CRISPR/Cas9 in yeast: a multi-week laboratory exercise for undergraduate students
Published online:
Abstract
Providing undergraduate life-science students with a course-based research experience that utilizes cutting-edge technology, is tractable for students, and is manageable as an instructor is a challenge. Here, I describe a multi-week lesson plan for a laboratory-based course with the goal of editing the genome of budding yeast, Saccharomyces cerevisiae. Students apply knowledge regarding advanced topics such as: CRISPR/Cas9 gene editing, DNA repair, genetics, and cloning. The lesson requires students to master skills such as bioinformatics analysis, restriction enzyme digestion, ligation, basic microbiology skills, polymerase chain reaction, and plasmid purification. Instructors are led through the technical aspects of the protocols, as well as the teaching philosophy involved throughout the laboratory experience. As it stands, the laboratory lesson is appropriate for 6-8 weeks of an upper-level undergraduate laboratory course, but may be adapted for shorter stints and students with less experience. Students complete the lesson with a more realistic idea of life science research and report significant learning gains. I anticipate this lesson to provide instructors and students in undergraduate programs with a hands-on, discovery-based learning experience that allows students to cultivate skills essential for success in the life sciences.
Citation
Ulbricht, R.J. 2019. CRISPR/Cas9 in yeast: a multi-week laboratory exercise for undergraduate students. CourseSource. https://doi.org/10.24918/cs.2019.19Society Learning Goals
Biochemistry and Molecular Biology
- Macromolecular Structure Determines Function and Regulation
- What factors determine structure?
- Information storage and flow are dynamic and interactive
- What is a genome?
- How do genomes transmit information from one generation to the next?
- Discovery requires objective measurement, quantitative analysis and clear communication
- What is the scientific process?
- What constitutes a scientific community of practice?
Genetics
- Nature of Genetic Material
- How is DNA organized?
Lesson Learning Goals
- Increase students' understanding of scientific sub-disciplines such as genetics, microbiology, and molecular biology.
- Increase students' understanding of CRISPR/Cas9 gene editing.
- Develop and refine practical skills related to work in a biological laboratory.
- Develop and refine scientific reasoning by:
- Designing scientific experiments and/or developing biological tools that apply to scientific problems.
- Critically examining scientific evidence and applying inferences to new scenarios.
- Communicate scientific knowledge, findings, and process.
- Appreciate the complexity and ambiguity of authentic scientific research.
Lesson Learning Objectives
Week 1: CRISPR design
- Locate the coding sequence, flanking sequence, protein product, and characteristics of a given gene from the Saccharomyces Genome Database (https://www.yeastgenome.org/).
- Design and defend the design of guide RNA and single stranded template for DNA repair in CRISPR/Cas9 gene editing studies to generate Saccharomyces cerevisiae auxotrophic mutants.
Week 3-4: Cloning
- Describe the qualities of the vector, pML104, that allow replication and selection in bacteria and yeast as well as allow expression of necessary factors in CRISPR/Cas9 genome editing, including Cas9 and sgRNA.
- Describe the rationale of and perform procedures necessary for cloning a small cassette (i.e., sgRNA gene) into a vector (i.e., pML104) including; restriction digest, annealing of DNA strands, removal of 5’ phosphates, ligation, and transformation.
- Recognize and design appropriate controls for cloning procedures such as ligation and transformation.
Week 5: Screening clones
- Describe the method of polymerase chain reaction (PCR), including the rationale for essential components of a reaction mixture and thermal-cycling conditions.
- Locate the binding sites of and design primers for PCR, then report the expected size of the amplification product.
- Describe and perform isolation of plasmid DNA from E. coli.
Week 6: Selection of clones and transformation of yeast
- Describe the rationale for and perform procedures to transform yeast, including the essential components of a transformation mixture and conditions necessary for transformation.
- Describe the basic conditions required for cultivating yeast.
- Describe the rationale for and perform agarose gel electrophoresis of a given size of DNA.
- Analyze DNA separated by agarose gel electrophoresis, including size estimation.
- Recognize and describe the qualities of a template for DNA repair that allows efficient DNA repair.
Week 7: Phenotyping
- Design an experiment to determine auxotrophic phenotypes.
- Predict the outcome of multi-step experiments.
Multiweek
- Recognize and describe conditions necessary for growth of E. coli and S. cerevisiae.
- Qualitatively and quantitatively analyze scientific data from scientific experiments, including bacterial and yeast transformation, agarose gel electrophoresis, extraction of plasmid DNA from bacteria, PCR, and auxotroph phenotypic analysis.
- Communicate science to peers through maintenance of a laboratory notebook, verbal communication with group members, and writing of a formal laboratory report written in a format acceptable for journal publication.
- Troubleshoot scientific protocols by identifying procedures that are prone to error, comparing recommended protocols to actual procedure, and using positive and negative controls to narrow the location of a potential error.
- Communicate specific potential or actual uses of CRISPR/Cas9 in science and/or medicine.
- Use various bioinformatics approaches to analyze macromolecular primary sequence and structure.
- Illustrate how DNA is replicated and genes are transmitted from one generation to the next in multiple types of organisms including bacteria, eukaryotes, viruses, and retroviruses.
- Define what a genome consists of and how the information in various genes and other sequence classes within each genome are used to store and express genetic information.
- Explain the meaning of ploidy (haploid, diploid, aneuploid etc.) and how it relates to the number of homologues of each chromosome.
- Predict the effects of mutations on the activity, structure, or stability of a protein and design appropriate experiments to assess the effects of mutations.
- Predict the growth behavior of microbes based on their growth conditions, e.g., temperature, available nutrient, aeration level, etc.
- Discuss the benefits of specific tools of modern biotechnology that are derived from naturally occurring microbes (e.g. cloning vectors, restriction enzymes, Taq polymerase, etc.)
- Accurately prepare and use reagents and perform experiments.
- When presented with an observation, develop a testable and falsifiable hypothesis.
- When provided with a hypothesis, identify the appropriate experimental observations and controllable variables.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Undergraduate biology students benefit from authentic research experiences. The benefits of research participation are clear: They include clarification of a career path and enhancement of conceptual learning, problem solving skills, laboratory skills, resilience, and independence. The definition of an "authentic" research experience is less clear: Most definitions include engagement in the process of science (experimental design, data collection and analysis, and technical skills), as well as communication of scientific principles (1-4). Both course-based undergraduate research experiences (CURE) and individual undergraduate research experiences (URE) can provide the opportunity to learn elements of the process of science. However, with exposure and experience, student views of the scientific process become more complex (5), representing "authentic" science. In practice, the scientific process starts with observations and inquiry, then proceeds to identification of a research question, design of experiments to answer the question, followed by collection and analysis of data. Then, rather than proceeding directly to drawing conclusions, the process typically diverts to a trouble-shooting stage before circling back to revisit the research question, requiring redesign of the experiments. The outcome, at times, is the generation of novel data for consumption by the scientific community. However, results are often inconclusive, and/or lacking in interest to the scientific community. Some educational authentic research experiences include the creation of novel, publishable data of interest to the scientific community to be a defining characteristic. Programs with the goal of data product often do not accomplish that goal, but still show significant learning outcomes (1,6). Moreover, students within some course-base laboratory programs designed without the intention to generate novel data still report their experiences as "authentic" (7). Altogether, the outcomes associated with product-centered research experiences may also be achieved with intentional design, even when they do not produce novel data (7).
I describe a laboratory activity that employs discovery-based learning to integrate molecular biology concepts and the process of science to maximize learning. Students are instructed in the concepts underlying advanced laboratory skills including molecular cloning, bacterial transformation, yeast genetics, and PCR. Students are expected to analyze and integrate this knowledge by contributing to experimental design and trouble-shooting unsuccessful attempts. The concepts and skills are divided into mini-goals that connect across multiple exercises to have the ultimate outcome of site-specific editing of a eukaryotic genome. While there are multiple opportunities for student contributions, the choices are constrained to minimize the load on instructors.
