Make It Stick: Teaching Gene Targeting with Ribbons and Fasteners
Published online:
Abstract
Manipulating gene expression is a commonly used tool to study the effect of a single gene or the hierarchy of gene networks in many different biological disciplines. When working with mice, the most commonly used techniques to manipulate genes include gene targeting via homologous recombination to achieve loss-of-function and gain-of-function mutations. Students often struggle with the concepts behind homologous recombination and the physical changes that happen at the target gene locus. Our activity uses different colored ribbons connected by hook-and-loop fasteners (e.g., those most popularly produced by the VELCRO Brand) that students use to design targeting constructs as well as to model the recombination between their constructs and the gene locus. This hands-on exercise helps students better understand the mechanisms of homologous recombination happening at the gene locus, enabling them to progress to higher-level cognition such as predicting experimental outcomes and designing their own gene targeting experiments.
Citation
Jakob S. and Anderson, W. J. 2019. Make it stick: Teaching gene targeting with ribbons and fasteners. CourseSource. https://doi.org/10.24918/cs.2019.9Society Learning Goals
Developmental Biology
- Gene Networks
- How do differences in regulation of gene expression explain the formation of atypical cells, tissue, organs and structures?
- Comparative Development and Evolution
- How do changes in expression patterns of existing genes, or genetic modifications of existing signaling pathways result in new phenotypes?
- Experimental Approaches
- How do the methods and tools of developmental biology help us understand cause and effect relationships during embryogenesis? (correlation, gain of function and loss of function; molecular, cellular, tissue, organ)
Genetics
- Methods & Tools in Genetics
- What experimental methods are commonly used to analyze gene structure and gene expression?
Lesson Learning Goals
Students will understand homologous recombination as a genetic tool to help study the effect of gene expression during embryonic development.Lesson Learning Objectives
- Students will be able to design targeting constructs.
- Students will be able to predict changes to the gene locus after homologous recombination.
- Students will be able to design experiments to answer a biological question (e.g., "Design an experiment to test if the expression of gene X is necessary for limb development").
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
In many biological disciplines, researchers study gene expression or the effect of changes in gene expression on a given phenotype. The most commonly used manipulations of gene expression are loss-of-function approaches in which a gene is deleted from the genome (knock-out) or gain-of-function approaches in which a gene is ectopically expressed in a place or at a time it is usually not expressed (for a review see 1,2). Subsequent phenotypic changes are then characterized in detail to better understand gene function and underlying mechanisms. These techniques have been used in mammalian systems (most notably in mice), as a vast variety of genetic tools have been developed to manipulate the mouse genome over the past few decades. However, similar approaches have also been used in other model organisms.
The traditional approach for these gene manipulations utilizes homologous recombination to specifically target a desired gene locus. During homologous recombination, nucleotide sequences are exchanged between two highly similar or identical DNA sequences. Researchers can control the precise region of recombination (targeting) and, depending on the chosen technique, where and when the genetic manipulation comes into effect (that is, the spatial and temporal specificity). For example, the Cre/lox system allows a researcher to delete a target gene only in a certain cell type or only at a certain time point (for review see 1,2). Generally, the researcher first designs a targeting construct that replaces endogenous sequences in the genome with a chosen exogenous sequence. This DNA construct is then electroporated into mouse embryonic stem cells. Homologous recombination between the targeting construct and the gene locus leads to the incorporation of the desired manipulation (Figure 1). In addition to loss- and gain-of-function manipulations, the principle of homologous recombination is also often used in describing the activity of Cre recombinase, which is a useful tool for indelible lineage tracing . In lineage tracing the Cre-mediated recombination results in the permanent labeling of a cell with a genetic marker, which will be inherited by all daughter cells. This allows the tracing of all cells descendant from the originally labeled cell (for a review see 3,4).

