DNA Detective: Genotype to Phenotype. A Bioinformatics Workshop for Middle School to College.
Published online:
Abstract
Advances in high-throughput techniques have resulted in a rising demand for scientists with basic bioinformatics skills as well as workshops and curricula that teach students bioinformatics concepts. DNA Detective is a workshop we designed to introduce students to big data and bioinformatics using CyVerse and the Dolan DNA Learning Center's online DNA Subway platform. DNA Subway is a user-friendly workspace for genome analysis and uses the metaphor of a network of subway lines to familiarize users with the steps involved in annotating and comparing DNA sequences. For DNA Detective, we use the DNA Subway Red Line to guide students through analyzing a "mystery" DNA sequence to distinguish its gene structure and name. During the workshop, students are assigned a unique Arabidopsis thaliana DNA sequence. Students "travel" the Red Line to computationally find and remove sequence repeats, use gene prediction software to identify structural elements of the sequence, search databases of known genes to determine the identity of their mystery sequence, and synthesize these results into a model of their gene. Next, students use The Arabidopsis Information Resource (TAIR) to identify their gene's function so they can hypothesize what a mutant plant lacking that gene might look like (its phenotype). Then, from a group of plants in the room, students select the plant they think is most likely defective for their gene. Through this workshop, students are acquainted to the flow of genetic information from genotype to phenotype and tackle complex genomics analyses in hopes of inspiring and empowering them towards continued science education.
Citation
Sternberger AL, Wyatt SE. 2019. DNA Detective: Genotype to Phenotype. A Bioinformatics Workshop for Middle School to College. CourseSource. https://doi.org/10.24918/cs.2019.34Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
With the advent of next-generation sequencing (NGS) and other high-throughput technologies, the way we study biology has drastically changed. The era of genomics has brought with it big data and an increasing need for scientists capable of analyzing extremely large, often unstructured, and computationally taxing data (1). In a 2017 survey funded by the National Science Foundation, nearly 90% of the 704 principal investigators questioned reported that they currently work with or plan to work with big data in the near future (2). Thus, there also exists a need to expand our educational programs to provide data analysis skills to our students (3,4). However, many instructors lack the resources to teach big data analysis (5,6). Thankfully, steadily increasing numbers of open-source genomic databases and software provide new teaching tools, research approaches, and opportunities for instructors of diverse disciplines to provide students of all ages invaluable bioinformatics expertise.
One such teaching tool is DNA Subway (https://dnasubway.cyverse.org) a free, online workflow for genome analysis. The Dolan DNA Learning Center and computer programmers developed DNA Subway as part of the CyVerse (formerly iPlant Collaborative) to make complex, genomic analyses more accessible to biology instructors and students. Through the DNA Subway pipeline, instructors and students can analyze next generation sequence data via a user-friendly interface and software that is actively used in the research community. The DNA Subway workspace uses the metaphor of a network of subway lines to transport users through the steps involved in gene annotation and genome analysis. The subway network consists of five lines including 1) the Red Line for gene prediction and annotation, 2) the Yellow Line to prospect genomes and build gene trees, 3) the Blue Line for determining sequence relationships, 4) the Green Line to analyze RNA-Seq data and differential gene expression, and 5) the Purple Line for microbiome and environmental DNA analysis. Instructors can select from one or multiple lines when designing their workshop/curriculum and have the option of uploading their own NGS data or choosing from provided examples.
While the DNA Subway platform is useful in any academic or research setting, we have incorporated its Red Line into a workshop for an undergraduate, introductory biology classroom, and have also used it for outreach with middle school girls. Although some educators have made an effort to include bioinformatics lessons in high school (7-10) and college classrooms (11-21), there are limited bioinformatics activities designed for middle school children (grades 6-8) (22,23). Exciting middle school children about the possibilities of big data analysis is critical to STEM education. They are at a particularly impressionable age and these activities can encourage their interest in STEM careers (23,24). Apart from inspiring students and teaching them about big data analysis, we also use DNA Detective to introduce broad biological concepts including DNA as the universal, genetic code and the correlation between genotype and phenotype. In this way, students learn not only how to use bioinformatics tools, but also where big data comes from, what it looks like, and what kinds of questions can be answered using big data and bioinformatics approaches. If instructors are unfamiliar with the process of gene annotation, Yandell and Ence’s “A beginner’s guide to eukaryotic genome annotation” (25) is a great introductory resource. Instructors can also provide this resource to upper-level students to provide context prior to the workshop.
OBJECTIVES
Our objectives for DNA Detective are to teach students about 1) big data and 2) the flow of genetic information from genotype to phenotype using bioinformatics tools and software. Additionally, we hope to inspire and empower students towards continued science education.