Student engagement in the activity is maximized in part due to use of technology that is on the cutting-edge of life science research. Clustered regularly interspaced short palindromic repeats (CRISPR) gene-editing technology is at the forefront of scientific inquiry. Several mentions of CRISPR in mainstream media have piqued the public curiosity. A large part of the excitement over CRISPR is its relative simplicity in design and use, which makes it an optimal tool for use in teaching. Institutions such as Rollins College and University of New Mexico have described CRISPR/Cas9-mediated engineering of zebrafish and Drosophila genomes, respectively, in undergraduate laboratories with positive outcomes (8,9). Here, I describe a laboratory activity using CRIPSR/Cas9 to modify the genome in baker's yeast, Saccharomyces cerevisiae, a eukaryotic model organism that is easy and inexpensive to maintain. Overall, the laboratory experience is tractable to undergraduate students and can be performed with limited materials and expertise.
INTRODUCTION TO CRISPR
CRISPR is a genome-editing technology that was initially discovered in bacteria, where it serves as an innate immune system. CRISPR-associated (Cas) proteins are double-stranded endonucleases that are guided to cleave DNA at sites specified by an antisense base-paired CRISPR RNA (crRNA). Trans-acting crRNA (tracrRNA) binds both crRNA and Cas protein, linking the two so that the crRNA can guide Cas proteins to a complementary sequence of DNA. The only constraints for the ability of Cas (commonly Cas9) to cleave the DNA is that it has a region complementary to the 20 nucleotide crRNA that is immediately upstream of an NGG protospacer-adjacent-motif (PAM) (10,11). Scientists have simplified the system even further by fusing tracrRNA and crRNA into a single guide RNA (sgRNA) (12).
Repair to the CRISPR-generated double-stranded break can occur through one of two mechanisms; precise homology-directed repair (HR) and error-prone non-homologous end joining (NHEJ). NHEJ can be harnessed to generate random insertion and/or deletions (indels) at the site of the break, while HR can be used to integrate targeted alterations to the genomic sequence near the break (13). HR requires a homologous strand of DNA to serve as a template for repair. In a diploid organism, the opposing allele may be used as a template, generating two similar alleles. In S. cerevisiae, double stranded genome break repair is performed almost exclusively through HR (14). In order for the double stranded break to be repaired in haploid yeast strains, a homologous donor sequence must be incorporated into the cell to facilitate homologous repair. A synthetic single stranded oligonucleotide (ssODNA) is often used as a template for homologous repair after genome cleavage (15), directing HR to incorporate specified insertions, deletions, or mutations to the affected region of the genome (Figure 1 and Figure 2).
THE STUDENT EXPERIENCE
In the pilot of this laboratory, I provided students with a strain of S. cerevisiae and guided them through the experimental design to use CRISPR/Cas system to mutate the TRP1 gene, and produced tryptophan auxotrophs. While mutation of this gene provides a tractable phenotyping regime that is practical, relatively easy for students to understand, and has low technical barriers, it also eliminates one potential element that could contribute to student engagement: novel data production. I argue that the intentional design of the activity provides significant learning outcomes in its current, relatively simple format. Novel data could be generated with only slight modifications to the current exercise. For instance, since the design of the crRNA and HR template is left largely up to the students, students are generating novel data regarding the qualities (i.e., sequence, genomic location, length, ect) of crRNA and template sequences that are optimal for CRISPR/Cas9 gene editing to occur in yeast, a subject that remains relatively unclear in the literature (reviewed in 16). Alternatively, instructors may adapt this protocol to disrupt any gene of interest in the yeast genome, and could then apply the produced yeast strain toward the production of novel data.
While students are provided with intentional instruction on the concepts required for understanding, and instructed on the considerations for design, implementation, interpretation, and troubleshooting of procedures independently, they are responsible for significant elements of the scientific process. The added activity of a comprehensive written report provides them with an opportunity to become more proficient and comfortable with scientific communication. Students report significant gains in the process of science, but also in their understanding of concepts related to this course and other life science courses. Overall, students come away with a more realistic understanding of the research and report significant learning outcomes (Figure 3).
INTENDED AUDIENCE
Participants were upper-level undergraduate or lower level graduate students in a molecular biology course at a primarily undergraduate, public four-year institution. The course is a required for completion of the degree in Cell and Molecular Biology from the Department of Biomedical Sciences, but is also open to Agriculture and Biology terminal master's students at the University. The number of students in each course laboratory section varied between 14 and 22.
REQUIRED LEARNING TIME
The course is a 15 week long semester. However, the described learning activities are accomplished in 6-8 weeks of the course. The laboratory class period is 2 h 50 min long, one day a week. Some student out-of-class time is used for preparation or data collection. The course was first taught in the Spring of 2016 and was run in three semesters, with at least three sections each semester.
PREREQUISITE STUDENT KNOWLEDGE
Prerequisites for the course for which this activity was designed include completion of genetics, biochemistry and molecular cell biology courses, all containing laboratory components. In the lecture component of the course before the start of the laboratory exercise described here, students receive content knowledge and are assessed on DNA replication (in eukaryotes and prokaryotes), DNA repair, and CRISPR/Cas9 technology.
PREREQUISITE TEACHER KNOWLEDGE
Instructors should have some skills and/or knowledge of basic molecular biology, including molecular cloning, and yeast and bacterial culturing. Content knowledge on DNA repair mechanisms (specifically double-stranded break repair) is required and can be obtained through most genetics, biochemistry, molecular cell or molecular biology text books (e.g., 17-19). Knowledge of the mechanisms and uses for CRISPR/Cas9 gene editing is also required and can be obtained readily (reviewed by (13,20,21).
SCIENTIFIC TEACHING THEMES
ACTIVE LEARNING
This exercise provides students with an undergraduate research opportunity, an experience that is the epitome of active learning.
ASSESSMENT
SURVEY
Self-reported learning gains were assessed through survey completion. Surveys were administered anonymously via Salgsite.org. Likert scale questions were offered with the following options: no gains, little gain, moderate gain, good gain, great gain, and not applicable. Separate prompts requested student comments. All study protocols were approved by institutional IRB (protocol identification number FY2018-784).
LAB NOTEBOOKS
Student notebooks are used to assess preparation for each laboratory, as well as observations, data collection, and data analysis. Before the start of each laboratory exercise, students submit an introduction, materials and methods, and expected results portion of the lab notebook. The one page referenced introduction explicitly states the research goal and provides information necessary to the understanding of the goals and processes in the laboratory. Prompts are provided in the lab handout to direct students on some elements that should be included in this introduction. Students are encouraged to write the introduction after the materials and methods, addressing specific questions they had about the protocol in the introduction. The introduction is designed as practice in scientific writing, in addition to assessment of the content (accuracy and completeness of background). Extensive feedback is provided on elements of scientific writing, including sentence and paragraph structure, formatting of the references and in-text citations, third person narrative, etc. Formatting is determined by the required text, "A Short Guide to Writing in Biology" (22).
The materials and methods section of the laboratory notebook is based on the protocol given in the lab handouts. However, there are often places where students need to provide additional details not provided in the handouts. For instance, the lab handout might say to "pour an agarose gel" whereas each student's materials and methods section should explicitly write out the directions for this, including the amount of agarose and the volume of buffer. This is intended to provide a framework for experimental protocols but also provide an opportunity for students to solve problems and apply their skills and knowledge. The students in this setting have experience with basic lab techniques from at least four previous laboratory courses, as well as in the early weeks of this course, and therefore, are more than capable of transferring those skills to this laboratory. Students with less experience will likely need to be provided with more resources and guidance during and immediately before each exercise. I have recently began providing some content on the course website several days before the materials and methods are due to provide additional guidance through areas that are troublesome for students (Supporting File S1: CRISPR in Yeast - Lectures). Additional mini-lectures by the instructor at the start of each lab often help students complete or correct misconceptions or errors in the protocol. Students are also encouraged to consult with peers to address any inconsistencies in the materials and methods, prior to performing experiments. To minimize time spent grading the materials and methods, the instructor may grade for simple completion or choose 1-3 elements (e.g., reaction conditions, culture volumes) within the materials and methods to assess.
