylogenetic trees that they will generate later in the semester based on DNA barcodes. These activities also help students understand that taxonomy should reflect the evolutionary history of an organism, which can also be depicted in phylogenetic trees. Detailed lab preparation notes are included in the supporting materials (Supporting File S3.5: DNA Barcoding - Week 03 - Phylogeny Prep Notes.docx) along with the aforementioned files.
Week 4 - Phylogeny 2 Caminalcules
Adapted from Gendron R.P. 2000. The Classification and Evolution of Caminalcules. American Biology Teacher 62:570-573. (Detailed outline - Supporting File S4.1: DNA Barcoding - Week 04 - Caminalcules Lesson Plan.docx)
In this activity, adapted from Gendron (24), students reconstruct the evolutionary history of a set of imaginary organisms called caminalcules. The lab itself consists of four activities (A-D) in which students use different types of data to interpret the relationships of these organisms: morphology of living caminalcules; morphology of fossil caminalcules, and DNA barcode sequences of both living and fossil caminalcules. The logistics of completing Parts A-D are explained in detail in the directions of the student handout included in the supporting materials (Supporting File S4.2: DNA Barcoding - Week 04 - Caminalcules Handout STUDENT.docx) and the supporting instructor's notes (Supporting File S4.3: DNA Barcoding - Week 04 - Caminalcules- Handout INSTRUCTOR KEY.docx).
Students begin by building a hierarchical classification of 14 living (aka extant) caminalcules based on similarities and differences in their morphology. They then must draw an unrooted cladogram (tree) that is consistent with their taxonomy. Next, students are presented with fossil morphological data they use to refine their previously hypothesized classification system. Lastly, DNA sequences, or barcodes (Supporting File S4.4: DNA Barcoding - Week 04 - Caminalcules DNA Barcodes.pptx), are provided for a subset of caminalcules which students use to build a molecular phylogeny based on sequence divergence (Supporting File S4.5: DNA Barcoding - Week 04 - Caminalcules - MYA alignment sheet.pptx). Students then compare their molecular tree to the tree they previously developed using morphological characteristics. This lab helps students understand how taxonomy is related to phylogeny (and vice versa). It also helps students understand how sequence divergence can be used to assess the evolutionary relationships of organisms, which will be important for students to understand how DNA barcodes work. Helpful lecture slides (Supporting File S4.6: DNA Barcoding - Week 04 - Caminalcules Lecture Slides.pptx) and detailed lab preparation notes (Supporting File S4.7: DNA Barcoding - Week 04 - Caminalcules Prep Notes.docx) are included in the supporting materials.
Unit 2: Ecology
Overview
In the second unit, Ecology, students spend weeks five and six learning about the value of biodiversity and how to quantify it. This section emphasizes concepts from ecology and experimental design, and skills in field work, data collection, using spreadsheet software, graphing, and writing. Students also collect, photograph, and geotag an organism to identify via DNA barcoding. Although these labs are designed to estimate and compare biodiversity at forest edges versus forest interiors, they could easily be adapted to examine different habitat types, taxa, or questions that are more relevant to faculty or regional conservation issues (see teaching discussion for more details and ideas for adaptations).
Week 5 - Biodiversity
(Detailed outline - Supporting File S5.1: DNA Barcoding - Week 05 - Biodiversity 1 Lesson Plan.docx)
This is week one of a two-week lab. In this first week students compare the biodiversity of forest edges vs. forest interiors. Each student also collects an organism to identify in subsequent labs using DNA barcoding. The class begins with students working in small groups to design a study to test the hypothesis that forest edges have reduced biodiversity in comparison to less disturbed forest interior habitats. They also identify key elements of their study including the hypothesis, independent and dependent variable(s). Following this activity, the class establishes a final study design and works through the logistics of how student groups will be divided and how data will be collected and shared. These details are provided in the week five supporting lab handout (Supporting File S5.2: DNA Barcoding - Week 05 - Biodiversity 1 Handout INSTRUCTOR KEY.docx) and prep notes (Supporting File S5.3: DNA Barcoding - Week 05 - Biodiversity 1 Prep Notes.docx), and lecture slides (Supporting File S5.6: DNA Barcoding - Week 05 - Biodiversity 1 Lecture Slides.pptx). Next, students and instructors travel to forest edge and forest interior sites and use transects to collect data on the number of different plant, invertebrate, and fungal species at each of these two locations on a data collection table we provide (Supporting File S5.4: DNA Barcoding - Week 05 - Biodiversity Data Collection Sheet.docx). During this time each student also collects a small piece (~1 cm diameter) of fresh tissue from a plant, invertebrate, or fungus of their choosing to identify using DNA barcoding (in subsequent labs). Detailed sample collection protocols are included (Supporting File S5.2: DNA Barcoding - Week 05 - Biodiversity 1 Handout INSTRUCTOR KEY.docx; Supporting File S5.3: DNA Barcoding - Week 05 - Biodiversity 1 Prep Notes.docx) but typically, any sample of fresh, soft, fleshy, non-desiccated leaf, invertebrate, or fungal tissue will work. Each student will photograph their organism in the field, then bring their tissue sample to the lab for storage in a freezer. In the following week, students analyze their biodiversity data with Excel software and write a lab report. In subsequent weeks they use DNA barcoding to identify the organism they collected and then upload it to a growing database of forest edge and forest interior species. Logistical details and teaching tips are available in the week five lab handout (Supporting File S5.5: DNA Barcoding - Week 05 - Biodiversity 1 Handout STUDENT.docx), lecture slides (Supporting File S5.6: DNA Barcoding - Week 05 - Biodiversity 1 Lecture Slides.pptx), and prep notes (Supporting File S5.3: DNA Barcoding - Week 05 - Biodiversity 1 Prep Notes.docx).
Week 6 - Biodiversity 2
(Detailed outline - Supporting File S6.1: DNA Barcoding - Week 06 - Biodiversity 2 Lesson Plan.docx)
This is week two of a two-week lab. In this second week, students use Excel software to analyze and graph the data they collected in the previous week (wk. 5). Students also take detailed, high quality photographs of the organisms they collected to DNA barcode. Students upload their photographs and data analyses into a shared, digital lab notebook (entry details included in: Supporting File S6.2: DNA Barcoding - Week 06 - Biodiversity 2 Handout - STUDENT) that will be graded according to a rubric (Supporting File S6.3: DNA Barcoding - Week 06 - Biodiversity Lab Notebook Rubric). We recommend using google docs to create digital notebooks for teams of four students to share online (a brief tutorial on creating and sharing google docs can be found at https://www.youtube.com/watch?v=haKzqSULaPs). Students are provided a brief lecture (Supporting File S6.4: DNA Barcoding - Week 06 - Biodiversity 2 Lecture Slides.pptx) on the structure and content of a scientific lab report that is due the next week (Supporting File S6.5: DNA Barcoding - Week 06 - Biodiversity Lab Report - Guidelines, Rubric, and Template.docx). In subsequent weeks students use DNA barcoding to identify the organism they collected. There is a post-lab worksheet included in the students' lab handout (Supporting File S6.2: DNA Barcoding - Week 06 - Biodiversity 2 Handout - STUDENT.docx). There is also a formal lab report that is due the following week (Supporting File S6.5: DNA Barcoding - Week 06 - Biodiversity Lab Report - Guidelines, Rubric, and Template.docx). Instructors should use student responses to the post-lab worksheet to address any lingering issues in their understanding. Additional logistical details and teaching tips are available in week six supporting files which include a data template (Supporting File S6.6: DNA Barcoding - Week 06 - Biodiversity 2 Data Template.xlsx), lab handout key (Supporting File S6.7: DNA Barcoding - Week 06 - Biodiversity 2 Handout - INSTRUCTOR KEY.docx) and prep notes (Supporting File S6.8: DNA Barcoding - Week 06 - Biodiversity 2 Prep Notes.docx).
Unit 3: Cell & Molecular Biology
Overview
The next unit, Cell & Molecular Biology, signifies the start of the DNA barcoding pipeline. In this unit students learn about DNA structure, pipetting, centrifugation, DNA extraction, polymerase chain reaction (PCR), gel electrophoresis, and DNA sequencing. Students apply this knowledge to extract and amplify DNA from their samples and send off successfully amplified samples for sequencing. Time is built in for two rounds of DNA extraction and PCR amplification to allow students to refine their technique and troubleshoot failed PCR reactions. Instructors who are only interested in incorporating DNA barcoding into their courses, and not the previously described labs, may skip the previous units and have students begin here. However, in this case, students will need to collect tissue samples on their own outside of class following the sampling recommendations in the week five lecture (Supporting File S5.4: DNA Barcoding - Week 05 - Biodiversity 1 Lecture Slides.pptx).