WORKSHOP PREP
Prep to Use Our Mystery Genes/Mutants
Prior to implementing DNA Detective, instructors select “mystery” genes that students will be analyzing. While genes from many organisms can be evaluated through the DNA Subway pipeline, we use three Arabidopsis thaliana plant genes: DIMINUTO 1 (DIM1), PHOSPHOGLUCOMUTASE (PGM1), and PHYTOENE DESATURASE (PDS). We chose these genes because their mutant phenotypes are obvious when compared to wildtype plants (Figure 1), and the mutant seed stocks are available for purchase through the Arabidopsis Biological Resource Center (ABRC, https://abrc.osu.edu). As per common nomenclature, Arabidopsis mutants are indicated in lowercase and italics. We provide more information on dim1, pgm1 and pds including their phenotypes, links to descriptions and genetic data through The Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org), and their ABRC stock IDs in Table 1.

Table 1. Descriptions of mutant phenotypes for DIM1, PGM1, and PDS, as well as links to their genetic data on The Arabidopsis Information Resource (TAIR) and stock IDs for mutant seed purchase through the Arabidopsis Biological Resource Center (ABRC).
For comparison purposes, instructors should additionally grow wildtype Arabidopsis, which can also be ordered from ABRC. Seed stocks can take up to two weeks to arrive, and growth of Arabidopsis from seed to flowering takes approximately one month, so instructors should order seed at least 1.5-2 months in advance of the class or workshop. Once delivered, spread wildtype, dim1, and pgm1 seeds on moist soil and cold stratify in a refrigerator (4˚C) for 2-4 days. Following stratification, move pots to a growth chamber (at 21˚C with a 12h light/12h dark cycle) or well-lit window (room temperature is adequate for growth of Arabidopsis). Water pots approximately every four days. Because pds plants are albino and cannot photosynthesize, their seed need to be sterilized (Supporting File S1. DNA Detective - Sterilization protocol for pds seeds) and plated on sucrose media (Supporting File S2. DNA Detective - 0.5 MS/1% sucrose media protocol for pds seeds) prior to stratification for 2-4 days at 4˚C. Following stratification, pds plates should be kept in the growth chamber or window with the other seeds but require no watering.
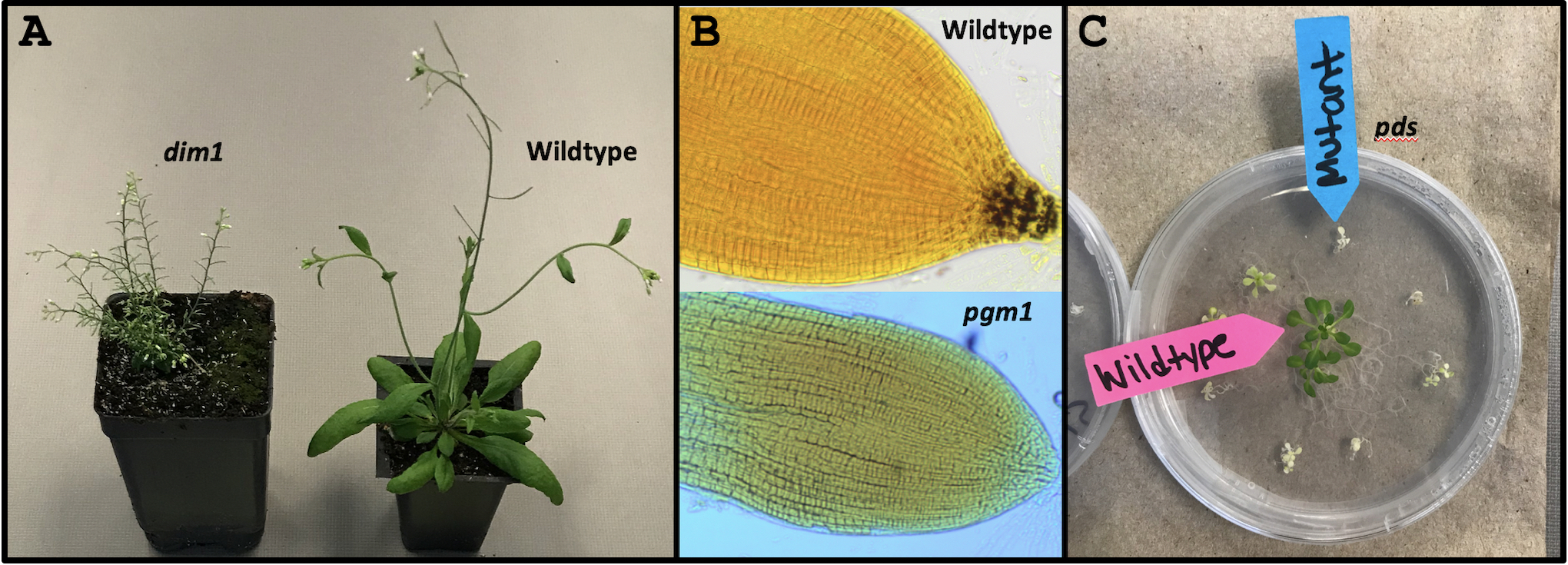
Figure 1. Phenotypes of wildtype Arabidopsis plants compared to (A) the dwarf dim1, (B) starch-less pgm1, and (C) albino pds mutants.