From the instructor perspective, the expected results portion of the lab notebook provides the most information on conceptual understanding. Students are expected to develop a hypothesis, a critical element for an authentic research experience, but also, to be able to visualize the product(s) of the assay based on this hypothesis. A common mistake is to state the hypothesized conclusion, but not actually describe the evidence that will support or reject the hypothesis. I have had success limiting the expected results to a few sentences, and even encouraging students sketch their anticipated data.
The final piece of notebook assessment is the results and data interpretation. The results are checked for completion (and feedback provided) in the week following the lab, but are collected and stringently graded twice during the semester. The format of results is standardized as much as possible according to the required writing text (22). For example, all figures must have a figure number, descriptive title, legend and be fully labeled. The results should be described in text, and the meaning or interpretation of the results discussed. When experiments fail, students are asked to hypothesize reasons for this, and describe a potential solution for this problem. Some students recognize this exercise of data interpretation and troubleshooting as helpful in development and practice of critical thinking skills: "I will carry with me confidence in troubleshooting experiments, drawing my own conclusions...", "I think this course helped me with critical thinking, especially if something in the experiment went wrong, I was required to think about what could have possibly gone wrong and how I could fix it if I was going to redo it."
EXAMS
The students are assessed for their understanding of topics with two formal exams. The first exam is given the week following the dry lab design exercise (Table 1 Lesson Plan Timeline). The topics included in this exam are discussed and practiced in the introductory laboratory exercises (i.e., sterile technique, DNA isolation, restriction enzyme digest, agarose gel electrophoresis, etc.) and, importantly, in the practical design of CRISPR-based experiments in yeast. The assessment is both practical and formal. For the practical portion, students are asked to describe how to do techniques, or to perform exercises parallel to those done in class. For example, they are asked to retrieve sequence information from genome databases, design guide RNA that will target CRISPR/Cas9 to knock-out gene expression from a given gene, describe how to streak a plate for individual colonies, etc. The exam also assesses the students' retention of concepts discussed in the lab, including their understanding of the purpose and/or principle behind techniques and solutions (Supporting File S2: CRISPR in Yeast - Sample Assessments). Students are assessed after the design of their CRISPR-based experiments, but before its application to allow the students and instructors to proceed confidently with the CRISPR experiments, but also to provide instructors with time to prepare and order materials necessary for the student-designed portion of the project.
A final practical exam assesses student understanding of laboratory methods, reagents, and practices relevant to the gene editing. This exam takes place the same week that formal lab reports are also due, with the goal of assessing assimilation of knowledge that was compiled in the formal report (Supporting File S2: CRISPR in Yeast - Sample Assessments).
FORMAL LAB REPORTS
Each student must write a formal lab report over the entire CRISPR gene editing in yeast laboratory exercise. These reports are modeled after a primary research article, with an introduction, materials and methods, results, and discussion. To encourage early writing, limited portions of the lab reports are turned into the instructor as early as week 2. The material assessed in these early drafts is limited (i.e., materials and methods from cloning week 1, or results from cloning lab 2), to limit load on the instructor and students. The drafts are low stakes (points awarded for completion) and feedback is designed to provide progressive guidance on scientific writing. Specific instruction in science writing is provided during portions of the laboratory with wait times (i.e., incubations). The lab report rubric (Supporting File S5: CRISPR in Yeast - Lab Report Rubric) is provided to the students during these discussions. Instructor feedback can be streamlined with the rubric. It is also possible to orchestrate peer-reviews of drafts during the laboratory section, or outside of class.
The lab reports provide an opportunity to assess the ability of students to communicate their understanding of the laboratory procedure, goals and outcomes to an audience of their peers. This communication is a pillar of the authentic research experience (1). Indeed, students report significant gains in their ability to and comfort with scientific writing (Figure 3B). Moreover, the lab report is an opportunity to compile the separate pieces of the exercise, making connections between the content and skills gained through the weeks. Students report significant gains in the ability to make these connections (Figure 3A), likely due, in large part, to lab reports. One student commented, "The lab report forced me to understand the bigger picture of what we did throughout the entire course of the semester."
INCLUSIVE TEACHING
As a CURE, this lesson provides students with an opportunity to participate in and interact with science in a way that overcomes significant barriers, giving a wider range of students the opportunity to see themselves and their peers as scientists (23). As student-scientists, they are required to prepare rationales and protocols (in the form of introduction, procedures, and expected results) ahead of individual experiments, giving time and framework to assemble their own questions and answers to problems. This strategy encourages active participation by students who are less likely to participate in discussion (24). The lesson and its structure also gives students the freedom to make decisions (and mistakes) regarding the process with minimal judgement or interference from instructor. Students are instead encouraged to utilize peer groups to validate or defend their own ideas. This provides a cooperative learning experience where the outcome of peer-discussions will determine students' performance in the lab (25). Weekly feedback on individual writing assignments provides another opportunity for inclusive teaching (24), but also can be used, along with formative assessments, to correct any misconceptions stemming from misguided student-led learning.
LESSON PLAN
The lesson plan is provided in modules: design, molecular cloning, screening of clones, yeast transformation, phenotyping, and genotyping (optional). Each module takes place in 1-2 class periods (or weeks). Each class period begins with a brief instructional period to clarify common points of misconception and provide more explicit instruction (i.e., where to find materials, how to operate equipment, etc.).
CRISPR DESIGN
Students are asked to design an experiment using CRISPR/Cas9 to generate yeast tryptophan auxotrophs by creating mutations in the TRP1 gene. Students have mastered the ability to design crRNA and recognize PAM sequences within a given gene or short stretch of DNA sequence in the lecture component. The student laboratory manual (Supporting File S3: CRISPR in Yeast - Laboratory Manual) guides students through: 1. Obtaining the sequence of a yeast gene, 2. Designing sgRNA recognition sites in the gene that potentially disrupt the gene, and 3. Designing a template for repair of the gene after cleavage. The activity can be completed in groups (I limit groups to a maximum of three students per group). This activity requires each student to have access to a computer with access to the internet and word processing. The laboratory begins by instructing students on concepts including an overview of yeast genetics and auxotrophy, DNA repair, Cas9 and guide RNA recognition are also reviewed. While students are familiar with homology-directed repair and that a homologous template is used in this repair pathway, they are unfamiliar with the specific qualities that a template must possess to be a template for repair, therefore, these concepts and data to support the specific qualities of an appropriate template for DNA repair are discussed in depth.
This dry bioinformatics exercise is entered into laboratory notebooks similar to other wet labs, where each entry (or exercise) has an introduction (with in-text citations and references), materials and methods, expected results, results/data interpretation. The results for the design lab should include the full TRP1 gene sequence from yeast, the location and sequence of at least three sgRNA binding sites on this gene, and the sequence of a single stranded template for DNA repair. All resources used to generate this data should be referenced in the laboratory notebook. The interpretation portion of the results should include a discussion about the expected phenotype and genotype of yeast that would be if the experiment were performed successfully.
In order to move forward with student-led experiments, I chose to collect all of the student-designed guide RNAs, then select the most common RNAs for further experimentation (many of the guide RNA recognition sites recur or overlap between lab groups). I limit the number of RNAs chosen for budgetary reasons and to reduce confusion on the part of instructors, however, it is possible to allow each individual group to proceed with their own chosen guide RNA. Hybridization of complementary oligonucleotide pairs yields double-stranded regions containing the sgRNA gene sequence, as well as ends that are compatible with direct cloning into SwaI and BclII site of the CRISPR/Cas9 expression vector, pML104 (26). Synthetic oligonucleotide pairs were ordered from Integrated DNA Technologies. It typically takes 2-3 days for standard processing and shipping.