Figure 1. General background on gene targeting and generating mutant mouse lines. (A) Overview strategy for generating a loss-of-function allele. The targeted gene (in this case, Fgf4) is removed from its genomic locus. (B) A targeting construct replaces the open reading frame for the Fgf4 gene. The 5’ and 3’ homology arms (corresponding in this case to sequences upstream and downstream of the open reading frame) in the targeting construct aid in aligning the construct to the correct location in the genome (in this example, the Fgf4 gene on chromosome 11 in mouse). Homologous recombination between the targeting construct and the genomic locus results in the incorporation of a positive selection cassette (in this example, the neomycin resistance gene abbreviated neoR) where the Fgf4 gene formerly resided. Elements in the targeting construct located outside of the homology arms (in this example, the negative selection cassette using HSV-tk) are not incorporated into the genomic locus. (C) Schematic of the entire approach from targeting embryonic stem cells to generating knockout mice. The targeting construct is electroporated into mouse embryonic stem cells (ESCs). The ESCs undergo positive selection for cells expressing the neoR gene (using G418) and negative selection for cells expressing the HSV-tk gene (using ganciclovir). Details are provided in Supporting File S1: Genetic Tools Review Slides. Targeting is confirmed by DNA analysis and then properly targeted ESCs are injected into blastocyst stage embryos. These embryos are then transferred to a foster mother to generate chimeras. Chimeras are then tested for germline transmission of the ESC genome to produce mice in which all cells harbor the mutation. These F1 mice are intercrossed to generate a heterozygous mouse line, which can then be further crossed to generate homozygous null embryos/animals for analysis.
While numerous reviews and videos exist that explain the mechanism of homologous recombination (for a selection see Table 1), we have found students continue to struggle with understanding the process. The failure to understand these underlying principles hampers higher-level student learning, where they become capable of making predictions and designing experiments.
The use of manipulatives in teaching has been documented to have positive effects on understanding, retention, and problem solving, and they transfer to different STEM disciplines (5,6). Here we developed an exercise in which students use ribbons to design targeting constructs and subsequently model homologous recombination leading to a gene knock-out at a specified locus. Teaching fellows and students commented that this hands-on three-dimensional activity helps facilitate student understanding better than conventionally used artistic representations. By solidifying this core concept, students seemed to progress more easily to a level of thinking that allowed them to make predictions about changes happening at the targeted gene locus, as well as designing experiments to answer questions about whether a gene is necessary and sufficient for a biological process (by using a loss-of-function and gain-of-function approach, respectively).
INTENDED AUDIENCE
The audience consisted of undergraduate students at a large private research university taking a developmental biology course. Prior to enrolling in the course, most students have completed introductory courses covering the basics of molecular biology and genetics during their freshman year. Students were familiar with the concepts of gene structure (e.g., exons, introns, promoters, enhancers), recombination, inheritance, transcription (e.g., transcriptional start and stop sites, transcription factors, untranslated regions, open reading frames), and translation, which aid in the exercise. As a result, students were mainly sophomores, with a few freshmen, juniors, and seniors. Enrollment of the course varies typically between 50 - 80 students. At the point the students started the course, they had not yet declared their major, but eventually almost all chose to major in a life science field by the end of the semester.
Students assembled collectively in lecture, which meets three times a week for 75 minutes. Additionally, the course included a 90-minute weekly discussion section consisting of groups of 14-18 students. The activity described here was designed for the discussion section following a lecture that introduced gene targeting, homologous recombination, the Cre/lox system, and lineage tracing. The activity can easily be broken up into parts and interspersed within the lecture. The activity can also be adapted to teach homologous recombination more generally (e.g., in the context of meiosis).
REQUIRED LEARNING TIME
The exercise takes approximately 60 minutes. Prior to the exercise, students watch an online video at their leisure outside of class that takes approximately 40 minutes.
PRE-REQUISITE STUDENT KNOWLEDGE
The pre-requisites are two integrated introductory biology/chemistry courses (Introduction to Chemistry, Molecular Biology, and Cell Biology; Introduction to Genetics, Genomics, and Evolution) that students generally take in their freshman year. The pre-requisite for the activity is the lecture that introduces gene targeting, homologous recombination, the Cre/lox system, and lineage tracing (concepts are included in Supporting File S1: Genetic Tools Review Slides).