Week 7 - Pipetting and DNA extraction
(Detailed outline - Supporting File S7.1: DNA Barcoding - Week 07 - Pipetting and DNA Extraction Lesson Plan.docx)
In this lab students learn how to use pipettes and then apply these skills to extract DNA from tissue samples they previously collected. Students are first provided with a general background lecture on pipetting and centrifugation (Supporting File S7.2: DNA Barcoding - Week 07 - Pipetting and DNA Extraction Lecture Slides.pptx). Following the prelab lecture, students first practice basic pipetting and centrifugation techniques. Next, the class comes back together for additional background on DNA extractions (Supporting File S7.2: DNA Barcoding - Week 07 - Pipetting and DNA Extraction Lecture Slides.pptx). Students work individually to extract DNA from the tissue of an organism they collected previously. These DNA samples are stored at -20?C until the following lab, when they will be amplified using PCR. After lab, each student will also complete a post-lab exercise assessing their pipetting and centrifugation skills (see Supporting File S7.3: DNA Barcoding - Week 07 - Pipetting and DNA Extraction Handout - STUDENT.docx for details). Additional logistical details and teaching tips are available in week 7 supporting files: lab handout key (Supporting File S7.4: DNA Barcoding - Week 07 - Pipetting and DNA Extraction Handout - INSTRUCTOR KEY.docx) and prep notes (Supporting File S7.5: DNA Barcoding - Week 07 - Pipetting and DNA Extraction Prep Notes.docx).
Week 8 - Polymerase Chain Reaction (PCR)
(Detailed outline - Supporting File S8.1: DNA Barcoding - Week 08 - PCR Lesson Plan.docx)
In this lab students learn how to perform a polymerase chain reaction using the DNA they extracted in the previous week. Students are first provided with a short lecture (Supporting File S8.2: DNA Barcoding - Week 08 - PCR lecture slides.pptx) on PCR, the primers used in DNA barcoding, and important details regarding lab protocols. Following the prelab lecture, students use a master mix spreadsheet (Supporting File S8.3: DNA Barcoding - Week 08 - Master Mix Calculator Template.xlsx) to calculate the volume of reagents needed for their PCRs. After completing this spreadsheet, students work in groups of four to prepare a PCR master mix using taxa specific primers, then PCR amplify each of their samples along with a positive and negative control (six total samples per group of four students). Students then complete a lab notebook entry, documenting which samples are in which tubes of their PCR strip, which primers were used, and what PCR cycling conditions were used (Supporting File S8.4: DNA Barcoding - Week 08 - Lab Notebook Grading Rubric.docx). The lab notebook entry should also include a screenshot of their Excel master mix calculations. Lastly, students complete a post-lab worksheet (Supporting File S8.5: DNA Barcoding - Week 08 - PCR Handout - STUDENT.docx) that assesses student understanding of master mixes, controls, and why certain genomic regions were chosen for creating DNA Barcodes. Logistical considerations and details for the preparation of laboratory materials and PCR reagents are included in supporting prep notes (Supporting File S8.6: DNA Barcoding - Week 08 - PCR Prep Notes.docx) along with an instructor key to the lab (Supporting File S8.7: DNA Barcoding - Week 08 - PCR Handout - INSTRUCTOR KEY.docx).
Week 9 - Gel electrophoresis
(Detailed outline - Supporting File S9.1: DNA Barcoding - Week 09 - Gel Electrophoresis Lesson Plan.docx)
In this lab students use gel electrophoresis to interpret the results of the previous week's PCR. At the start of this lab, students are given an introductory presentation on how gel electrophoresis works and important logistical details of the lab protocols (Supporting File S9.2: DNA Barcoding - Week 09 - Gel Electrophoresis Lecture slides.pptx). As most of the students have not run a gel before, instructors explain appropriate gel loading technique and then give students the opportunity to practice until they feel confident in loading. Once students are ready to load their gel, instructors demonstrate how to assemble the gel apparatus and cast gels. While the gel solidifies students prepare their PCR products for loading. Once the gels have solidified, instructors demonstrate how to load samples into a gel, followed by students taking turns loading their PCR products into the gel. Students then work in groups of 3-4 to run, stain, and image their gels. While the gels are running, students are quizzed on how to interpret their results (Supporting File S9.2: DNA Barcoding - Week 09 - Gel Electrophoresis Lecture slides.pptx). Then students use a provided template (Supporting File S9.3: DNA Barcoding - Week 09 - Gel Image Template.pptx) to label their gel images, which get transferred into their lab notebooks (Supporting File S9.4: DNA Barcoding - Week 09 - Gel Electrophoresis Handout - STUDENT.docx). After they complete the lab, students work in pairs to write up the results of their gels (Supporting File S9.5: DNA Barcoding - Week 09 - Gel Electrophoresis Rubric.docx). This assignment gives students practice formatting figures, reporting results, and determining appropriate analysis and interpretation of a gel. Logistical considerations and details for the preparation of laboratory materials and chemicals are included in supporting prep notes (Supporting File S9.6: DNA Barcoding - Week 09 - Gel Electrophoresis Prep Notes.docx) along with an instructor key to the lab (Supporting File S9.7: DNA Barcoding - Week 09 - Gel Electrophoresis Handout - INSTRUCTOR KEY.docx).
Week 10 - DNA extraction and PCR 2
(Detailed outline - Supporting File S10.1: DNA Barcoding - Week 10 - DNA Extraction and PCR II Lesson Plan.docx)
In this lab students who were not able to successfully amplify DNA in previous weeks are given a second opportunity to extract and PCR amplify DNA from samples they've collected. Students can use the results from the previous week's gel electrophoresis to troubleshoot and decide if they'd like to collect a new organism to DNA barcode or try a new DNA extraction protocol for their original sample (Supporting File S10.2: DNA Barcoding - Week 10 - Alternative Plant DNA Extraction Protocol.docx). Students with new samples will photograph their specimens and then all students work individually to extract DNA from their samples. Students then work in teams to create a PCR master mix using an electronic spreadsheet (Supporting File S10.3: DNA Barcoding - Week 10 - Master Mix Calculator Template.xlsx). Students then prepare a PCR master mix as a group, and PCR amplify their samples along with a positive and negative control. Students document their PCR setup, photos, and extraction techniques in their lab notebook (Supporting File S10.4: DNA Barcoding - Week 10 - DNA Extractions and PCR II Handout - STUDENT.docx) which can be graded according to the rubric we provide (Supporting File S10.5: DNA Barcoding - Week 10 - DNA Extractions and PCR II Lab Notebook Grading Rubric.docx). Logistical considerations and details for the preparation of laboratory materials and reagents are included in supporting prep notes (Supporting File S10.6: DNA Barcoding - Week 10 - DNA Extractions and PCR 2 Prep Notes.docx) along with an instructor key to the lab (Supporting File S10.7: DNA Barcoding - Week 10 - DNA Extractions and PCR II Handout - INSTRUCTOR KEY.docx).
Week 11 - Gel 2 and DNA Sequencing
(Detailed outline - Supporting File S11.1: DNA Barcoding - Week 11 - Gels 2 and DNA Sequencing Lesson Plan.docx)
In this lab students complete a second round of gel electrophoresis and then send successfully PCR amplified products off to a facility to be sequenced. Students repeating gel electrophoresis analysis come to class at the regular time and immediately assemble casting trays and pour the melted agarose. While agarose is solidifying, students prep their PCR products, then load, run, stain, and image their gels. The remainder of the lab period is available for all students to set up their DNA sequencing reactions. At this point, all students should be present in the lab to make accounts on the DNA Subway website and receive a brief introduction to the DNA Subway blue line (Supporting File S11.2: DNA Barcoding - Week 11 - Gel 2 & DNA Sequencing Lecture.pptx), a user-friendly web-based tool that will be used in the following week's lab to create and analyze students' DNA barcode sequences.
Upon completing their gels, students are instructed on how to prepare successfully amplified samples for Sanger sequencing (Supporting File S11.2: DNA Barcoding - Week 11 - Gel 2 & DNA Sequencing Lecture.pptx). Students use their gel electrophoresis results to determine which amplified PCR products are appropriate to send for sequencing, then load their samples into prelabeled sequencing tubes, which are overnighted to a sequencing facility which electronically returns sequence data files within 24 hours. Students who were not able to successfully amplify DNA in either of the two previous rounds of PCR help other group members by reading protocols, recording which samples were sequenced, and recording sequencing tube serial numbers in their lab notebooks. With the remaining class time, students who need to can annotate gel photos in their lab notebooks. Following lab, students who ran a second gel must submit a completed lab notebook entry that details the results of their second gel and DNA sequencing (Supporting File S11.3: DNA Barcoding - Week 11 - Gel 2 & DNA Sequencing Handout - STUDENT.docx). Logistical considerations and details for the preparation of laboratory materials and reagents are included in supporting prep notes (Supporting File S11.4: DNA Barcoding - Week 11 - Prep Notes.docx) along with an instructor key to the lab (Supporting File S11.5: DNA Barcoding - Week 11 - Gel 2 & DNA Sequencing Handout - INSTRUCTOR KEY.docx).