Possible modifications to shorten the timeline of the overall exercise is to instead provide students with instructor-designed sgRNAs. Instructors could use this design exercise as practice or a demonstration on how the guide RNAs were designed, or perhaps reverse the activity and ask them to map the gene that the given RNA targets, the Cas9 cut site and a possible DNA repair template.
MOLECULAR CLONING
I chose the plasmid pML104 for expression of the essential components for CRISPR/Cas9 gene editing: Cas9 endonuclease and sgRNA (26). pML104 has a two restriction sites for directional cloning of small dsDNA cassette that allows production of an sgRNA from an RNA Polymerase III promoter. I allow students to purify their own plasmids from dam mutant E. coli in the practice portion of the laboratory, prior to discussion of CRISPR. Alternatively, the instructor may provide prepared pML104 to students. Since one of the restriction enzymes necessary for cloning (BclI) is blocked by Dam3 methylation, it is important to consider the type of E. coli strain used to amplify pML104. Two separate oligonucleotides are required for cloning of each student-designed sgRNA gene cassette (Figure 1). I order these oligonucleotides with the proper adapter sequences for cloning into pML104, however, it is possible to allow more advanced students to design these oligonucleotides. The sequences of the oligonucleotides are shared with students, as well as the reasoning behind the design. They are expected to map the location of sgRNA gene cassette cloning on pML104, and then map the location in the yeast genome targeted by their chosen guide RNA (if not already mapped in previous exercises).
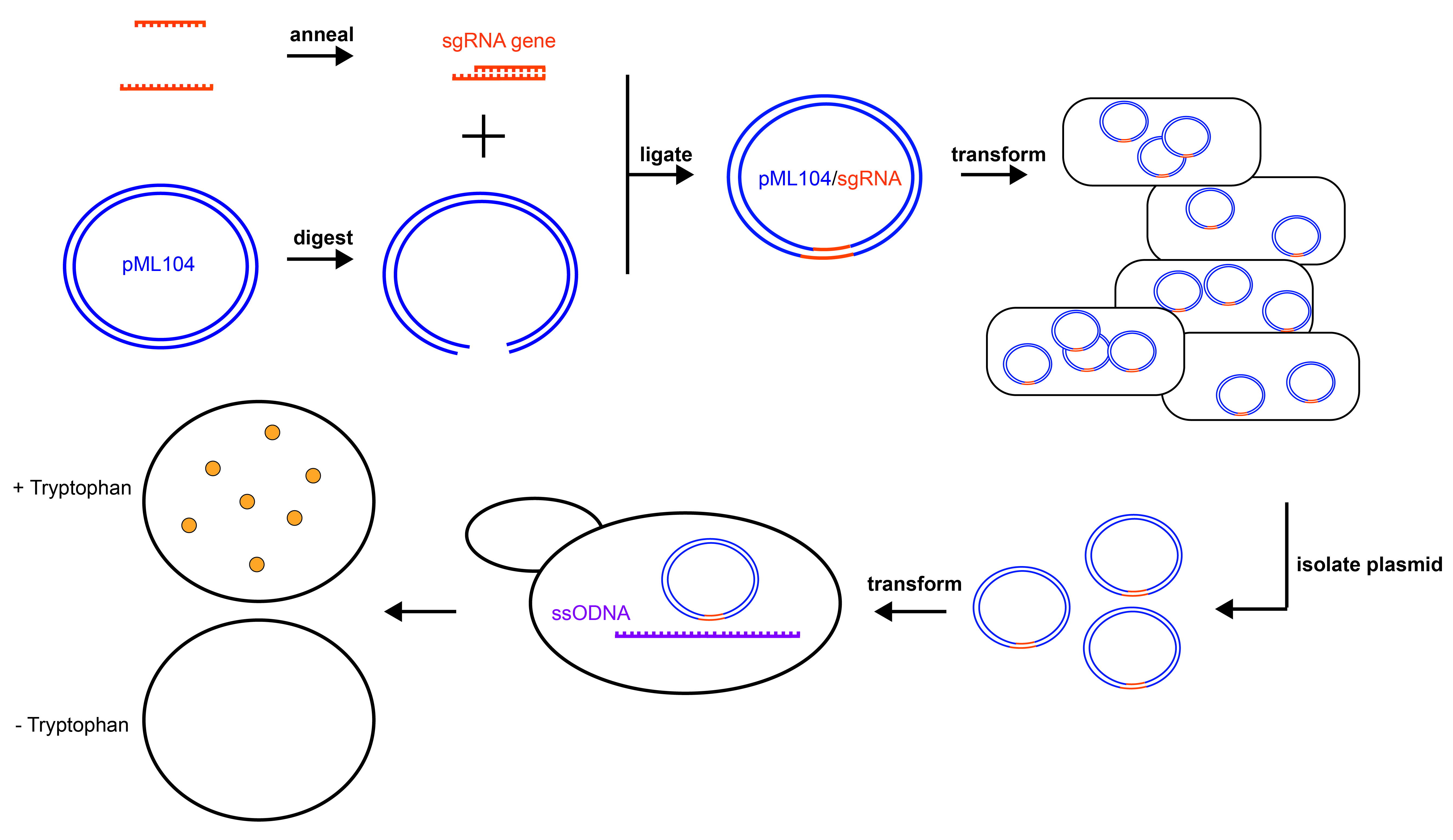
Figure 1. Experimental Overview. Graphical summary of the experiments to disrupt the TRP1 gene in yeast. pML104 (blue) is digested to generate asymmetrical DNA ends, while the sgRNA gene cassette (orange) is generated by annealing synthetic DNA oligonucleotides. Digested pML104 and the sgRNA gene cassette are ligated to form circular pML104/sgRNA, then transformed into E. coli for selection and amplification. pML104/sgRNA is isolated from E. coli and then transformed into yeast along with a ssODNA (purple), which will serve as the template for DNA repair. Mutants can be detected by lack of growth on media lacking tryptophan (- Tryptophan) but growth on media containing tryptophan (+ Tryptophan).
The cloning of the double-stranded sgRNA gene cassette into pML104 is divided between two class periods. First, the plasmid is digested and oligonucleotides are annealed to generate the cassette for cloning (Figure 1). The published protocol for cloning in pML104 suggests digestion and gel purification of pML104 (26). In the interest of time, I modified the protocol to dephosphorylate the vector and purify by phenol/chloroform extraction instead. Either technique will prevent recircularization of the vector. In the second period, the sgRNA gene cassette generated from annealed oligonucleotides is ligated into digested pML104 using a rapid DNA ligation kit, then transformed into E. coli (Figure 1). I chose to purchase chemically competent E. coli cells for transformation (Invitrogen, sub-cloning efficiency DH5alpha), however, protocols to generate chemically or electro-competent cells in-house are widely available (27). Students are expected to conduct appropriate control experiments, such as a positive control for bacterial transformation and a negative control for ligation. A description of each control and its purpose can be provided by the instructor, however, I chose to guide students through the reasoning behind the selection of controls and then allow them to decide on the controls they will conduct. The results from the cloning portion of the lab should include an estimation of transformation efficiency (formula and description provided in Supporting File S3: CRISPR in Yeast - Laboratory Manual) from both experimental and control plates. Students should speculate the reason for growth or no growth on selective experimental plates, based on control plates.
Providing students with an instructor-cloned sgRNA is one way to shorten the timeline of the overall experiment by at least two weeks. This modification would also lower the barriers for less experienced students, and provide a more direct path from design of CRISPR/Cas9 experiments to implementation. With this modification, the next steps to screen the clones would be optional.
SCREENING CLONES
Screening for insertion of the sgRNA gene cassette into pML104 is accomplished using PCR. But first, students select colonies of transformed E. coli and isolate plasmid DNA. Students are responsible for starting their own selective cultures (as well as a negative control) the afternoon before their class is scheduled. Students who forget to start cultures or fail to see growth can either opt to perform the procedure during another lab section, on their own time, or obtain a portion of a culture from another group. We have tried an alkaline lysis/ethanol precipitation mini-prep plasmid isolation protocol (27,28), as well as a kit (Promega SV miniprep). The results from the kit are substantially more reliable, and result in fewer errors by students. I take care to discuss the contents and purpose of all materials in the kit, as well as alternative DNA purification methods.