PRE-REQUISITE TEACHER KNOWLEDGE
The instructor should be familiar with gene structure, transcription, homologous recombination, and gene targeting. Table 1 contains a list of papers and videos that are excellent resources for gene structure and the mechanism behind both homologous recombination and the Cre/lox system. Review slides for these topics are also included in the supporting material (Supporting File S1: Genetic Tools Review Slides).
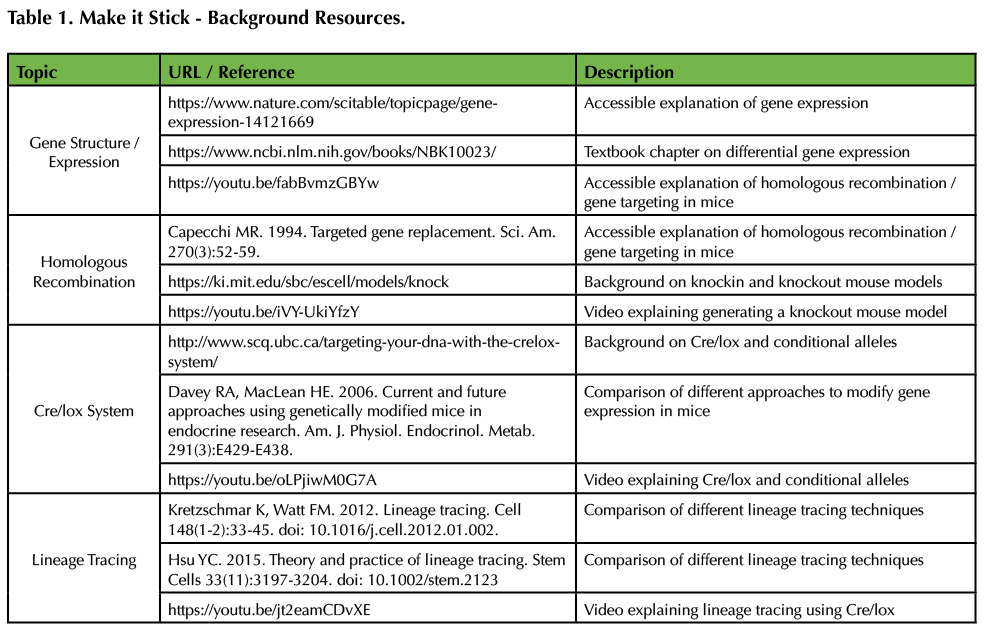
Table 1. Make it Stick - Background Resources. The listed resources are suggestions for preparation on gene structure, gene targeting via homologous recombination, the Cre/lox system, and lineage tracing.
SCIENTIFIC TEACHING THEMES
ACTIVE LEARNING
The activity included hands-on manipulations of material (modeling) as students put together the correct parts of ribbons to make their target construct ribbon and subsequently modeled the homologous recombination at a specified gene locus. Students performed this activity in pairs or small groups, which added a think-pair-share element. After all students completed part one of the exercise with their partner(s), the instructor brought the entire group back together (Table 2). Students shared their solution in peer discussion, as well as potential problems they encountered. The instructor facilitated this discussion, allowing students to answer their peers' questions and to bounce potential questions back to the students to encourage a student-centered discussion and to foster an inclusive classroom.
ASSESSMENT
Formative assessment mainly consisted of student feedback during and after the exercise as well as teaching fellow observations during class. Summative feedback included homework and exams, which included questions that build on this exercise (see Supporting File S5: Example Examination Questions).
INCLUSIVE TEACHING
Students were first taught the concepts and theory of gene targeting in a classroom active learning lecture setting. Students watched a pre-recorded lecture video that taught them the principles behind gene targeting before going through examples in class. This was followed-up with a discussion section meeting in which the activity was performed. The activity gave the students an opportunity to go through the process step-by-step, using tactile manipulation. The activity appealed to students who appreciate visual learning tools and tactile manipulations to visualize what is happening at the gene locus. Students were given the opportunity to think about the posed question first, then to discuss it with their partner(s), and finally to be part of a small group discussion. Engaging students via think-pair-share has been shown to reduce anxiety and increase student learning (7,8). The activity is designed to help students understand a common lab technique that is used in labs by "real" researchers. As a result, they felt they are learning something applicable and relevant, which they found exciting and engaging.