Unit 4: Bioinformatics
Overview
In the final unit, Bioinformatics, students spend weeks 12-13 analyzing their DNA sequences using a free, user-friendly web-based software platform called DNA Subway Blue Line. This software allows students to taxonomically identify the specimens they collected in week five by comparing their sequences to others in an extensive online database. Students then upload their sample photos, DNA barcode sequence, location/habitat information, and taxonomic identity to a publicly available online database called iNaturalist (www.inaturalist.org). To finish their project, students create a poster summarizing their results. The last week of the laboratory course ends with a comprehensive final exam that covers the concepts and techniques learned throughout the semester.
Week 12 - DNA Subway
(Detailed outline - Supporting File S12.1: DNA Barcoding - Week 12 - Bioinformatics - DNA Subway Lesson Plan.docx)
The aim of this assignment is for students to use bioinformatic analyses in combination with photos, range maps, and common sense to draw conclusions about the taxonomic identity of the specimen they collected in week five. Students use the DNA Subway Blue Line web-based suite of bioinformatics software (https://dnasubway.cyverse.org/) to analyze DNA Barcodes generated from the specimens they collected previously. Before analyzing their own sequences, students are given a user-friendly, instructor-guided PowerPoint (Supporting File S12.2: DNA Barcoding - Week 12 - DNA Subway Lecture.pptx) that demonstrates how to analyze two demo-barcode sequences (Supporting File S12.3: DNA Barcoding - Week 12 - 1-plant-pos-fall15F_PREMIX_J43474_3, Supporting File S12.4: DNA Barcoding - Week 12 - 1 - plant - pos - fall15R _ PREMIX _ J43475 _ 4) using the DNA Subway Blue Line. Following this demonstration, students follow directions in their lab manual (Supporting File S12.5: DNA Barcoding - Week 12 - DNA Subway Handout - STUDENT.docx) to analyze their own sequences using the Blue Line. This includes: 1) Cleaning and assembling their raw sequence data into a properly formatted DNA barcode; 2) Determining the identity of their DNA sequence using what is called a BLAST analysis; and 3) Exploring the relationship of their DNA barcode to other DNA sequences using a MUSCLE alignment and phylogenetic trees that are generated in DNA Subway. These analyses are logged by students by answering questions in their lab handout (Supporting File S12.5: DNA Barcoding - Week 12 - DNA Subway Handout - STUDENT.docx) as well as recording information in their lab notebook (see prompts in Supporting File S12.5: DNA Barcoding - Week 12 - DNA Subway Handout - STUDENT.docx). Students then use these data along with photos and range maps to determine if there is sufficient evidence to identify their specimen's taxonomic identity to the genus or species level. Following lab, students complete a post-lab worksheet (see end of: Supporting File S12.5: DNA Barcoding - Week 12 - DNA Subway Handout - STUDENT.docx) about their DNA Subway Blue Line analyses, which is collected by instructors at the end of class to ensure that students made accurate conclusions regarding the identity of their sample.
This is one of the more difficult and information dense labs for both students and instructors, so we have gone to great measures to include lots of helpful materials that are designed to be accessible to even the most uninitiated users with little to no experience in bioinformatics. One of the most useful of these is the supplemental PowerPoint (Supporting File S12.2: DNA Barcoding - Week 12 - DNA Subway Lecture.pptx), which includes extensive animations and notes below each slide that explain each step of DNA Subway in very user-friendly terms. We highly recommend that instructors spend time before class using these slides in conjunction with the demo sequences (Supporting File S12.3: DNA Barcoding - Week 12 - 1-plant-pos-fall15F_PREMIX_J43474_3, Supporting File S12.4: DNA Barcoding - Week 12 - 1 - plant - pos - fall15R _ PREMIX _ J43475 _ 4) before meeting with students. A DNA subway video tutorial is available at https://www.youtube.com/watch?v=7WF--Ba2P10. Additional tutorials and resources are also available at https://www.dnabarcoding101.org. Other logistical considerations and details for the preparation of laboratory materials are included in supporting prep notes (Supporting File S12.6: DNA Barcoding - Week 12 - Prep Notes.docx) along with an instructor key to the lab (Supporting File S12.7: DNA Barcoding - Week 12 - DNA Subway Handout - INSTRUCTOR KEY.docx) and a grading rubric for student lab notebooks (Supporting File S12.8: DNA Barcoding - Week 12 - DNA Subway lab notebook rubric.docx).
Week 13 - iNaturalist and Posters
(Detailed outline - Supporting File S13.1: DNA Barcoding - Week 13 - iNaturalist and DNA Subway Posters Lesson Plan.docx)
Following a short PowerPoint presentation that outlines lab goals and protocols (Supporting File S13.2: DNA Barcoding - Week 13 - iNaturalist and Posters - Lecture.pptx), students upload photos, DNA barcodes, and taxonomic information of the organism they collected and identified to iNaturalist - a free, user-friendly, web-based database designed to share location and taxonomic data on organisms photographed in nature. These observations are then added to an ongoing course project within iNaturalist that compiles all the organisms collected from forest edges and interior habitats. The course project we've created on iNaturalist is specific to JMU and our research question, so other institutions will need to create their own iNaturalist project(s) specific to their region and/or research question(s) they are addressing. We've included a supplemental worksheet (Supporting File S13.8: DNA Barcoding - Week 13 - Quantify Biodiversity in iNaturalist.docx) that teaches students how to mine data from the iNaturalist database. Students (and instructors) can use this exercise to compare the overall biodiversity of forest edge versus interior habitat types or make other comparisons that are more relevant to their course's research questions (see final page of Supporting File S13.8: DNA Barcoding - Week 13 - Quantify Biodiversity in iNaturalist.docx for details). Students then use their lab handout (Supporting File S13.3: DNA Barcoding - Week 13 - iNaturalist and Posters Handout - STUDENT.docx) and an internet-connected computer to complete in class research on the natural history of the organism they barcoded. Once they've completed this activity, students study the basics for formatting a poster for a scientific presentation (Supporting File S13.4: DNA Barcoding - Week 13 - iNaturalist and Posters - Links to Poster Design Readings.docx), then download a DNA barcoding poster template (Supporting File S13.5: DNA Barcoding - Week 13 - iNaturalist and Posters - Poster Template.pptx) which they use for the rest of the lab or as homework to complete their DNA barcoding poster. This poster will be submitted for a grade (Supporting File S13.6: DNA Barcoding - Week 13 - DNA Barcoding Poster Grading Rubric.docx). Logistical considerations and details for the preparation of this week's laboratory materials are included in supporting prep notes (Supporting File S13.7: DNA Barcoding - Week 13 - iNaturalist and Posters - Prep notes.docx).
Week 14: Final Exam
The final week of lab is reserved for a cumulative final exam. For security purposes our exam is not included in this publication, but we do include a study guide that outlines each major topic that our final entails (Supporting File S14.1: DNA Barcoding - Week 14 - Final Exam Study Guide.docx).
TEACHING DISCUSSION
The benefits of CUREs have been well-documented, but the ability to scale these course activities to meet the needs of large enrollment, first-year undergraduate courses can be difficult. The semester long series of DNA barcoding lab modules presented here is the first half of a year-long DNA barcoding CURE that has been successfully implemented in our large-enrollment first-year Foundations of Biology course at James Madison University. The first semester of this course enrolls approximately 500 students per semester with labs capped at 24 students per section. As of September 2018, the course is in its fifth semester of implementation. Students in these labs have created over 1,500 unique DNA barcode sequences and documented over 300 unique species of plants, fungi, and invertebrates. These labs have served approximately 2,500 JMU students with varying academic majors including biology, biotechnology, health sciences, economics, chemistry, and others, demonstrating the ability of these DNA barcoding labs to meet the needs of large enrollment first year courses (Figure 3). Furthermore, most sections of this lab have been led by part-time teachers and master's students with limited to no prior teaching or research experience, demonstrating that these lessons can be taught successfully by instructors without initial technical or instructional expertise.