Prep to Use Instructor-Selected Genes/Mutants
If instructors wish to use genes other than the three presented in this workshop, they can identify additional genes with easily visible phenotypes through the RIKEN Arabidopsis Genome Encyclopedia (RARGE II), a public web-database with integrated phenotype data for 66,209 mutant Arabidopsis lines (26). From the RARGE II homepage (http://rarge-v2.psc.riken.jp), search for Arabidopsis mutants by clicking the “Phenotypes” link. Then, select the box next to the plant part/developmental stage (e.g. whole plant, seedling, flower, root) to be mutated. An instructor can further refine this selection by clicking the “+” sign next to the selected plant part or growth stage. For example, if an instructor selects “Whole plant” followed by “+”, a drop-down menu will appear with options including mutant colors, variegation, decreased height, increased height, etc. Once you make final selections, click “Search”, and a new page will load with information on Arabidopsis genes whose mutants fit those criteria. This information includes gene names and TAIR accession numbers (e.g. AT2G01900).
Instructors should copy and paste each selected mystery gene’s genomic sequence and save it in FASTA format. To do so, access the TAIR homepage and enter the gene name or accession number (provided through RARGE II) in the search bar at the top of the page. Before searching, ensure that “Gene” is selected in the drop-down menu next to the search bar. The page will reload with search results, and under the “Locus” column click the corresponding gene accession number. Scroll down to the “Sequence” header and click on “full length genomic”. Next, select “Send to WU-BLAST”, copy the DNA sequence from the “Input” box, and paste the sequence into a text editor capable of saving in FASTA format (we use BBEdit, https://www.barebones.com/support/bbedit/). FASTA format consists of a header line starting with a “>” sign followed by the gene name or accession. The subsequent lines contain the gene sequence. Examples are provided in Supporting File S3: DNA Detective – DIM1 genomic sequence, Supporting File S4: DNA Detective – PGM1 genomic sequence, and Supporting File S5: DNA Detective – PDS genomic sequence. If mutant germplasms (seeds) exist for the gene and are in stock at ABRC, order them from the TAIR page accessed by clicking on the gene accession number under the “Locus” column, as previously described. Scroll down to the “Germplasm” header and click the link under the “stock name” subheading. The resulting page contains detailed information on the mutant phenotype including any special growth conditions and an “Order from ABRC” link. If instructors do not already have an ABRC account, they will need to register online (https://www.arabidopsis.org/community/abrc-new-register.faces) prior to ordering seed. This process should be repeated for each gene that is to be analyzed during the workshop.
Data and Software
Genomic sequences often contain repetitive DNA that researchers need to mask (i.e. remove) to build the final gene model. So, one of the first steps in the DNA Subway Red Line pipeline is finding and masking repeats. Full length genomic sequences with repeats added for DIM1, PGM1, and PDS are available in Supporting File S3: DNA Detective – DIM1 genomic sequence, Supporting File S4: DNA Detective – PGM1 genomic sequence, and Supporting File S5: DNA Detective – PDS genomic sequence. To ensure that repeats are identified if using custom genes, instructors may want to artificially add a few (e.g. ATCATCATCATCATCATCATCATCATCATC) to the FASTA file of each gene. Prior to the workshop or class, upload FASTA files to classroom or students’ personal computers. If students are going to use the DNA Subway platform on multiple occasions during the course or workshop, it is beneficial to have them create personal logins (https://user.cyverse.org/register), as only work from registered users can be saved. For the DNA Detective workshop, a guest login is adequate.
WORKSHOP APPROACH
Introduction
For DNA Detective, a 45-minute workshop (recommended timeline provided in Table 2), we have hosted 8-12 middle school girls at a time. The girls were in grades 6-8 and ranged in age from 11 to 14. We have also scaled the workshop to accommodate a non-majors, introductory biology course of 60 undergraduates, which are split among three labs of ~20 students each. However, when implementing DNA Detective, outreach or class size is only dependent on the number of computers available, and there is no prerequisite student knowledge or skill required for students to successfully complete the workshop. To promote inclusive, collaborative learning, students are divided into groups of 2-3, and each group is assigned one mystery FASTA file. Before students embark on the DNA Subway Red Line, we give a brief, introductory lecture (Supporting File S6: DNA Detective - Lecture Presentation Slides) to acquaint them with DNA as the genetic code, gene structure/modeling, and the software that will be used as part of the Red Line analyses. During the intro, we discuss the roles of different cells (e.g. muscle and nerve cells in animals or vascular tissue in plants) and how DNA dictates these roles. We describe the content of DNA as a metaphor of an alphabet and explain that the four letters (i.e. bases: ATGC) are arranged into "words" (i.e. sequences), and collectively these words make up "sentences" called genes.