LESSON PLAN
The activity is designed for a discussion section meeting after the basics of gene manipulation via homologous recombination and Cre/lox techniques have been discussed in lecture (concepts are shown in Supporting File S1: Genetic Tools Review Slides).
Pre-class Preparation
The instructor needs to be familiar with homologous recombination and Cre/lox techniques. Relevant resources explaining gene structure, homologous recombination, and the Cre/lox system are listed in Table 1.
Material (Ribbon) Preparation
The instructor needs to prepare the genome ribbon consisting of:
- 5' upstream genomic sequence homology arms
- open reading frame (ORF)
- 3' downstream genomic sequence homology arms
and the ten construct ribbons:
- 5' homology arm
- 3' homology arm
- positive selection cassette / neo(R)
- negative selection cassette / HSV-tk
- ubiquitous promoter
- transcriptional termination sequence / STOP
- reporter / GFP
- open reading frame (ORF)
- loxP site
- loxP site
Before class, the instructor needs to prepare the following: cut the different colored ribbons into individual pieces, attach hook-and-loop fastener stickers to the ends of each ribbon so ribbon parts can be linked to one another, and write the name of each ribbon part (e.g., “GFP” or “neoR”) on the respective ribbons (see Figure 2 and Supporting File S2: Ribbon Preparation).
One set of genome and construct ribbons is necessary for each group of students. We usually have students work in pairs. Due to the group discussion in the end, the activity works best for 12-18 students. This small class or section size allows one member of the teaching staff (e.g., one teaching assistant leading a section) to walk around and interact with all students in the given time frame, as well as keep summarizing discussions short enough to fit into the proposed time frame. However, the activity can easily be adapted to larger class sizes with more teaching staff.
If desired, the instructor can also prepare an instructor ribbon, essentially the same parts as for the student ribbons (genome ribbon and ten construct ribbons) but using a larger ribbon (both in terms of width and length). This instructor ribbon can then be used to demonstrate the correct answers in front of the class for all students to see (Supporting File S2: Ribbon Preparation).
Progression Through Activity in Class
At the beginning of class, student groups are created, with each group receiving the genome ribbon and the ten individual construct ribbon parts.
The students are given the exercise handout (Supporting File S3: Student Handout) that lists four exercises to work through using their ribbons (the number of exercises can be adapted as needed). The instructor follows the instructor guidelines, which provide the detailed walk-through and answers for each exercise (Supporting File S4: Instructor Guide to Student Handout).
Students are given several (2 to 3) minutes to read through Exercise 1, which is aimed at understanding homologous recombination at the target gene locus (genome ribbon). Students are asked to design the targeting construct from the ten available ribbon parts and subsequently to model how the open reading frame would be removed from the genome via homologous recombination between the gene locus and the targeting construct (Figure 3).
Students work on the exercise in pairs for 10 minutes while the instructor walks around to check-in and answer any questions. Once the students complete Exercise 1, the instructor brings the entire group back together and asks students to share their solutions with everyone (Figure 3). The instructor then demonstrates the correct answers with the instructor ribbons (details are provided in the instructor guidelines in Supporting File S4: Instructor Guide to Student Handout). Once everyone understands the correct answers and rationale, students move to Exercise 2.
The students are given several (2 to 3) minutes to read Exercise 2, which is aimed at understanding the generation of a floxed gene (a gene flanked by loxP sites, short sequences that are recognized by Cre recombinase that allow the gene to be excised out of the genome via homologous recombination). Students are asked to design the targeting construct from the ten available ribbon parts and subsequently to model how the target gene would be removed from the genome via homologous recombination between the gene locus and the targeting construct. Students work on the exercise in pairs for 10 minutes while the instructor walks around to check-in and answer any questions. Once students complete Exercise 2, the instructor brings the entire group back together and asks students to share their solutions with everyone. The instructor then demonstrates the correct answers with the instructor ribbons. Once everyone understands the correct answers and rationale, students can move on to the next exercise.