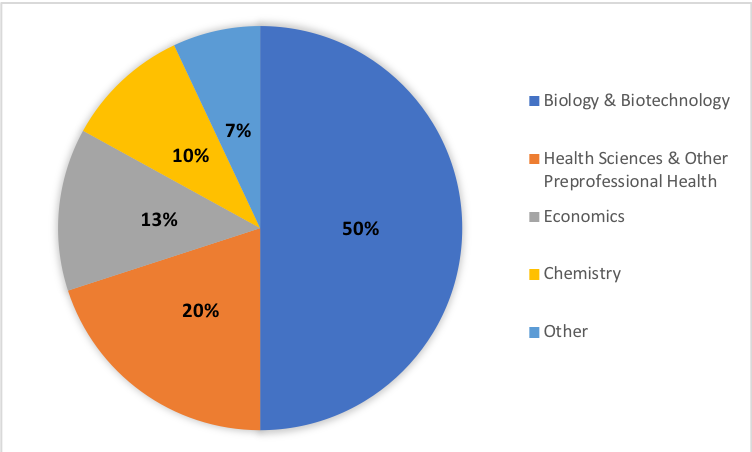
Figure 3: Distribution of student majors who have completed this 14-week lab series, demonstrating the adaptability of this course for students with differing background and interests
We used multiple methods to assess how well these labs met our learning objectives in our first cohort of students in the fall semester of 2016 (n=474; research approved by JMU Institutional Review Board for Human Subjects #17-0307). First, course effectiveness was evaluated using student scores on major assignments, which were mapped to the learning objectives. Students across all 20 sections (n=474) averaged 78.9% (SD = 9.1) on a cumulative final exam (Supporting File S14.1: DNA Barcoding - Final Exam Study Guide.docx) designed to assess conceptual and technical understanding of each course learning objective (Supporting File S0.2: DNA Barcoding - Alignment of objectives and assessments.docx). Student learning was also assessed using the rubric-scored (Supporting File S13.6: DNA Barcoding - Week 13 - DNA Barcoding Poster Grading Rubric.docx) final posters, in which students averaged 86.2% (SD = 9.6, n = 474). These data provide initial evidence of satisfactory knowledge of course content in meeting our learning objectives, however in the future, further assessment comparing pre- and post-course performance on these assignments will be needed to more definitively determine if our lessons improve student performance above their baseline level prior to completing these labs. Second, in a voluntary survey administered on the last day of the course, >60% of students self-reported an increased level of comfort with concepts from ecology, molecular biology, and bioinformatics as compared to the start of the semester, while <4% of students reported less comfort (Figure 4, n = 430).
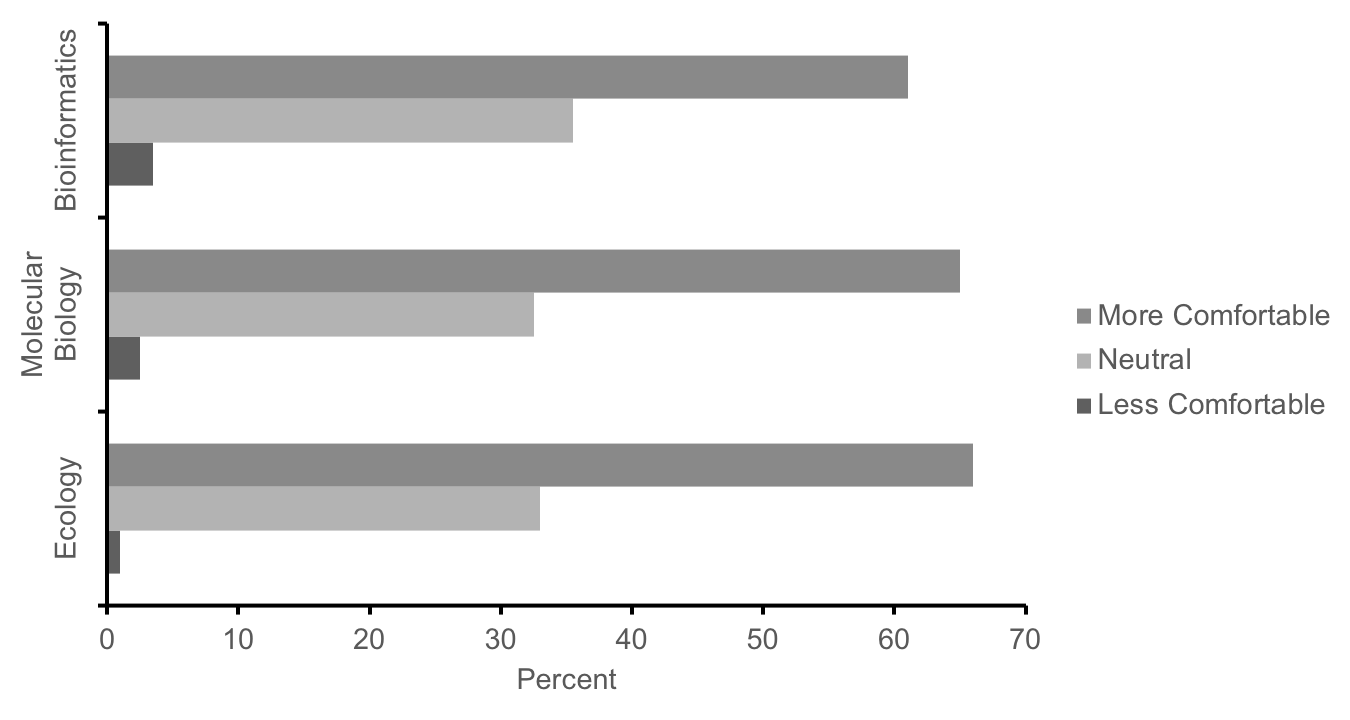
Figure 4: Summary responses of our first cohort of students (n = 430) to the following prompt: In comparison to the start of the term, how has the lab experience changed your level of comfort with concepts from ecology, molecular biology, and bioinformatics? Students report increased comfort with concepts from these fields after completing these labs.
Likewise, 70% of students self-reported increased comfort with methods and techniques related to ecology, molecular biology, and bioinformatics at the completion of the course, while <4% of students reported less comfort (Figure 5, n = 430). Lastly, in the same survey, 90% of students agreed that these labs were a good way to learn science, and 80% indicated that the experience increased their interest in science. These responses and performance measures suggest that this series of labs is effective in teaching concepts and methods in ecology, molecular biology, and bioinformatics as well as motivating first-year students' interest in science. While a more in-depth investigation as to how students benefit from this research experience is currently underway (25-26), the convergence of self-report survey data with the aforementioned direct measures (i.e. performance on the final exam and posters) provide initial evidence that this course is meeting learning objectives (Supporting File S0.2: DNA Barcoding - Alignment of objectives and assessments.docx).
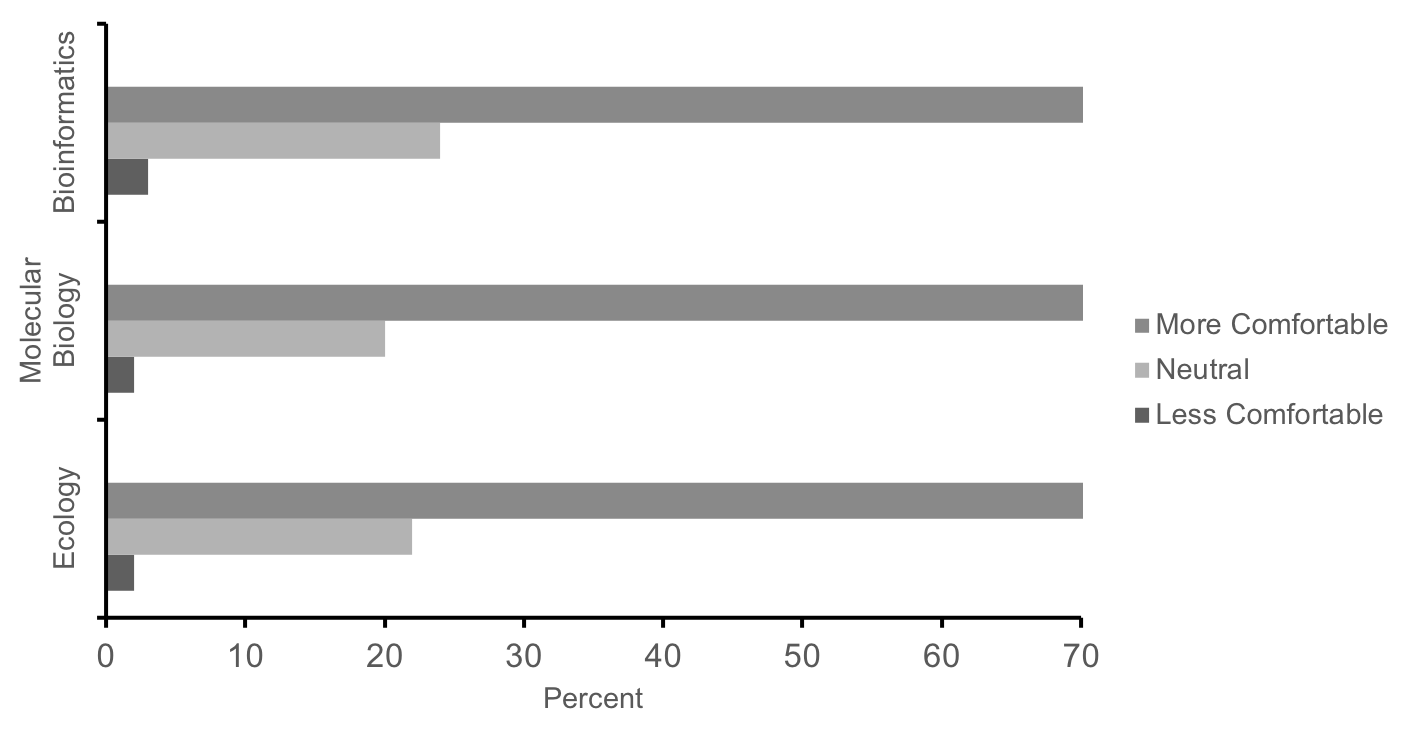
Figure 5: Summary responses of our first cohort of students (n = 430) to the following prompt: In comparison to the start of the term, how has the lab experience changed your level of comfort with techniques and methods from ecology, molecular biology, and bioinformatics? Students report increased comfort with techniques in these fields after completing these labs.