Next, we introduce the concept of genetic variation and discuss the human gene TAS2R38, which encodes a protein receptor that affects the ability to taste bitter-tasting compounds (27). The two common forms of TAS2R38 confer either a "taster" phenotype, those who can strongly taste bitter compounds, or a "non-taster" phenotype, individuals that detect little to no bitter taste. After introducing this concept, we pass out paper strips containing phenylthiocarbamide (PTC - a synthetic compound used to test whether students are tasters or not) and encourage students to place the strips in their mouth to identify if they are tasters or non-tasters. While PTC is harmless, the flavor can be very intense for tasters. Be sure to have a trash can nearby for students to spit out the paper strips. Next, we explain that DNA sequencing technology "reads" genes and that one needs to process the data from sequencing (strings of ATGCs) bioinformatically to identify gene regions. Finally, we describe the general structure of a gene (e.g. coding sequences, exons, introns) and introduce the activities that students will be doing in order to be a DNA Detective. These activities include using bioinformatics software to identify and mask repeats in their mystery DNA sequence, predict gene regions, search sequence databases to identify the name of their gene, build a representative gene model, and select mutant Arabidopsis phenotypes that correspond to their gene.
DNA Subway Red Line
After the introduction, students use guest logins to access the DNA Subway homepage (https://dnasubway.cyverse.org) where they select the "Annotate a Genomic Sequence" tab followed by the "Classic" Red Line option to enter the Red Line project page (Supporting File S6: DNA Detective - Lecture Presentation Slides, slide 13). We prompt students to create a new project by uploading their mystery FASTA file and take turns being group leader to complete every "stop" (analysis). Group leaders alternate at each stop of the Red Line and are responsible for running the analysis, deciphering the output of that analysis, and discussing it with their partner(s). At each Red Line stop, we walk around the room and have group leaders summarize their results out loud to the rest of the workshop/class. Following their summation, we instruct group leaders to pass the computer mouse or laptop to the next group member to ensure that every student gets a chance to lead.
Stop 1 – Find/Mask Repeats
After creating a project, we direct students to the Red Line analysis page (Supporting File S6: DNA Detective - Lecture Presentation Slides, slide 14). During the first stop on the Red Line, students find and mask repeats in their sequences using RepeatMasker software. Repeats are locations in a DNA sequence that contain repetitive nucleotide bases (e.g. CTCA at bases 1-37). The central dogma of molecular biology states that transcription is the first step in gene expression during which DNA is copied into messenger RNAs (mRNAs), which are then translated into proteins. Only a portion of a DNA sequence, the coding sequence (CDS), is represented in the final transcribed and processed mRNA that undergoes translation. Because repeats are seldom found in the CDS of DNA, it is important to mask repeats so that gene prediction software used in later Red Line stops will not search for genes in repeat locations. To initiate RepeatMasker, group leaders click on the button labeled "RepeatMasker." Once the software is running, the status circle next to the RepeatMasker button will turn yellow and then into a green "V" when the analysis is complete. To view the RepeatMasker results, leaders click again on the RepeatMasker button. Results relevant to the DNA Detective workshop are highlighted in a red box on slide 15 of Supporting File S6: DNA Detective - Lecture Presentation Slides and include a list of identified repeats, the length of the repeats, and the start and end positions of repeats within the sequence. If instructors need more information about RepeatMasker results including detailed descriptions of each column, they should refer to the DNA Subway manual (https://cyverse-dnasubway-guide.readthedocs-hosted.com/en/latest/). Once group leaders have summarized their repeat results to the classroom, they close out of the RepeatMasker window and return to the Red Line analysis page.
Stop 2 – Predict Genes
During the second Red Line stop, students predict genes via Augustus and SNAP gene prediction software using their masked DNA sequences as input. Gene prediction is the process of identifying gene's coding regions (i.e. the beginning and end positions of the exons of genes) in DNA sequences. Eukaryotic genes, like the ones we use for DNA Detective, have defining features such as introns located between exons and untranslated regions (UTRs) at the beginning (5') and end (3') of the gene sequence (Supporting File S6: DNA Detective - Lecture Presentation Slides, slide 11). Unlike repeats, these features help guide gene prediction software in discerning gene structure and location within a DNA sequence. When group leaders are ready, they click on both the "Augustus" and "SNAP" buttons to run them simultaneously. They then view results of Augustus and SNAP by clicking again on their corresponding buttons. Output of both programs includes a list of the types of coding regions identified, lengths of the regions in base pairs, and start and end coordinates. For example, slide 16 in Supporting File S6: DNA Detective - Lecture Presentation Slides highlights relevant results from the Augustus output for DIM1. If one plots the coordinates for each region on a number line, you can infer other genomic features such as introns and UTRs (Supporting File S6: DNA Detective - Lecture Presentation Slides, slide 16). This coordinate plotting method is very similar to how Augustus and SNAP algorithms work. Group members discuss the output of each program, any differences between the programs' results, and group leaders again summarize their findings to the class before returning to the Red Line analysis page.