PCR is used to screen for the presence of the sgRNA gene cassette within pML104 backbone. I use a PCR master mix that contains all necessary PCR components, except template and primers. The use of this mix helps prevent common student errors (pipetting, missing reagents, etc.), but care should be taken to fully explain the contents of this mix. The primer sequences are provided (Supporting File S3 CRISPR in Yeast - Laboratory Manual). In the preparation for the lab, students are asked to locate the primer binding sites on the plasmid map, and predict results. Some suggestions that can help students locate primer binding sites are provided in a PowerPoint file available to students before class (Supporting File S1: CRISPR in Yeast - Lectures). The primer design may be delegated to individual students or groups. However, I chose to use instructor-validated primers to minimize ambiguity of the results. I break the screening up into class sessions: in the first week, DNA is isolated and PCR amplification is started. The next week, agarose gel electrophoresis is used to visualize the PCR products. Results will include estimations of plasmid concentration and purity from spectrophotometer readings, an annotated photograph of the agarose gel, a standard curve generated from the molecular size markers, and an estimation of all PCR product lengths (using the standard curve). From these results, students should speculate whether each chosen colony contains the desired DNA plasmid.
If no colonies appear on experimental plates or no positive clones are observed, there are a few options: Students may proceed using instructor cloned sgRNA, students may proceed using sgRNA cloned by other student groups, and/or students may repeat cloning steps on their own time, or during class. Having students repeat failed experiments is advantageous in that they receive extra practice, and the process of assessing each step to determine which may need alteration can help students understand and retain information about the purpose of each individual step. The advantage to students proceeding, despite failed cloning, is that everyone stays on the same schedule, easing the instructor's load.
YEAST TRANSFORMATION
With their pML104/sgRNA clones in hand, students proceed with transformation of haploid, wild-type (TRP1) yeast (Figure 1). The pML104/sgRNA is transformed into yeast along with an instructor-designed, single stranded, synthetic piece of DNA (ssODNA, Figure 1). The ssODNA is about 90 nucleotides long and has arms of homology, which are similar or identical to the genomic DNA on either side of the Cas9 cleavage site. After Cas9 cleaves the genomic DNA, the ssODNA will serve as a template for homologous repair (Figure 2). The ssODNA should deviate from the TRP1 genomic sequence so that the repaired gene will have a frameshift and/or premature stop codon near the Cas9 cleavage site. Moreover, the PAM sequence of the guide RNA should be disrupted to prevent repeated cleavage (and cell death) of the trp1 gene after repair (13,16,26). The ssODNA can be ordered from IDT but should be PAGE purified (instead of standard desalting) to ensure that the full length ssODNA is delivered.
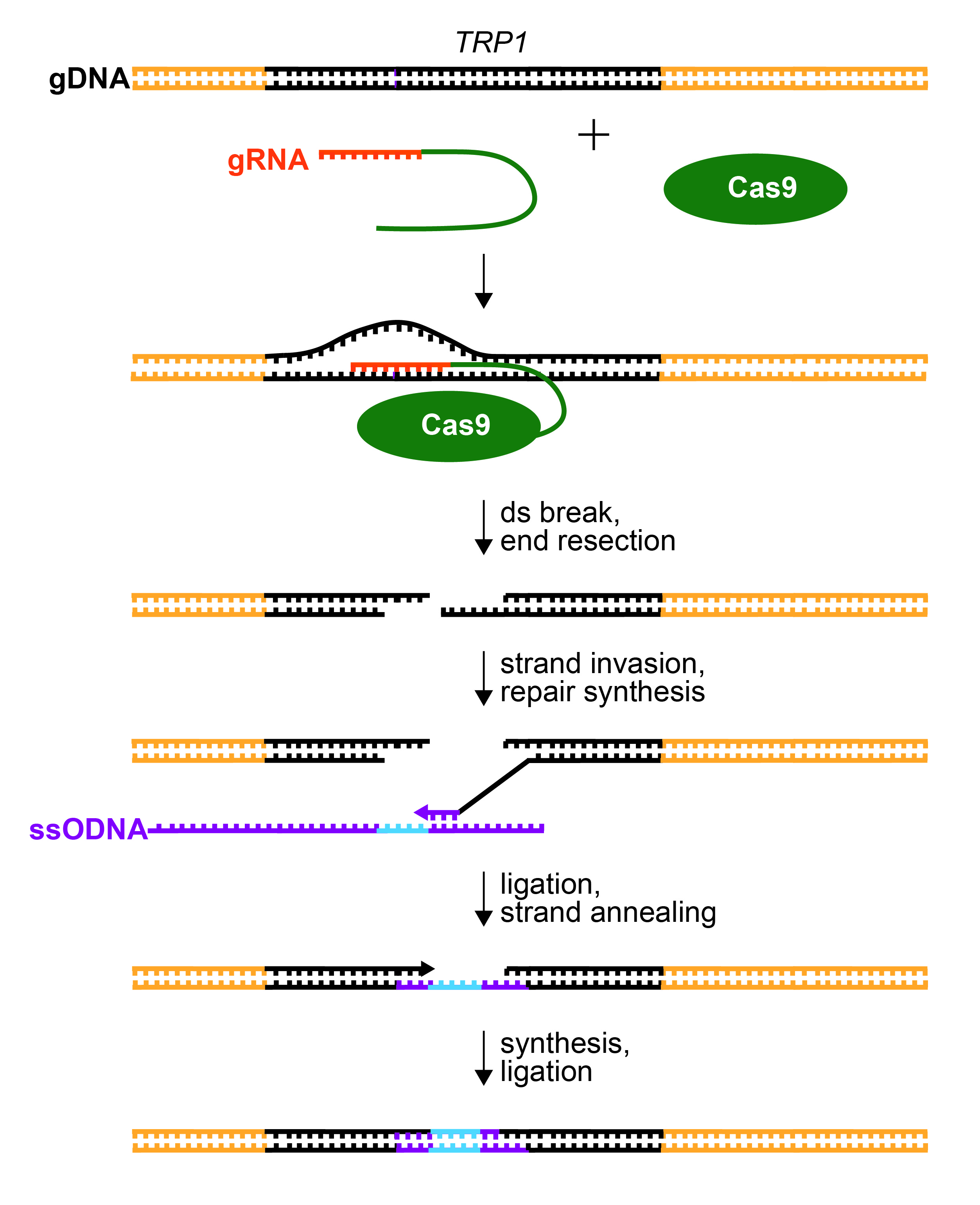
Figure 2. Repair of CRISPR/Cas9-Mediated dsDNA Break. gRNA (orange) recognizes the TRP1 gene (black) by base pairing to one strand immediately upstream of a PAM sequence. The gRNA is fused to tracrRNA (green) to form a sgRNA. Cas9 endonuclease is guided to cleave the TRP1 gene by sgRNA recognition. The ends of cleaved DNA are resected. A 3’ overhang will base pair to homologous sequence (purple) within the provided ssODNA template (strand invasion). The ssODNA also contains the desired mutation to disrupt the TRP1 gene (insertion, deletion, and/or substitution; blue). The 3’ end of the genomic DNA is extended by DNA polymerase (repair synthesis), copying the sequence from the ssODNA. The extended 3’ end ligates with the resected 5’ end in the TRP1 gene. The gap in the opposite strand is filled by DNA synthesis and ligation.
Controls for yeast transformation and gene editing are decided by students. They are encouraged to include a negative control (no expected growth), a positive control for transformation and an experimental plate that contains the template for homologous repair as well as their cloned pML104/sgRNA. In early versions of the exercise, students made their own plates for selection of transformed yeast, including mixing media, autoclaving the media, and pouring plates. I felt this activity would contribute to critical thinking about the purpose of the plates, selection, and yeast auxotrophs; however, the number of mistakes in making solutions and misunderstanding (regarding the amino acids to add in or leave out) led to an overwhelming number of ambiguous results. In the final version, the plates for selection of pML104 positive yeast (synthetic complete, lacking uracil) were provided by the instructors. Student results/data analysis should include descriptions of control and experimental growth as well as mathematical estimation of transformation efficiency.