Challenges
Expenses related to molecular techniques such as PCR amplification and DNA sequencing can be a major hurdle to the implementation of these labs, however by limiting each student to barcoding a single sample, we have been able to keep the costs of consumables including pipette tips, PCR and gel reagents, and DNA sequencing at levels equal to our previous budget prior to implementation of these labs. We estimate costs of ~$13 per student per semester for molecular lab related expenses (wk. 7-12) including sequencing costs which are $4.50 per read (see Supporting File S0.3: DNA Barcoding - DNA barcoding equipment list.xls). These estimates do not include initial investments in durable equipment, such as micropipettes, microcentrifuges, vortexers, PCR cyclers, gel boxes, and gel visualization equipment (see Supporting File S0.3: DNA Barcoding - DNA barcoding equipment list.xls for a comprehensive material list). If cost is an issue, there are a variety of ways to adapt portions of these labs to cut costs. For example, instructors could supply students with previously obtained DNA barcode sequences, and focus primarily on the bioinformatics aspects of these labs, thereby drastically reducing costs. Options for alternative implementations are discussed further below.
The likelihood of students successfully creating unique DNA barcodes is another important issue. Although our protocols have proven robust across a variety of taxa and sample types, there is variability in student success extracting and amplifying DNA from the specimens they collect. Some of this variability may be a result of student inexperience performing molecular techniques, however, results from student gels suggest that difficulty with DNA extraction and purification appears to be one of the greatest hurdles to generating original barcode sequences. Variability in success rates may also be attributed to specimen selection. Desiccated, pulpy, and/or waxy specimens are challenging to extract and purify DNA from no matter the skill level of the student. To help enhance student success in creating DNA barcodes, we provide detailed instructions on sample collection that can increase the likelihood of successful DNA isolation and amplification (Supporting File S5.6: DNA Barcoding - Week 05 - Biodiversity 1 Lecture Slides.pptx). Experienced instructors with well-equipped labs and access to microvolume spectrophotometers (e.g. ThermoFisher Nanodrop) may want to measure DNA content and purity of student's DNA extracts prior to PCR in weeks eight and ten. This could help reduce costs spent on failed PCR reactions. More importantly, there is allotted lab time (weeks 10-11) for students to conduct a second set of DNA extractions, PCRs, and gel electrophoresis in the case their first attempts fail at any of these steps. We estimate that with iteration ~80% of our students are able to successfully create a DNA barcode sequence from the organisms they collect. Students who are unable to successfully amplify DNA on their first try (weeks 7-9) are typically directed to collect a new or different sample for their second attempt in week 10 and/or try a new DNA extraction protocol (Supporting File S10.2: DNA Barcoding - Week 10 - Alternative Plant DNA Extraction Protocol.docx). Those who are unable to successfully amplify DNA on both attempts are provided with back up unknown sequences (https://tinyurl.com/ybnwn9gp) or another student's sequence to analyze. Thus, the ability of a student to complete the lab activities does not depend solely upon their creation of a unique DNA barcode from their own sample. Furthermore, we see the occasional failure to successfully create DNA barcodes as a strength of the lab (26). It provides opportunities for iteration, troubleshooting, and refinement of molecular techniques. It also demonstrates that research and techniques don't always work as planned, while simultaneously providing a safety net of backup sequences to ensure that all students can complete each lab activity.
Alternative Implementations
We have outlined a 14-week series of lab modules conducted as a semester-long lab experience targeted for first-year undergraduate life science students. While a host of literature demonstrates the positive impact of semester-long CUREs in first-year biology courses (5), we recognize that implementation of these modules at this scale is not always practical as it requires significant financial and ideological support from university administration and faculty as well as extensive effort from the instructors implementing the course. Furthermore, weeks five and six of these labs require access to a forest, which will not be practical for many schools. In light of these barriers that may preclude a full-scale implementation of the above modules; alternative or shortened implementations of these labs would undoubtedly be of use in a variety of contexts.
One solution is to selectively implement a limited subset of these lab modules to emphasize particular concepts or methodologies based on differing course needs and student populations. For example, an instructor could choose to implement only the DNA barcoding pipeline (Wk. 7 - Wk. 12) in their course. Alternatively, the full DNA barcoding pipeline (Wk. 7 - Wk. 12) can be adjusted to be run in as few as two to three class meetings. For example, previous iterations of these DNA barcoding workflows were successfully condensed for two to three-day JMU-hosted workshops and summer camps aimed at high school and undergraduate students. In order to streamline accelerated modules, students typically bring a previously collected specimen to class or previously collected specimens are provided by lab instructors. The latter approach can be particularly effective in concentrating a class effort on analysis of a single group of specimens. For example, this approach has been successfully developed into an ongoing JMU Center for Genome & Metagenome (CGEMS) longitudinal study characterizing native mosquito populations on the JMU campus and the surrounding Shenandoah Valley. In addition, an entirely computer-based module emphasizing bioinformatic analysis of previously obtained DNA barcode data is available for implementation into a single 120-minute module (27). This module is configured specifically for ubiquitous usage requiring only a computer and internet connection for student participation in a lecture or lab-based setting.
If comparing biodiversity at forest edges versus interiors is not feasible or of interest, the central question and introduction to the ecology labs in weeks five and six could be changed to allow students to compare almost any two habitats. For example, students could compare biodiversity on north vs. south facing slopes of hills, high vs. low traffic urban parks, ponds with vs. without fish, or any two habitats where living organisms can be collected. Students could even decide on a comparison that they'd like to make. Alternatively, the ecology labs (week five and six) could be adapted or replaced to prepare students to sample specific taxonomic groups (e.g. mosquitos) or address questions relevant to research programs at your institution. For example, honors sections of our labs at JMU have been involved in identifying species of dung beetles on farms, identifying fungi collected by the Mycological Society of America, and identifying cryptic species of tadpoles, while other programs have been developed to look at mislabeling of sushi (28) or herbal products (29). Given the broad scope of DNA barcoding analyses, the modules presented here can be implemented in a number of alternative versions depending on resource availability, faculty research interests, forest access, student populations, and time.
Conclusion
There is mounting evidence supporting the benefits that undergraduate research experiences can provide to STEM students (5-9), especially at the beginning of their undergraduate careers (10). Unfortunately, many traditional undergraduate research experiences lack the ability to scale to meet the needs of large enrollment, first year courses. Here we've provided a classroom-tested, scalable, and transferable semester-long undergraduate research experience based on DNA barcoding that has shown promise as a model to teach core concepts in biology as well as engage students in authentic, open-ended research documenting species diversity using DNA barcoding. These labs also provide opportunities for faculty interested in documenting local biodiversity to incorporate first-year students into their research programs. We hope that these labs can serve as a model for institutions to implement authentic DNA barcoding-based research in their own first-year undergraduate courses.
SUPPORTING MATERIALS
Supporting materials are organized by week. Each week has a lesson plan that provides an overview of the lesson, detailed prep notes, lecture slides, worksheet(s), a lab manual printout, and any additional files required to successfully complete the lesson.