Stop 3 – Search Databases
Once students have deciphered the structure of their mystery genes, they use BLASTN and BLASTX to annotate them. Gene annotation is the process of attaching biological information to a gene such as its name (e.g. DIM1) and molecular function. BLAST (Basic Local Alignment Search Tool) is an algorithm that searches sequence databases for published nucleotide (BLASTN) and amino acid (BLASTX) sequences with high similarity to a queried sequence. In this case, the queried sequences are the masked, mystery DNA sequences from the first Red Line stop. Students run BLASTN and BLASTX simultaneously by clicking their labeled buttons, and the analyses are complete once the program status symbols display a green "V". Students can view results by clicking again on BLASTN and BLASTX. For both BLASTN and BLASTX results, the "Type" column lists database matches (match) and/or partial matches (match_part) to the queried, mystery DNA sequence (Supporting File S6: DNA Detective - Lecture Presentation Slides, slide 17). The "Length" column reflects the total number of base pairs included in matches, and the "Start" and "End" columns depict the match boundaries. Once students have briefly scanned the results of both programs, we instruct group leaders to open the BLASTN output, find the full match with the highest score (i.e. highest similarity to their mystery gene), and locate "description =" within the "Attributes" column for that match (Supporting File S6: DNA Detective - Lecture Presentation Slides, slide 17). The text following the description header is the name of a database gene with high or perfect homology to their mystery gene. After discussing their results, group leaders close BLASTN to return to the Red Line analysis page.
Stop 4 – Build Gene Models
At the final Red Line stop, students build a model of their mystery gene. Because gene predictors and sequence databases do not always agree with each other, it is important to manually compare/merge the August and SNAP gene predictions with results from RepeatMasker, BLASTN, and BLASTX. First, group leaders click the Apollo button on the Red Line analysis page. A window opens prompting students to save a .jnlp (Java Network Launching Protocol) file. We instruct students to save the file to their computer desktop and then double-click the desktop shortcut to launch Apollo. As Apollo loads, two error messages may appear on-screen: "Failed to set style apollo.dataadapter.ensj.EnsJAdapter--couldn't find style file. This could be a problem!", and "Failed to set style apollo.dataadapter.synteny.SyntenyAdapter--couldn't find style file. This could be a problem!". These errors do not affect Apollo functionality, and group leaders click "OK" to proceed to the program. Once Apollo opens, we direct students to click the "View" tab and to deselect "Show reverse strand". Viewing only the forward DNA strand is sufficient for the workshop and makes the screen less congested. Next, we guide students to the "Tiers" tab and to click on "Show types panel". A "Types" window opens, and we tell students to select the "Label" boxes next to BLASTN, BLASTX, SNAP, Augustus, and Repeats (Supporting File S6: DNA Detective - Lecture Presentation Slides, slide 18). This adds software labels to indicate the origin of each predicted gene model. To build their own gene models, students first click on a predicted model in the results panel (Supporting File S6: DNA Detective - Lecture Presentation Slides, slide 19). Single-clicking selects only the portion of the model the cursor is touching (e.g. a single exon), while double-clicking selects the entire model (e.g. a transcript with two exons and an intron).
Next, students move the selected model portion(s) into the user-created annotations panel (Supporting File S6: DNA Detective - Lecture Presentation Slides, slide 19) by physically dragging with their cursor or right-clicking and selecting "Add as gene transcript" from the pop-up menu. Once there are results from multiple models in the user-created annotations panel, the results are merged into a contiguous model. To do so, group leaders 1) double-click to select the first result, 2) press the shift key and double-click the second result, 3) and right-click and select "Merge transcripts" from the menu. Students repeat this process until groups create the most-likely model based on all of the evidence in the results panel. Group leaders then click the "File" tab on the top menu bar and select "Upload to DNA Subway" before closing out of Apollo and returning to the Red Line analysis page. Finally, students click on "Local Browser" to open GBrowse, a graphical interface that displays the results generated at previous Red Line stops and user-created gene models (Supporting File S6: DNA Detective - Lecture Presentation Slides, slide 20). Similar to the third Red Line stop, GBrowse annotates BLASTN results with the predicted gene's name, and we encourage students to write it down for the final activity.