This general flow can be repeated with as many exercises as planned for the lesson. The included exercise handout gives some example exercises as well as instructor guidelines (Supporting File S3: Student Handout and Supporting File S4: Instructor Guide to Student Handout). A recommended timeline for the activity is provided in Table 2.
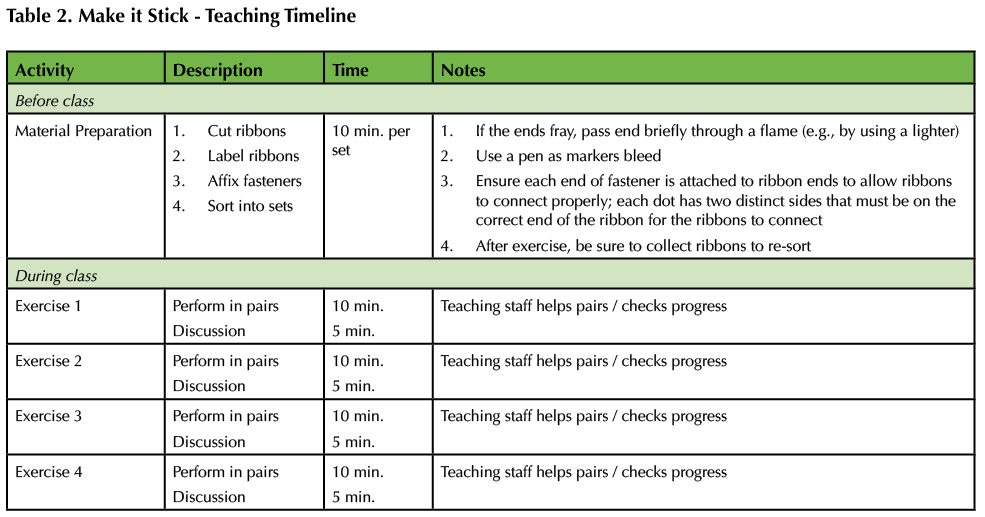
Table 2. Make it Stick - Teaching Timeline.
TEACHING DISCUSSION
Teaching assistants observed high student engagement with the activity. Those teaching assistants who taught the same class in the previous academic year (i.e., without the ribbon activity) mentioned that the hands-on activity was much more effective in teaching the principle of homologous recombination than drawing at the board. According to the teaching fellows, students seemed to have an easier time grasping three-dimensional rearrangements using the ribbons. Students were more vocal participants in section and more engaged in discussion with their peers. Students grasped the concept earlier than the prior year and were able to answer questions in section more correctly.
One teaching fellow commented:
"[The ribbon activity] was one of my favorite exercises in the year. I think it was much more effective than drawing out the Cre/lox system on the board or using printed out cards because a lot of students didn't quite understand how the recombination part happened. In the previous year, when I would ask the same question on the board, only a couple of students would actually be able to give me the correct answer in that point in the section. I would say more students understood the concept out of the group, and they understood it earlier on in the section (before we even got to the [practice] questions). And when we took up the [practice] questions, there was more participation and correct answers."
Below are two representative student feedback comments about the ribbon activity collected after the course concluded:
"The tangibility of the ribbon exercise was extremely helpful in understanding how homologous recombination works. By physically moving the ribbon 'DNA,' I found that I could visualize and conceptualize the process of DNA construct incorporation into the genome better than I could have had I merely listened to a lecture or seen it illustrated in a book or drawn on a chalkboard. I found that this activity especially helped me understand how the HSV-tk cassette was not incorporated via homologous recombination and how it could consequently be used to select for constructs that had integrated properly. The exercise also reinforced why each component of a construct (i.e. homology arms, neomycin resistance, HSV-tk cassette) was necessary. Overall, this activity made the concept of construct construction and integration into the genome more memorable - when I approached problems later on in the class that drew upon this topic, I found that I was able to easily generate constructs with the proper components and visualize maneuvers taking place inside the nucleus of cells in order to properly incorporate those constructs into the genome."