PHENOTYPING
Finally, in the last weeks of the lab, the students pick yeast colonies from control and experimental transformed yeast to determine if the TRP1 gene has been disrupted (Figure 1). While there are several methods to assess yeast growth for phenotyping, I chose a spotting protocol where yeast colonies are suspended in water, and a small volume from a series of dilutions is spotted on a control (rich media or synthetic complete media), and phenotyping (synthetic media, lacking tryptophan) plate. The number of colonies selected for phenotyping is largely dependent on the transformation efficiency achieved. Students with low transformation efficiency may "borrow" colonies from instructor-transformed plates, or from the plates of fellow students. Results from this portion will include photographs of the yeast growth on control and experimental plates. Conclusions should be drawn as to whether each selected yeast colony contains a functional TRP1 gene, or if the gene has been disrupted by CRISPR/Cas9 gene editing.
GENOTYPING (OPTIONAL)
Because of the time devoted to other exercises in the course, I have not had time to genotype the mutant yeast. Some students requested to genotype their mutant yeast on their own time. However, the overall results were inconclusive for different reasons from each student or group. The genotyping protocol developed includes placing a suspension with yeast cells directly into PCR reactions to amplify the targeted region of the TRP1 gene. The PCR reaction may be assembled in week 8, or assembled during week 7 and analyzed during week 8. Primer sequences are provided in the laboratory manual (Supporting File S3: CRISPR in Yeast - Laboratory Manual), however, students may design their own primers for amplifying the affected region. The ssDNA template co-transformed with the pML104/sgRNA is designed to generate a frameshift mutation in the modified trp1 gene. Ideally, in yeast that contain the desired mutation. However, CRISPR/Cas9 gene editing often occurs with significant errors (29), making predictions of the exact genotypic sequence unreliable.
PCR samples are purified and directly sequenced by Sanger sequencing. Samples can be sequenced by vendors such as Genewiz, for about $7 a sample. Genewiz typically turns around samples in 1-2 business days, however, time must be allowed for sample preparation and shipping. For an extra fee, vendors like Genewiz will also purify PCR products for sequencing to save students and instructors time. Overall, analysis of these sequences would provide practical exposure to Sanger sequencing, simultaneously providing additional information about samples that appear to be TRP1 knockouts.
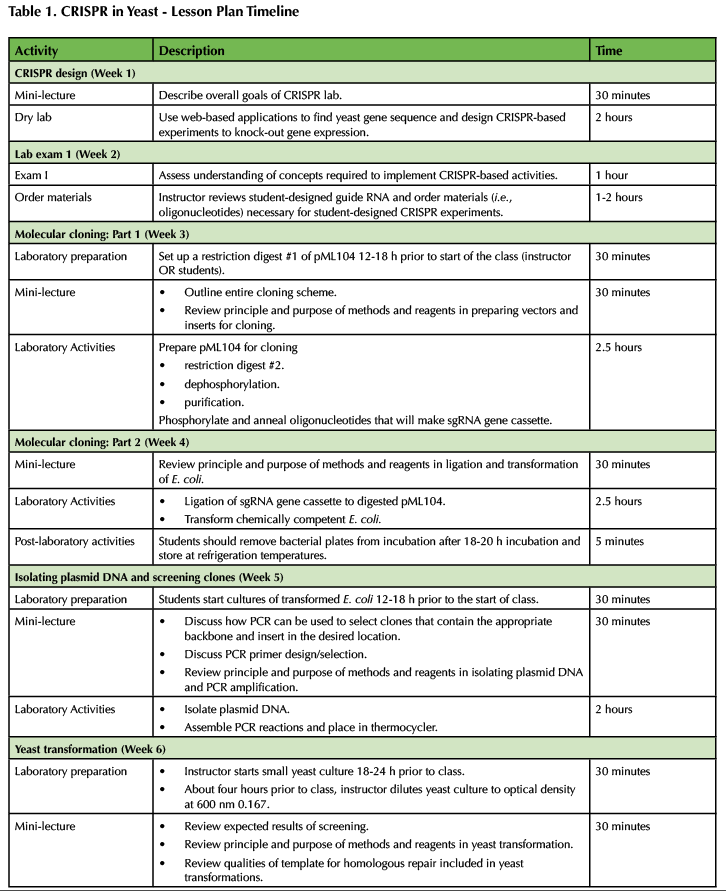
Table 1. CRISPR in Yeast - Lesson Plan Timeline
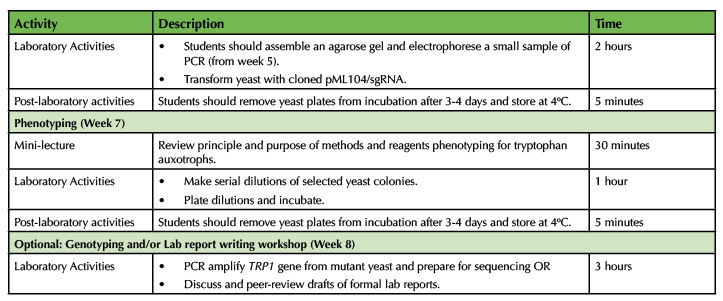
Table 1. CRISPR in Yeast - Lesson Plan Timeline (continued)
ADDITIONAL RESOURCES AND INFORMATION
Outlines for content covered in mini-lectures, as well as activity-specific learning objectives are found in Supporting File S4: CRISPR in Yeast - Instructor's Notes. Preparatory activities, and materials/equipment necessary to accomplish each lab activity are also present in these instructor's notes. Pre-laboratory narrated and non-narrated PowerPoint lectures for weeks 3-5 are a recent addition to the laboratory (not included with the surveyed laboratory sections). These lectures have been combined and included in Supporting File S1: CRISPR in Yeast - Lectures. A rubric for the final laboratory report is provided (Supporting File S5: CRISPR in Yeast - Lab Report Rubric).
POTENTIAL MODIFICATIONS
Some potential timeline modifications to individual laboratory session are discussed within the lesson activities, including providing instructor-cloned sgRNA, eliminating the two weeks of cloning, but also potentially forgoing screening of the clones. In this situation, it may even be possible to bypass at least some of the experimental design, though the design is an essential piece for student understanding. The timeline may also be condensed by performing multiple procedures in one week (as opposed to a single experiment each week). For example, cloning is described here to take place over two weeks, but could be performed using multiple days in the same week. The only procedure that might pose a problem in this situation is the yeast transformation. In my experience, it can take the yeast 5-7 days to show any significant growth after transformation. Therefore, this procedure could not be combined with phenotyping in the same week.
The gene targeted for modification here is TRP1, but this lesson could be modified to disrupt any gene in yeast with a measurable phenotype. For example, I have piloted MET17 disruption and have recently had success targeting ADE2. The exercise is easily amenable to other targets that are interesting or desirable to the instructor, or to individual students. While having several different genes targeted within one course can be overwhelming for many instructors, this could be manageable for smaller courses.
TEACHING DISCUSSION
In summary, I describe a multi-week, laboratory module that provides undergraduate students with learning opportunities that will increase understanding of scientific principles, improve skills necessary to perform and communicate science, and provide more realistic knowledge about how research is accomplished. Moreover, the laboratory is malleable and practical, providing instructors with the flexibility to introduce concepts or practice them at high-levels.