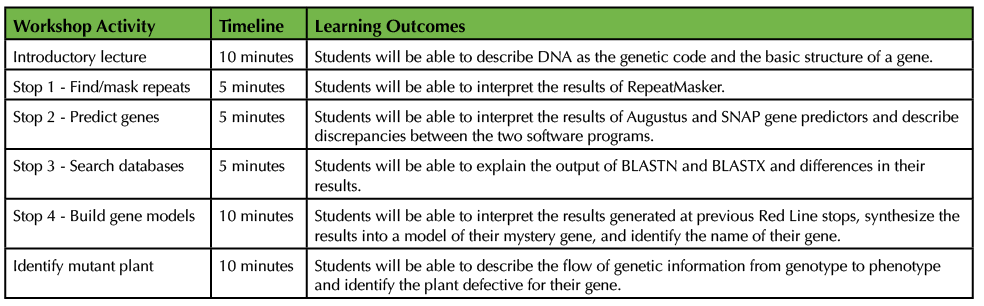
Table 2. Recommended timeline for 45-minute DNA Detective workshop and learning outcomes for each activity.
Gene Function and Mutant Plant Identification
Once every group has built a gene model and identified the name of their gene, we instruct students to open a new web browser tab and access the TAIR homepage (https://www.arabidopsis.org). On the homepage, students type the name of their gene (e.g. DIM1) into the search bar at the top right corner, click search, and then select the "Locus" link (e.g. AT3G19820) for the gene with the same name as theirs. This is usually (but not always) the first result listed. Clicking the "Locus" link takes students to detailed descriptions of their mystery gene (the same information as TAIR links provided in Table 1) including its function. Then, based on their gene's function and hypothesized phenotype, students choose from the selection of plants the one they think is defective for their mystery gene. We place folders labeled mystery gene 1-3 in front of the corresponding mutant plants. Students can open these to reveal the defective gene's name and function (Supporting File S6: DNA Detective - Lecture Presentation Slides, slides 21-24), allowing students to check their hypotheses. Because pgm1 has no external phenotype, we create wet mounts of wildtype and pgm1 roots and stain for starch using Lugol's solution prior to the workshop. When viewed under a compound microscope, students can see that wildtype roots are stained at the starch-rich tips, whereas pgm1 roots are starchless and do not stain (Figure 1B).
PROBLEMS ENCOUNTERED
The DNA Subway Red Line is currently provided in two versions: Classic and Web Apollo. The Classic version requires Java installation in both the browser and system. Web Apollo does not have Java requirements but has limited functionality (including the ability to upload custom sequences) compared to the Classic version. Thus, DNA Detective requires the Classic version and Java installation. For outreach with middle school students, we were able to use classroom computers and download Java ahead of time. However, when classroom computers are not available, students need to download the software on their own laptop. Some students have problems with proper installation or don't complete the installation, causing the workshop to run past its 45-minute allotment. A second problem encountered is the run time of some analyses. While some stops on the Red Line take only a few seconds to run, others can take a few minutes (computer/browser dependent), and it is important to keep students engaged during these pauses. Lastly, deciphering the results of each Red Line stop with little or no genetic/bioinformatics experience can be daunting to students. To circumvent these concerns, we include slides in the introductory lecture that contain examples of the project upload/Red Line analysis page and output for each analysis (Supporting File S6: DNA Detective - Lecture Presentation Slides; slides 13-20). While each Red Line analysis is running, we use these slides to walk through the results with students so they are prepared to discuss them with their group members once the programs finish.
WORKSHOP EFFECTIVENESS AND BENEFITS
DNA Detective workshop is a low cost and user-friendly workshop to teach middle school to college-aged students about basic biological concepts, big data, and bioinformatics. An instructor could easily scale the workshop to accommodate large classroom sizes. Students work together in groups to enhance their problem solving skills and promote active learning, a teaching approach shown to benefit students over traditional, lecture-based teaching (28,29). We believe that this is particularly beneficial to middle school girls. Bioinformatics remains a male-dominated field, and by hosting our DNA Detective workshop for middle school girls, we hope to counter gender barriers and encourage young women to consider a STEM profession. Overall, students really like the activity. Having the live plants available, although not necessary, is an advantage. Students have a visible goal (choosing the plant defective in their gene) and are motivated by the game-like format of DNA Detective. During the workshops, students engage with each other, collaboratively complete the puzzle-like, drag and drop gene model, and are excited to take ownership of their plant. Even in our middle school workshops, 100% of the groups completed all the stops and selected the correct mutant plant.
SCIENTIFIC TEACHING THEMES
Active Learning
Students engage in bioinformatics analysis of big data to identify defects in genes that lead to phenotypic variation. Given a "mystery" DNA sequence, students use computers and bioinformatics software available online to 1) find and mask DNA repeats, 2) predict genes, 3) search gene and protein databases, 4) build and annotate gene models, and 5) select plants defective for their gene from a group of plants provided. This entire pipeline is carried out directly by the students, giving them ownership of the project and increasing student motivation and participation.