"The ribbon exercise made the concept of homologous recombination tangible. Being able to physically move the pieces to discover how the system works made a complicated process simple and fun to learn."
Overall student feedback showed that the students liked the ribbon activity. The teaching staff noted the increased effectiveness of teaching the concept of homologous recombination compared to board drawings in prior years, which is in line with literature on the positive effects of manipulatives (5,6). We believe that this activity is an easy way to enhance the understanding of a complicated and crucial core concept, which is necessary to foster student progress towards higher levels of thinking. The activity can be easily adapted to a variety of different question types, making it a useful tool to teach homologous recombination in developmental biology or other disciplines (for some suggestions see Supporting File S4: Instructor Guide to Student Handout).
SUPPORTING MATERIALS
- S1. Teaching Gene Targeting - Genetic Tools Review Slides. A review presentation summarizing the background needed to understand homologous recombination, gene targeting, designing targeting constructs, and the Cre/lox system.
- S2. Teaching Gene Targeting - Ribbon Preparation. A detailed description on how to prepare the different ribbon components needed for this activity.
- S3. Teaching Gene Targeting - Student Handout. A suggestion of exercises for which the ribbon activity can be used within a 60-minute class period.
- S4. Teaching Gene Targeting - Instructor Guide to Student Handout. The instructor guide for S3: Student Handout with detailed steps and answers accompanying the student handout. Additional suggestions for exercises are also included.
- S5. Teaching Gene Targeting - Example Examination Questions.
ACKNOWLEDGMENTS
We would like to thank the teaching assistants who taught this exercise and the students who enrolled in the course during the Fall 2017 semester for their comments, enthusiasm, and feedback on the section activity. We thank Amie Holmes and Lisa Fountain for feedback on the manuscript. Susanne Jakob was a participant in the 2018 CourseSource Writing Workshop in Minneapolis, MN. We are grateful to the organizers of the workshop for the excellent feedback and advice.
References
- Bouabe H, Okkenhaug K. 2013. Gene targeting in mice: a review. Methods Mol. Biol. 1064:315-336. doi: 10.1007/978-1-62703-601-6_23.
- Carter M, Shieh J. 2015. Guide to Research Techniques in Neuroscience. Waltham, MA: Academic Press.
- Hsu YC. 2015. Theory and practice of lineage tracing. Stem Cells 33(11):3197-3204. doi: 10.1002/stem.2123
- Kretzschmar K, Watt FM. 2012. Lineage tracing. Cell 148(1-2):33-45. doi: 10.1016/j.cell.2012.01.002.
- Carbonneau KJ, Marley SC, Selig JP. 2013. A meta-analysis of the efficacy of teaching mathematics with concrete manipulatives. J. Educ. Psychol. 105(2):380-400. doi: 10.1037/a0031084.
- Krontiris-Litowitz J. 2003. Using manipulatives to improve learning in the undergraduate neurophysiology curriculum. Adv. Physiol. Educ. 27(1-4):109-119. doi: 10.1152/advan.00042.2002.
- Kwok AP, Lau A. 2015. An exploratory study on using the think-pair-share cooperative learning strategy. J. Math. Sci. 2:22-28.
- Kaddoura M. 2013. Think pair share: a teaching learning strategy to enhance students' critical thinking. E. R. Q. 36(4):3-24.
Article Files
Login to access supporting documents
Make It Stick: Teaching Gene Targeting with Ribbons and Fasteners(PDF | 1 MB)
S1.Teaching Gene Targeting - Genetic Tools Review Slides.pptx(PPTX | 480 KB)
S2. Teaching Gene Targeting - Ribbon Preparation.docx(DOCX | 23 KB)
S3. Teaching Gene Targeting - Student Handout.docx(DOCX | 18 KB)
S4. Teaching Gene Targeting - Instructor Guide to Student Handout.docx(DOCX | 1 MB)
S5.Teaching Gene Targeting - Example Examination Questions.docx(DOCX | 27 KB)
- License terms

Comments
Comments
There are no comments on this resource.