COMMON STUDENT MISCONCEPTIONS
Students are commonly confused about the types of controls needed for different portions of the overall experiment. This confusion seems to stem from student perception of the exercise as one continuous experiment, as opposed to a series of experiments to accomplish a larger goal. With the distorted view, the term "control" tends to be standardized. For example, the positive control for yeast transformation is transformation of the vector, pML104, while yeast containing the vector pML104 are the negative control in phenotyping/genotyping. It is important to discuss, at each step, what variables require a control. I encourage students to avoid use of the terms "negative control" or "positive control", but instead use terms surrounding the word "control" that are more descriptive. For example, a more descriptive term for a negative PCR control is a control reaction in which no template DNA was added.
Students in this setting have little experience with science writing, and are still learning how to read a primary research article. Therefore, writing the lab report is, perhaps, the biggest challenge. It is surprising for students to hear that scientific writing and reading often does not occur from the first page to last page, but often starts with the figures. The figures assembled in the laboratory notebook can be directly applied to the report as an essential element in the results. They are encouraged to write the materials and methods section of the paper first, followed by results, discussion then introduction. Moreover, instead of writing these sections as one unit, writing small sections of materials and methods, results, and discussion for one particular experiment is often an easier format to follow.
OVERCOMING INSTITUTIONAL LIMITATIONS
The described multi-week laboratory tool occurs in a widely available, tractable model. Yeast strains are available for small fee, and stocks can be stored for years as glycerol stocks at -80°C. Maintaining active S. cerevisiae requires minimal equipment (incubator capable of 30°C) and space. Moreover, reagents for growth are relatively inexpensive and easy to prepare, compared to other model systems such as mammalian cell culture. The plasmid for the CRISPR/Cas9 gene editing (pML104) is available through Addgene for the relatively low cost of $65. Other materials and equipment required for this protocol are provided in Supporting File S4: CRISPR in Yeast - Instructors' Notes.
INTRODUCTORY LABORATORY SESSIONS
The CRISPR/Cas9 lab is explicitly started during week six of the semester. The five weeks before this are used to review, practice and/or introduce concepts and techniques that are required for successful completion of the CRISPR/Cas9 lab. Incoming students are familiar with practices common to molecular biology, such as micropipetting, sterile technique, making solutions, and agarose gel electrophoresis. However, they practice performing each of these techniques independently during weeks 2-3 of the laboratory to ensure confidence and accuracy.
I encourage students to take ownership of the lessons and materials provided by this lab during these introductory weeks. For instance, each group makes solutions (including bacterial growth media, electrophoresis running buffer, sterilizing double-deionized water) for use through the course of the semester. Should these solutions run out or appear contaminated, the groups are responsible for replenishing the stocks in their own time. Ownership is encouraged throughout the course of the semester, whenever possible. For example, groups that do not get acceptable amounts or purity of plasmid DNA, mistakenly dispose of their DNA (rather than store it), or fail to appropriately label their DNA stocks have to repeat the plasmid isolation before they can proceed with the CRISPR cloning lab.
HOW BENEFICIAL WAS THIS UNDERGRADUATE LABORATORY EXPERIENCE?
Students report significant gains in content knowledge and understanding of the material, suggesting that the experience is beneficial to learning (Figure 3). However, the goal of the lab was to provide an authentic laboratory experience, therefore, certain pillars that help define authenticity were surveyed. Significant gains in laboratory skills were reported (Figure 3), likely due to the wide variety of opportunities to practice and master skills related to science in general (i.e., gathering information from scientific journals, keeping a lab notebook, making solutions), as well as skills specific to molecular biology and microbiology (cloning, pipetting, plasmid DNA isolation, etc.). The opportunities provided for students to participate in experimental design, troubleshooting experiments, and interpretation of data also proved to improve skills and comfort with experimental design and data analysis (Figure 3). The largest reported gains were in this area of experimental design and data analysis (Figure 3A).
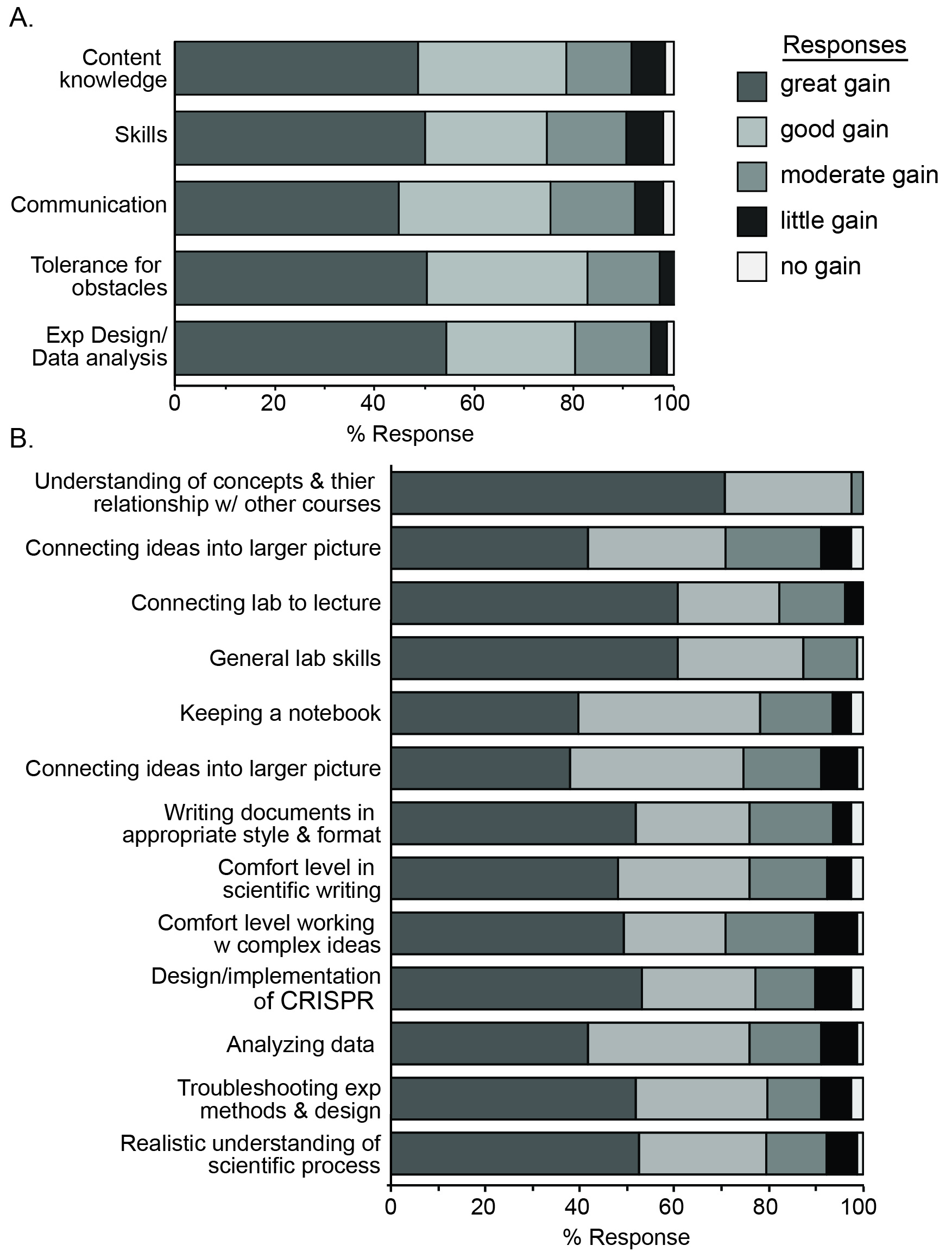
Figure 3. Student-reported learning gains. Students were surveyed for learning gains in the indicated pooled categories (A) or individual responses (B). The frequency of each Likert scale response (no gains, little gain, moderate gain, good gain, great gain) is presented.
One student said, "Experimental design was easily the biggest gain in this course. Of course information is nice, but critical thinking and using it to design experiments and gain new information is much more valuable."