- S0.1: DNA Barcoding - Sample Syllabus.docx
- S0.2: DNA Barcoding - Alignment of objectives and assessments.docx
- S0.3: DNA Barcoding - DNA barcoding equipment list.xls
Week 01 - Process of Science
- S1.1: DNA Barcoding - Week 01 - Process of Science Lesson Plan.docx
- S1.2: DNA Barcoding - Week 01 - Process of Science Handout INSTRUCTOR KEY.docx
- S1.3a: DNA Barcoding - Week 01 - Process of Science B&W FLOWCHART.pdf
- S1.3b: DNA Barcoding - Week 01 - Process of Science FLOWCHART.pdf
- S1.4: DNA Barcoding - Week 01 - Process of Science Handout STUDENT.docx
- S1.5: DNA Barcoding - Week 01 - Process of Science Prep Notes.docx
- S1.6: DNA Barcoding - Week 01 - Process of Science Lecture Slides.pptx
Week 02 - Hypothesis Testing
- S2.1: DNA Barcoding - Week 02 - Hypothesis Testing Lesson Plan.docx
- S2.2: DNA Barcoding - Week 02 - Hypothesis Testing Handout INSTRUCTOR KEY.docx
- S2.3: DNA Barcoding - Week 02 - Hypothesis Testing Lecture Slides.pptx
- S2.4: DNA Barcoding - Week 02 - Hypothesis Testing Prep Notes.docx
- S2.5: DNA Barcoding - Week 02 - Hypothesis Testing Handout STUDENT.docx
Week 03 - Phylogeny 1
- S3.1: DNA Barcoding - Week 03 - Phylogeny 1 Lesson Plan.docx
- S3.2: DNA Barcoding - Week 03 - Phylogeny Handout INSTRUCTOR KEY.docx
- S3.3: DNA Barcoding - Week 03 - Phylogeny Handout STUDENT.docx
- S3.4: DNA Barcoding - Week 03 - Phylogeny Lecture Slides.pptx
- S3.5: DNA Barcoding - Week 03 - Phylogeny Prep Notes.docx
Week 04 - Phylogeny 2 Caminalcules
- S4.1: DNA Barcoding - Week 04 - Caminalcules Lesson Plan.docx
- S4.2: DNA Barcoding - Week 04 - Caminalcules Handout STUDENT.docx
- S4.3: DNA Barcoding - Week 04 - Caminalcules- Handout INSTRUCTOR KEY.docx
- S4.4: DNA Barcoding - Week 04 - Caminalcules DNA Barcodes.pptx
- S4.5: DNA Barcoding - Week 04 - Caminalcules - MYA alignment sheet.pptx
- S4.6: DNA Barcoding - Week 04 - Caminalcules Lecture Slides.pptx
- S4.7: DNA Barcoding - Week 04 - Caminalcules Prep Notes.docx
Week 05 - Biodiversity 1
- S5.1: DNA Barcoding - Week 05 - Biodiversity 1 Lesson Plan.docx
- S5.2: DNA Barcoding - Week 05 - Biodiversity 1 Handout INSTRUCTOR KEY.docx
- S5.3: DNA Barcoding - Week 05 - Biodiversity 1 Prep Notes.docx
- S5.4: DNA Barcoding - Week 05 - Biodiversity Data Collection Sheet.docx
- S5.5: DNA Barcoding - Week 05 - Biodiversity 1 Handout STUDENT.docx
- S5.6: DNA Barcoding - Week 05 - Biodiversity 1 Lecture Slides.pptx
Week 06 - Biodiversity 2
- S6.1: DNA Barcoding - Week 06 - Biodiversity 2 Lesson Plan.docx
- S6.2: DNA Barcoding - Week 06 - Biodiversity 2 Handout - STUDENT.docx
- S6.3: DNA Barcoding - Week 06 - Biodiversity Lab Notebook Rubric.docx
- S6.4: DNA Barcoding - Week 06 - Biodiversity 2 Lecture Slides.pptx
- S6.5: DNA Barcoding - Week 06 - Biodiversity Lab Report - Guidelines, Rubric, and Template.docx
- S6.6: DNA Barcoding - Week 06 - Biodiversity 2 Data Template.xlsx
- S6.7: DNA Barcoding - Week 06 - Biodiversity 2 Handout - INSTRUCTOR KEY.docx
- S6.8: DNA Barcoding - Week 06 - Biodiversity 2 Prep Notes.docx
Week 07 - Pipetting and DNA Extraction
- S7.1: DNA Barcoding - Week 07 - Pipetting and DNA Extraction Lesson Plan.docx
- S7.2: DNA Barcoding - Week 07 - Pipetting and DNA Extraction Lecture Slides.pptx
- S7.3: DNA Barcoding - Week 07 - Pipetting and DNA Extraction Handout - STUDENT.docx
- S7.4: DNA Barcoding - Week 07 - Pipetting and DNA Extraction Handout - INSTRUCTOR KEY.docx
- S7.5: DNA Barcoding - Week 07 - Pipetting and DNA Extraction Prep Notes.docx
Week 08 - PCR
- S8.1: DNA Barcoding - Week 08 - PCR Lesson Plan.docx
- S8.2: DNA Barcoding - Week 08 - PCR lecture slides.pptx
- S8.3: DNA Barcoding - Week 08 - Master Mix Calculator Template.xlsx
- S8.4: DNA Barcoding - Week 08 - Lab Notebook Grading Rubric.docx
- S8.5: DNA Barcoding - Week 08 - PCR Handout - STUDENT.docx
- S8.6: DNA Barcoding - Week 08 - PCR Prep Notes.docx
- S8.7: DNA Barcoding - Week 08 - PCR Handout - INSTRUCTOR KEY.docx
Week 09 - Gel Electrophoresis
- S9.1: DNA Barcoding - Week 09 - Gel Electrophoresis Lesson Plan.docx
- S9.2: DNA Barcoding - Week 09 - Gel Electrophoresis Lecture slides.pptx
- S9.3: DNA Barcoding - Week 09 - Gel Image Template.pptx
- S9.4: DNA Barcoding - Week 09 - Gel Electrophoresis Handout - STUDENT.docx
- S9.5: DNA Barcoding - Week 09 - Gel Electrophoresis Rubric.docx
- S9.6: DNA Barcoding - Week 09 - Gel Electrophoresis Prep Notes.docx
- S9.7: DNA Barcoding - Week 09 - Gel Electrophoresis Handout - INSTRUCTOR KEY.docx
Week 10 - DNA Extraction and PCR 2
- S10.1: DNA Barcoding - Week 10 - DNA Extraction and PCR II Lesson Plan.docx
- S10.2: DNA Barcoding - Week 10 - Alternative Plant DNA Extraction Protocol.docx
- S10.3: DNA Barcoding - Week 10 - Master Mix Calculator Template.xlsx
- S10.4: DNA Barcoding - Week 10 - DNA Extractions and PCR II Handout - STUDENT.docx
- S10.5: DNA Barcoding - Week 10 - DNA Extractions and PCR II Lab Notebook Grading Rubric.docx
- S10.6: DNA Barcoding - Week 10 - DNA Extractions and PCR 2 Prep Notes.docx
- S10.7: DNA Barcoding - Week 10 - DNA Extractions and PCR II Handout - INSTRUCTOR KEY.docx
Week 11 - Gel 2 and DNA Sequencing
- S11.1: DNA Barcoding - Week 11 - Gels 2 and DNA Sequencing Lesson Plan.docx
- S11.2: DNA Barcoding - Week 11 - Gel 2 & DNA Sequencing Lecture.pptx
- S11.3: DNA Barcoding - Week 11 - Gel 2 & DNA Sequencing Handout - STUDENT.docx
- S11.4: DNA Barcoding - Week 11 - Prep Notes.docx
- S11.5: DNA Barcoding - Week 11 - Gel 2 & DNA Sequencing Handout - INSTRUCTOR KEY.docx
Week 12 - Bioinformatics: DNA Subway
- S12.1: DNA Barcoding - Week 12 - Bioinformatics - DNA Subway Lesson Plan.docx
- S12.2: DNA Barcoding - Week 12 - DNA Subway Lecture.pptx
- S12.3: DNA Barcoding - Week 12 - 1-plant-pos-fall15F_PREMIX_J43474_3
- S12.4: DNA Barcoding - Week 12 - 1-plant-pos-fall15R_PREMIX_J43475_4
- S12.5: DNA Barcoding - Week 12 - DNA Subway Handout - STUDENT.docx
- S12.6: DNA Barcoding - Week 12 - Prep Notes.docx
- S12.7: DNA Barcoding - Week 12 - DNA Subway Handout - INSTRUCTOR KEY.docx
- S12.8: DNA Barcoding - Week 12 - DNA Subway Lab Notebook Rubric.docx
Week 13 - iNaturalist and Posters
- S13.1: DNA Barcoding - Week 13 - iNaturalist and DNA Subway Posters Lesson Plan.docx
- S13.2: DNA Barcoding - Week 13 - iNaturalist and Posters - Lecture.pptx
- S13.3: DNA Barcoding - Week 13 - iNaturalist and Posters Handout - STUDENT.docx
- S13.4: DNA Barcoding - Week 13 - iNaturalist and Posters - Links to Poster Design Readings.docx
- S13.5: DNA Barcoding - Week 13 - iNaturalist and Posters - Poster Template.pptx
- S13.6: DNA Barcoding - Week 13 - DNA Barcoding Poster Grading Rubric.docx
- S13.7: DNA Barcoding - Week 13 - iNaturalist and Posters - Prep notes.docx
- S13.8: DNA Barcoding - Week 13 - Quantify Biodiversity in iNaturalist.docx
Week 14 - Final Exam
- S14.1. DNA Barcoding - Week 14 - Final Exam Study Guide.docx
ACKNOWLEDGMENTS
The authors would like to thank all of the JMU Department of Biology faculty and staff that participated in the design, implementation, and instruction of the DNA barcoding laboratory course. We thank JMU undergraduate Emily Miller (advised by J. Harsh) for her role in the analysis of the student survey data. We also thank David Micklos and the support staff at the Cold Spring Harbor Laboratory DNA Learning Center for their advice on large-scale implementation of DNA barcoding methodologies. Additionally, we thank Jason Williams and CyVerse support staff for on-site and remote training using DNA Subway tools. This work was supported by James Madison University 4-VA funding as well as Burroughs Wellcome Fund Grant #1017506 awarded to R.A.E, a JMU Geospatial Minigrant awarded to R.A.E, E.A.D. and O.J.H, National Science Foundation, Improving Undergraduate STEM Education Grant #1821657 awarded to R.A.E and O.J.H, and the JMU College of Science and Mathematics. All student surveys were conducted with prior approval from the JMU Institutional Review Board for Human Subjects #17-0307.