Assessment
We have not used any formal assessment rubric to evaluate effectiveness of the DNA Detective workshop. We assess the level of student excitement by gauging the interaction and enthusiasm of group members, and assess student interest by noting their persistent engagement with material and questions asked. We also track student learning by verifying successful identification of the group's gene and its mutant, and through students' post workshop comments.
Inclusive Teaching
Students work in small groups to foster collaborative discussion and prevent anyone feeling isolated. We provide all students, regardless of their background or experience, with the opportunity to engage with the material and support other students. We assign each student a task, and rotate tasks among group members, giving each student an opportunity to be a leader. As group leader, students share their hypotheses and data analysis process with their peers, providing a problem-based, shared learning experience. This collaborative learning teaches students to value the diverse opinions and problem-solving strategies of their peers. Additionally, as workshop leaders, instructors ensure a safe and welcoming learning environment where all ideas are valued and discussed.
SUPPORTING MATERIALS
- S1. DNA Detective - Sterilization protocol for pds seeds
- S2. DNA Detective - 0.5 MS/1% sucrose media protocol for pds seeds. Protocol used to make 0.5 MS/1% sucrose media for plating pds seeds prior to stratification
- S3. DNA Detective - DIM1 genomic sequence - MysteryGene1.fasta
- S4. DNA Detective - PGM1 genomic sequence - MysteryGene2.fasta
- S5. DNA Detective - PDS genomic sequence - MysteryGene3.fasta
- S6. DNA Detective - Lecture Presentation Slides
ACKNOWLEDGMENTS
The authors would like to thank Jason Williams for introducing us to the DNA Subway platform as well as the other CyVerse and Dolan DNA Learning Center staff for making our DNA Detective workshop possible.
References
- Marx V. 2013. Biology: The big challenges of big data. Nature. 498:255-260. https://doi.org/10.1038/498255a
- Barone L, Williams J, Micklos D. 2017. Unmet needs for analyzing biological big data: A survey of 704 NSF principal investigators. PLoS Comput Biol. 13:e1005755. https://doi.org/10.1371/journal.pcbi.1005755
- Weisstein AE, Gracheva E, Goodwin Z, Qi Z, Leung W, Shaffer C, Elgin SCR. 2016. A hands-on introduction to hidden Markov models. CourseSource. https://doi.org/10.24918/cs.2016.8.
- Smith TM, Emmeluth DS. 2002. Introducing bioinformatics into the biology curriculum: exploring the National Center for Biotechnology Information. Am Biol Teach. 64(2):93-99. https://doi.org/10.2307/4451250
- Madlung A. 2018. Assessing an effective undergraduate module teaching applied bioinformatics to biology students. PLoS Comput Biol. 14:e1005872. https://doi.org/10.1371/journal.pcbi.1005872
- Cummings MP, Temple GG. 2010. Broader incorporation of bioinformatics in education: opportunities and challenges. Brief Bioinform. 11(6):537-543. https://doi.org/10.1093/bib/bbq058
- Form D, Lewitter F. 2011. Ten simple rules for teaching bioinformatics at the high school level. PLoS Comput Biol. 10:e1002243. https://doi.org/10.1371/journal.pcbi.1002243
- Machluf Y, Yarden A. 2013. Integrating bioinformatics into senior high school: design principles and implications. Brief Bioinform. 14(5):648-660. https://doi.org/10.1093/bib/bbq030
- Wefer SH, Sheppard K. 2008. Bioinformatics in high school biology curricula: a study of state science standards. CBE Life Sci Educ. 7(1):155-162. https://doi.org/10.1187/cbe.07-05-0026
- Wefer SH. 2003. Name that gene: an authentic classroom activity incorporating bioinformatics. Am Biol Teach. 65(8):610-613. https://doi.org/10.1662/0002-7685(2003)065[0610:NTG]2.0.CO;2
- Peterson MP, Malloy JT, Buonaccorsi VP, Marden JH. 2015. Teaching RNAseq at undergraduate institutions: a tutorial and R package from the genome consortium for active teaching. CourseSource. https://doi.org/10.24918/cs.2015.14.
- Laakso MM, Paliulis LV, Croonquist P, Derr B, Gracheva E, Hauser C, Howell C, Jones CJ, Kagey JD, Kennell J, Silver Key SC, Mistry H, Robic S, Sanford J, Santisteban M, Small C, Spokony R, Stamm J, Van Stry M, Leung W, Elgin SCR. 2017. An undergraduate bioinformatics curriculum that teaches eukaryotic gene structure. CourseSource. https://doi.org/10.24918/cs.2017.13.