Importantly, students report a realistic understanding of research in my survey (Figure 3B): "The course activities helped me understand how a larger scientific research is performed.", "The activities performed in this lab gave me a much better understanding of the way certain research is performed", "I feel like I am an actual scientist after this class because we performed all the essentials for an experiment from design to execution." Students also felt the lab provided an authentic research.. "This lab mimicked more of a real life scenario and I enjoyed the challenge." It also seems they have a more authentic understanding of the challenges of research "I ...realize scientific experiments do not always yield usable data." "It takes a loooong time to get things done in scientific research. The ideas can seem to work on paper but making it happen at the bench can be a different story...", "There are times when things does not go according to plan so the activities really help us grasp the concept of each lab before diving into the actual experiment." Students recognized specific traits that aid in overcoming obstacles "It is important to be persistent and to carry out an experiment even if it goes wrong in some places", but also report significant gains in their tolerance of these obstacles (Figure 3A), a quality that reflects the authenticity of the experience (5).
It was anticipated that using popular and current technology in the course would be a motivating factor for students. Indeed, it had a positive impact on student attitudes: "All parts of this lab are very educational in terms of research being done in this day and age. It is extremely pleasing to be taught how to do something that is still prevalent today and hasn't been replaced with newer technology."
ACKNOWLEDGMENTS
I would like to thank the Missouri State University, Department of Biomedical Science for their support. I appreciate proofreading and minor manuscript revisions by Lisa Bonner and Courtney Baxter.
SUPPORTING MATERIALS
- S1. CRISPR in Yeast: Lecture. PowerPoint file containing lecture materials provided to introduce concepts for activities in weeks 3-5.
- S2. CRISPR in Yeast: Sample Assessments. Sample questions from exams distributed during week 2 and week 7. Includes answer key.
- S3. CRISPR in Yeast: Laboratory Manual. Student handouts for laboratory activities
- S4. CRISPR in Yeast: Instructor's Notes. Knowledge objectives, materials, lecture outline and other considerations for activities
- S5. CRISPR in Yeast: Lab Report Rubric. Used for peer-feedback and grading of formal lab reports.
- S6. CRISPR in Yeast: Data Analysis. Description of materials and methods used to obtain and analyze data presented in Figure 3.
References
- Spell RM, Guinan JA, Miller KR, Beck CW. 2014. Redefining authentic research experiences in introductory biology laboratories and barriers to their implementation. CBE Life Sci Educ 13:102-110.
- Lopatto D. 2003. The essential features of undergraduate research. Counc Undergrad Res Q 24.
- Rahm J, Miller HC, Hartley L, Moore JC. 2003. The value of an emergent notion of authenticity: Examples from two student/teacher-scientist partnership programs. J Res Sci Teach 40:737-756.
- Auchincloss LC, Laursen SL, Branchaw JL, Eagan K, Graham M, Hanauer DI, Lawrie G, McLinn CM, Pelaez N, Rowland S, Towns M, Trautmann NM, Varma-Nelson P, Weston TJ, Dolan EL. 2014. Assessment of course-based undergraduate research experiences: a meeting report. CBE Life Sci Educ 13:29-40.
- Linn MC, Palmer E, Baranger A, Gerard E, Stone E. 2015. Undergraduate research experiences: impacts and opportunities. Science 347:1261757.
- National Academies of Sciences, Engineering, and Medicine, Division on Earth and Life Studies, Board on Life Sciences, Division of Behavioral and Social Sciences and Education, Board on Science Education, Committee on Strengthening Research Experiences for Undergraduate STEM Students. 2017. Undergraduate Research Experiences for STEM Students: Successes, Challenges, and Opportunities. National Academies Press.
- Rowland S, Pedwell R, Lawrie G, Lovie-Toon J, Hung Y. 2016. Do We Need to Design Course-Based Undergraduate Research Experiences for Authenticity? CBE Life Sci Educ 15.
- Walsh S, Becker A, Sickler PS, Clarke DG, Jimenez E. 2017. An Undergraduate Laboratory Manual for Analyzing a CRISPR Mutant with a Predicted Role in Regeneration. https://scholarship.rollins.edu/cgi/viewcontent.cgi?article=1300&context=as_facpub
- Adame V, Chapapas H, Cisneros M, Deaton C, Deichmann S, Gadek C, Lovato TL, Chechenova MB, Guerin P, Cripps RM. 2016. An undergraduate laboratory class using CRISPR/Cas9 technology to mutate drosophila genes. Biochem Mol Biol Educ 44:263-275.
- Sander JD, Joung JK. 2014. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32:347-355.
- Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262-1278.
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816-821.
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281-2308.
- Vyas VK, Bushkin GG, Bernstein DA, Getz MA, Sewastianik M, Barrasa MI, Bartel DP, Fink GR. 2018. New CRISPR Mutagenesis Strategies Reveal Variation in Repair Mechanisms among Fungi. mSphere 3:e00154-18.
- Liang X, Potter J, Kumar S, Ravinder N, Chesnut JD. 2017. Enhanced CRISPR/Cas9-mediated precise genome editing by improved design and delivery of gRNA, Cas9 nuclease, and donor DNA. J Biotechnol 241:136-146.
- Zhang J-H, Adikaram P, Pandey M, Genis A, Simonds WF. 2016. Optimization of genome editing through CRISPR-Cas9 engineering. Bioengineered 7:166-174.
- Weaver R. 2011. Molecular Biology. McGraw-Hill Higher Education.
- Krebs JE, Goldstein ES, Kilpatrick ST. 2017. Lewin's GENES XII. Jones & Bartlett Learning.
- Alberts B. 2017. Molecular Biology of the Cell. Garland Science.
- Wang H, La Russa M, Qi LS. 2016. CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem 85:227-264.
- Barrangou R, van der Oost J. 2012. CRISPR-Cas Systems: RNA-mediated Adaptive Immunity in Bacteria and Archaea. Springer Science & Business Media.
- Pechenik JA. 2015. A Short Guide to Writing about Biology. Longman Publishing Group.
- Bangera G, Brownell SE. 2014. Course-based undergraduate research experiences can make scientific research more inclusive. CBE Life Sci Educ 13:602-606.
- Tanner KD. 2013. Structure matters: twenty-one teaching strategies to promote student engagement and cultivate classroom equity. CBE Life Sci Educ 12:322-331.
- Tanner K, Chatman LS, Allen D. 2003. Approaches to cell biology teaching: cooperative learning in the science classroom--beyond students working in groups. Cell Biol Educ 2:1-5.
- Laughery MF, Hunter T, Brown A, Hoopes J, Ostbye T, Shumaker T, Wyrick JJ. 2015. New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae. Yeast 32:711-720.
- 2001. Introduction of Plasmid DNA into Cells, p. 2796. In Ausubel, FM, Brent, R, Kingston, RE, Moore, DD, Seidman, JG, Smith, JA, Struhl, K (eds.), Current Protocols in Molecular Biology. John Wiley & Sons, Inc., Hoboken, NJ, USA.
- Green MR, Sambrook J. 2016. Preparation of Plasmid DNA by Alkaline Lysis with Sodium Dodecyl Sulfate: Minipreps. Cold Spring Harb Protoc 2016.
- Zhang J-P, Li X-L, Li G-H, Chen W, Arakaki C, Botimer GD, Baylink D, Zhang L, Wen W, Fu Y-W, Xu J, Chun N, Yuan W, Cheng T, Zhang X-B. 2017. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol 18:35.
Article Files
Login to access supporting documents
CRISPR/Cas9 in yeast: a multi-week laboratory exercise for undergraduate students(PDF | 854 KB)
S1. CRISPR in Yeast. Lectures_0.pptx(PPTX | 24 MB)
S2. CRISPR in Yeast. Sample Assessments.docx(DOCX | 211 KB)
S3. CRISPR in Yeast. Laboratory Manual.docx(DOCX | 1 MB)
S4. CRISPR in Yeast. Instructor notes.docx(DOCX | 39 KB)
S5. CRISPR in Yeast. Lab report rubric.docx(DOCX | 22 KB)
S6. CRISPR in yeast. Data Analysis_0.docx(DOCX | 21 KB)
- License terms

Comments
Comments
There are no comments on this resource.