References
- American Association for the Advancement of Science. 2011. Vision and Change: A Call to Action, Final Report. Washington, DC: AAAS.
- Boyer Commission on Educating Undergraduates in the Research University, S. S. Kenny (chair). 1998. Reinventing Undergraduate Education: A Blueprint for America's Research Universities. State University of New York-Stony Brook.
- National Academy of Sciences (NAS). 2007. Rising above the gathering storm: Energizing and employing America for a brighter economic future. Washington, DC: National Academy Press.
- President's Council of Advisors on Science and Technology. 2012. Report to the President--Engage to Excel: Producing One Million Additional College Graduates with Degrees in Science, Technology, Engineering, and Mathematics.
- Corwin, L. A., Graham, M. J., & Dolan, E. L. 2015. Modeling Course-Based Undergraduate Research Experiences: An Agenda for Future Research and Evaluation. CBE Life Sciences Education, 14(1):es1. http://doi.org/10.1187/cbe.14-10-0167
- National Academies of Sciences, Engineering, and Medicine. 2017. Undergraduate Research Experiences for STEM Students: Successes, Challenges, and Opportunities. Washington, DC: The National Academies Press. doi: 10.17226/24622.
- Denofrio, L.A., Russell, B., Lopatto, D., & Lu, Y. 2007. Linking student interests to science curricula. Science, 318, 1872-1873.
- Lopatto, D., et al. 2008. Genomics Education Partnership. Science, 322, 684-685.
- Bangera, G., & Brownell, S. E. 2014. Course-based undergraduate research experiences can make scientific research more inclusive. CBE-Life Sciences Education, 13(4):602-606.
- Rodenbusch, S.E., Hernandez, P.R., Simmons, S.L., and Dolan, E.L. 2016. Early engagement in course-based research increases graduation rates and completion of science, engineering, and mathematics degrees. CBE-Life Sciences Education, 15(2):ar20.
- Hebert, P. D., Cywinska, A., & Ball, S. L. 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society of London B: Biological Sciences, 270(1512): 313-321.
- Savolainen, V., Cowan, R. S., Vogler, A. P., Roderick, G. K., & Lane, R. 2005. Towards writing the encyclopedia of life: an introduction to DNA barcoding. Philosophical Transactions of the Royal Society of London B: Biological Sciences, 360(1462), 1805-1811.
- Telfer, A. C., Young, M. R., Quinn, J., Perez, K., Sobel, C. N., Sones, J. E., ... & Thevanayagam, A. 2015. Biodiversity inventories in high gear: DNA barcoding facilitates a rapid biotic survey of a temperate nature reserve. Biodiversity data journal, (3).
- Hebert, P. D., Penton, E. H., Burns, J. M., Janzen, D. H., & Hallwachs, W. 2004. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proceedings of the National Academy of Sciences, 101(41), 14812-14817.
- Wong, E. H. K., & Hanner, R. H. 2008. DNA barcoding detects market substitution in North American seafood. Food Research International, 41(8), 828-837.
- Harris, S. E., & Bellino, M. 2013. DNA barcoding from NYC to Belize. Science, 342(6165), 1462-1463.
- Santschi, L., Hanner, R. H., Ratnasingham, S., Riconscente, M., & Imondi, R. 2013. Barcoding Life's Matrix: Translating biodiversity genomics into high school settings to enhance life science education. PLoS biology, 11(1), e1001471.
- Steinke, D., Breton, V., Berzitis, E., & Hebert, P. D. 2017. The School Malaise Trap Program: coupling educational outreach with scientific discovery. PLoS biology, 15(4), e2001829.
- Henter, H. J., Imondi, R., James, K., Spencer, D., & Steinke, D. 2016. DNA barcoding in diverse educational settings: five case studies. Phil. Trans. R. Soc. B, 371(1702), 20150340.
- Auchincloss, L. C., Laursen, S. L., Branchaw, J. L., Eagan, K., Graham, M., Hanauer, D. I., ... Dolan, E. L. 2014. Assessment of Course-Based Undergraduate Research Experiences: A Meeting Report. CBE Life Sciences Education, 13(1):29-40.
- Alimoglu, M. K., Gurpinar, E., Mamakli, S., & Aktekin, M. 2011. Ways of coping as predictors of satisfaction with curriculum and academic success in medical school. Advances in physiology education, 35(1):33-38.
- Harsh, J.A., R. Balliet, Maltese A.V., and R.H. Tai. 2015. Essential features and benefits of undergraduate research experiences: perspectives of student researchers and practicing scientists. Annual Meeting of the National Association for Research in Science Teaching (NARST), Chicago, IL.
- Lopatto, D. 2003. The essential features of undergraduate research. Council on Undergraduate Research Quarterly, 24:139-142.
- Gendron R.P. 2000. The Classification and Evolution of Caminalcules. American Biology Teacher 62:570-573.
- Harsh, J.A., Miller, E., Hyman, O., Boyce. R., Coleman, C., Cummings, J., O'Neill, T., Nguyen, J., Popham, J. 2018. Assessing the effectiveness and impact of a large-scale two-semester course based undergraduate research experience focused on DNA-Barcoding for introductory biology students. Annual Meeting of the Society for the Advancement of Biology Education Research (SABER). Minneapolis, MN.
- Corwin, L., Harsh, J., Ellis, S., Gustafson, N., Woolner, E. 2018. Research-based courses expose students to scientific obstacles and offer opportunities to practice scientific coping. Annual Meeting of the Society for the Advancement of Biology Education Research (SABER). Minneapolis, MN.
- Williams, J., Enke, R.A., Hyman O.J., Lescak E., Donovan S.S., Tapprich W., Ryder E.F. 2018. Using DNA Subway to Analyse Sequence Relationships. (Version 2.0). QUBES. doi:10.25334/Q4J111
- Dybas, C. L. 2017. RIPPLE MARKS--THE STORY BEHIND THE STORY: Sushi Bait and Switch: What Fish Are You Really Eating? Oceanography, 30(2), 12-14.
- Newmaster, S. G., Grguric, M., Shanmughanandhan, D., Ramalingam, S., & Ragupathy, S. 2013. DNA barcoding detects contamination and substitution in North American herbal products. BMC medicine, 11(1), 222.
Article Files
Login to access supporting documents
CURE-all: Large Scale Implementation of Authentic DNA Barcoding Research into First-Year Biology Curriculum(PDF | 807 KB)
S0.1. DNA Barcoding - Sample Syllabus.docx(DOCX | 27 KB)
S0.2. DNA Barcoding - Alignment of objectives and assessments.docx(DOCX | 28 KB)
S0.3. DNA Barcoding - DNA barcoding equipment list.xls.xlsx(XLSX | 20 KB)
S1.1. DNA Barcoding - Week 01 - Process of Science Lesson Plan.docx(DOCX | 24 KB)
S1.2. DNA Barcoding - Week 01 - Process of Science Handout INSTRUCTOR KEY.docx(DOCX | 489 KB)
S1.3a. DNA Barcoding - Week 01 - Process of Science BXW FLOWCHART.pdf(PDF | 308 KB)
S1.3b. DNA Barcoding - Week 01 - Process of Science FLOWCHART.pdf(PDF | 269 KB)
S1.4. DNA Barcoding - Week 01 - Process of Science Handout STUDENT.docx(DOCX | 363 KB)
S1.5. DNA Barcoding - Week 01 - Process of Science Prep Notes.docx(DOCX | 40 KB)
S1.6. DNA Barcoding - Week 01 - Process of Science Lecture Slides.pptx(PPTX | 1 MB)