- Buonaccorsi VP, Hamlin D, Fowler B, Sullivan C, Stickler A. 2017. An introduction to eukaryotic genome analysis in non-model species for undergraduates: A tutorial from the Genome Consortium for Active Teaching. CourseSource. https://doi.org/10.24918/cs.2017.1.
- Drew JC, Triplett EW. 2008. Whole genome sequencing in the undergraduate classroom: outcomes and lessons from a pilot course. J Microbiol Biol Educ. 9(1):3-11. https://doi.org/10.1128/jmbe.v9.89
- Hertweck KL. 2016. Making toast: using analogies to explore concepts in bioinformatics. CourseSource. https://doi.org/10.24918/cs.2016.11.
- Holtzclaw JD, Eisen A, Whitney EM, Penumetcha M, Hoey JJ, Kimbro KS. 2006. Incorporating a new bioinformatics component into genetics at a historically black college: outcomes and lessons. CBE Life Sci Educ. 5(1):52-64. https://doi.org/10.1187/cbe.05-04-0071
- Lewitter F, Bourne PE. 2011. Teaching bioinformatics at the secondary school level. PLoS Comput Biol. 7(10):e1002242. https://doi.org/10.1371/journal.pcbi.1002242
- Maloney M, Parker J, LeBlanc M, Woodard CT, Glackin M, Hanrahan M. 2017. Bioinformatics and the undergraduate curriculum. CBE Life Sci Educ. 9(3):141-377. https://doi.org/10.1187/cbe.10-03-0038
- Musante S. 2004. Using bioinformatics in the undergraduate classroom. BioScience. 54(7):625. https://doi.org/10.1641/0006-3568(2004)054[0625:UBITUC]2.0.CO;2
- Porter SG, Smith TM. 2000. Bioinformatics in the community college. J Ind Microbiol Biotechnol. 24(5):314-318.
- Weisman D. 2010. Incorporating a collaborative web-based virtual laboratory in an undergraduate bioinformatics course. Biochem Mol Biol Educ. 38:4-9. https://doi.org/10.1002/bmb.20368
- Cooper K, McGraw A, Khazanchi D. 2017. Bioinformatics for middle school aged children: activities for exposure to an interdisciplinary field. 2017 IEEE Integrated STEM Education Conference (ISEC). 1-9. https://doi.org/10.1109/ISECon.2017.7910217
- Shuster M, Claussen K, Locke M, Glazewski K. 2016. Bioinformatics in the K-8 classroom: designing innovative activities for teacher implementation. Int J Des Learn. 7:60-70.https://doi.org/10.14434/ijdi.v7i1.19406
- Eccles JS. 1999. The development of children ages 6 to 14. Future Child. 9(2):30-44.
- Yandell M, Ence D. 2012. A beginner's guide to eukaryotic genome annotation. Nature Rev Genet. 13:329-342. https://doi.org/10.1038/nrg3174
- Akiyama K, Kurotani A, Iida K, Kuromori T, Shinozaki K, Sakurai T. 2014. RARGE II: an integrated phenotype database of Arabidopsis mutant traits using a controlled vocabulary. Plant Cell Physiol. 55(1):84. https://doi.org/10.1093/pcp/pct165
- Lipchock SV, Mennella JA, Spielman AI, Reed DR. 2013. Human bitter perception correlates with bitter receptor messenger RNA expression in taste cells. Am J Clin Nutr. 98(4):1136-1143. https://doi.org/10.3945/ajcn.113.066688
- Singer S, Smith KA. 2013. Discipline-based education research: understanding and improving learning in undergraduate science and engineering. J Eng Educ. 102:468-471. https://doi.org/10.1002/jee.20030
- Freeman S, Eddy SL, Mcdonough M, Smith MK, Okoroafor N, Jordt H, Pat M. 2014. Active learning increases student performance in science, engineering, and mathematics. Proc Natl Acad Sci. 111:8410-8415. https://doi.org/10.1073/pnas.1319030111
Article Files
Login to access supporting documents
DNA Detective: Genotype to Phenotype. A Bioinformatics Workshop for Middle School to College.(PDF | 554 KB)
S1. DNA Detective - Sterilization protocol for pds seeds.docx(DOCX | 16 KB)
S2. DNA Detective - MS_sucrose media protocol for pds seeds.docx(DOCX | 16 KB)
S3. DNA Detective - DIM1 genomic sequence-MysteryGene1.txt(TXT | 3 KB)
S4. DNA Detective - PGM1 genomic sequence -MysteryGene2.txt(TXT | 5 KB)
S5. DNA Detective - PDS genomic sequence-MysteryGene3.txt(TXT | 5 KB)
S6. DNA Detective Lecture Presentation Slides.pptx(PPTX | 21 MB)
- License terms

Comments
Comments
There are no comments on this resource.