S2.1. DNA Barcoding - Week 02 - Hypothesis Testing Lesson Plan.docx(DOCX | 27 KB)
S2.2. DNA Barcoding - Week 02 - Hypothesis Testing Handout INSTRUCTOR KEY.docx(DOCX | 117 KB)
S2.3. DNA Barcoding - Week 02 - Hypothesis Testing Lecture Slides.pptx(PPTX | 4 MB)
S2.4. DNA Barcoding - Week 02 - Hypothesis Testing Prep Notes.docx(DOCX | 23 KB)
S2.5. DNA Barcoding - Week 02 - Hypothesis Testing Handout STUDENT.docx(DOCX | 40 KB)
S3.1. DNA Barcoding - Week 03 - Phylogeny 1 Lesson Plan.docx(DOCX | 27 KB)
S3.2. DNA Barcoding - Week 03 - Phylogeny Handout INSTRUCTOR KEY.docx(DOCX | 394 KB)
S3.3. DNA Barcoding - Week 03 - Phylogeny Handout STUDENT.docx(DOCX | 508 KB)
S3.4. DNA Barcoding - Week 03 - Phylogeny Lecture Slides.pptx(PPTX | 5 MB)
S3.5. DNA Barcoding - Week 03 - Phylogeny Prep Notes.docx(DOCX | 173 KB)
S4.1. DNA Barcoding - Week 04 XXX Caminalcules Lesson Plan.docx(DOCX | 32 KB)
S4.2. DNA Barcoding - Week 04 - Caminalcules Handout STUDENT.docx(DOCX | 238 KB)
S4.3. DNA Barcoding - Week 04 - Caminalcules- Handout INSTRUCTOR KEY.docx(DOCX | 332 KB)
S4.4. DNA Barcoding - Week 04 - Caminalcules DNA Barcodes.pptx(PPTX | 62 KB)
S4.5. DNA Barcoding - Week 04 - Caminalcules - MYA alignment sheet.pptx(PPTX | 45 KB)
S4.6. DNA Barcoding - Week 04 - Caminalcules Lecture Slides.pptx(PPTX | 431 KB)
S4.7. DNA Barcoding - Week 04 - Caminalcules Prep Notes.docx(DOCX | 24 KB)
S5.1. DNA Barcoding - Week 05 - Biodiversity 1 Lesson Plan.docx(DOCX | 23 KB)
S5.2. DNA Barcoding - Week 05 - Biodiversity 1 Handout INSTRUCTOR KEY.docx(DOCX | 2 MB)
S5.3. DNA Barcoding - Week 05 - Biodiversity 1 Prep Notes.docx(DOCX | 24 KB)
S5.4. DNA Barcoding - Week 05 - Biodiversity Data Collection Sheet.docx(DOCX | 26 KB)
S5.5. DNA Barcoding - Week 05 - Biodiversity 1 Handout STUDENT.docx(DOCX | 2 MB)
S5.6. DNA Barcoding - Week 05 - Biodiversity 1 Lecture Slides.pptx(PPTX | 6 MB)
S6.1. DNA Barcoding - Week 06 - Biodiversity 2 Lesson Plan.docx(DOCX | 23 KB)
S6.2. DNA Barcoding - Week 06 - Biodiversity 2 Handout - STUDENT.docx(DOCX | 637 KB)
S6.3. DNA Barcoding - Week 06 - Biodiversity Lab Notebook Rubric.docx(DOCX | 18 KB)
S6.4. DNA Barcoding - Week 06 - Biodiversity 2 Lecture Slides.pptx(PPTX | 1 MB)
S6.5. DNA Barcoding - Week 06 - Biodiversity Lab Report - Guidelines Rubric and Template.docx(DOCX | 90 KB)
S6.6. DNA Barcoding - Week 06 - Biodiversity 2 Data Template.xlsx(XLSX | 14 KB)
S6.7. DNA Barcoding - Week 06 - Biodiversity 2 Handout - INSTRUCTOR KEY.docx(DOCX | 674 KB)
S6.8. DNA Barcoding - Week 06 - Biodiversity 2 Prep Notes.docx(DOCX | 19 KB)
S7.1. DNA Barcoding - Week 07 - Pipetting and DNA Extraction Lesson Plan.docx(DOCX | 24 KB)
S7.2. DNA Barcoding - Week 07 - Pipetting and DNA Extraction Lecture Slides.pptx(PPTX | 3 MB)
S7.3. DNA Barcoding - Week 07 - Pipetting and DNA Extraction Handout - STUDENT.docx(DOCX | 234 KB)
S7.4. DNA Barcoding - Week 07 - Pipetting and DNA Extraction Handout - INSTRUCTOR KEY.docx(DOCX | 253 KB)
S7.5. DNA Barcoding - Week 07 - Pipetting and DNA Extraction Prep Notes.docx(DOCX | 227 KB)
S8.1. DNA Barcoding - Week 08 - PCR Lesson Plan.docx(DOCX | 25 KB)
S8.2. DNA Barcoding - Week 08 - PCR lecture slides.pptx(PPTX | 4 MB)
S8.3. DNA Barcoding - Week 08 - Master Mix Calculator Template.xlsx(XLSX | 13 KB)
S8.4. DNA Barcoding - Week 08 - Lab Notebook Grading Rubric.docx(DOCX | 19 KB)
S8.5. DNA Barcoding - Week 08 - PCR Handout - STUDENT.docx(DOCX | 508 KB)
S8.6. DNA Barcoding - Week 08 - PCR Prep Notes.docx(DOCX | 27 KB)
S8.7. DNA Barcoding - Week 08 - PCR Handout - INSTRUCTOR KEY.docx(DOCX | 510 KB)
S9.1. DNA Barcoding - Week 09 - Gel Electrophoresis Lesson Plan.docx(DOCX | 26 KB)
S9.2. DNA Barcoding - Week 09 - Gel Electrophoresis Lecture slides.pptx(PPTX | 5 MB)
S9.3. DNA Barcoding - Week 09 - Gel Image Template.pptx(PPTX | 274 KB)
S9.4. DNA Barcoding - Week 09 - Gel Electrophoresis Handout - STUDENT.docx(DOCX | 48 KB)
S9.5. DNA Barcoding - Week 09 - Gel Electrophoresis Rubric.docx(DOCX | 18 KB)
S9.6. DNA Barcoding - Week 09 - Gel Electrophoresis Prep Notes.docx(DOCX | 29 KB)
S9.7. DNA Barcoding - Week 09 - Gel Electrophoresis Handout - INSTRUCTOR KEY.docx(DOCX | 48 KB)
S10.1. DNA Barcoding - Week 10 - DNA Extraction and PCR II Lesson Plan.docx(DOCX | 28 KB)
S10.2. DNA Barcoding - Week 10 XXX Alternative Plant DNA Extraction Protocol.docx(DOCX | 94 KB)
S10.3. DNA Barcoding - Week 10 - Master Mix Calculator Template.xlsx(XLSX | 13 KB)
S10.4. DNA Barcoding - Week 10 - DNA Extractions and PCR II Handout - STUDENT.docx(DOCX | 246 KB)
S10.5. DNA Barcoding - Week 10 - DNA Extractions and PCR II Lab Notebook Grading Rubric.docx(DOCX | 17 KB)
S10.6. DNA Barcoding - Week 10 - DNA Extractions and PCR 2 Prep Notes.docx(DOCX | 30 KB)
S10.7. DNA Barcoding - Week 10 - DNA Extractions and PCR II Handout - INSTRUCTOR KEY.docx(DOCX | 247 KB)
S11.1. DNA Barcoding - Week 11 - Gels 2 and DNA Sequencing Lesson Plan.docx(DOCX | 23 KB)
S11.2. DNA Barcoding - Week 11 - Gel 2 X DNA Sequencing Lecture.pptx(PPTX | 77 MB)
S11.3. DNA Barcoding - Week 11 - Gel 2 X DNA Sequencing Handout - STUDENT.docx(DOCX | 208 KB)
S11.4. DNA Barcoding - Week 11 - Prep Notes.docx(DOCX | 108 KB)
S11.5. DNA Barcoding - Week 11 - Gel 2 X DNA Sequencing Handout - INSTRUCTOR KEY.docx(DOCX | 209 KB)
S12.1. DNA Barcoding - Week 12 - Bioinformatics - DNA Subway Lesson Plan.docx(DOCX | 22 KB)
S12.2. DNA Barcoding - Week 12 - DNA Subway Lecture.pptx(PPTX | 7 MB)
S12.3. DNA Barcoding - Week 12-1-plant-pos-fall15F_PREMIX_J43474_3.txt(TXT | 228 KB)
S12.4. DNA Barcoding - Week 12 - 1-plant-pos-fall15R_PREMIX_J43475_4.txt(TXT | 228 KB)
S12.5. DNA Barcoding - Week 12 - DNA Subway Handout - STUDENT.docx(DOCX | 8 MB)
S12.6. DNA Barcoding - Week 12 - Prep Notes.docx(DOCX | 24 KB)
S12.7. DNA Barcoding - Week 12 - DNA Subway Handout - INSTRUCTOR KEY.docx(DOCX | 8 MB)
S12.8. DNA Barcoding - Week 12 - DNA Subway Lab Notebook Rubric.docx(DOCX | 22 KB)
S13.1. DNA Barcoding - Week 13 - iNaturalist and DNA Subway Posters Lesson Plan.docx(DOCX | 25 KB)
S13.2. DNA Barcoding - Week 13 - iNaturalist and Posters - Lecture.pptx(PPTX | 3 MB)
S13.3. DNA Barcoding - Week 13 - iNaturalist and Posters Handout - STUDENT.docx(DOCX | 1 MB)
S13.4. DNA Barcoding - Week 13 - iNaturalist and Posters - Links to Poster Design Readings.docx(DOCX | 19 KB)
S13.5. DNA Barcoding - Week 13 - iNaturalist and Posters - Poster Template.pptx(PPTX | 488 KB)
S13.6. DNA Barcoding - Week 13 - DNA Barcoding Poster Grading Rubric.docx(DOCX | 22 KB)
S13.7. DNA Barcoding - Week 13 - iNaturalist and Posters - Prep Notes.docx(DOCX | 24 KB)
S13.8. DNA Barcoding - Week 13 - Quantify Biodiversity in iNaturalist.docx(DOCX | 935 KB)
S14.1. DNA Barcoding - Week 14 - Final Exam Study Guide.docx .docx(DOCX | 24 KB)
- License terms

Comments
Comments
There are no comments on this resource.