Investigating Gene Expression and Cell Specialization in Axolotl Embryos
Editor: Stanley Lo
Published online:
Abstract
The process of cell specialization is critical to the formation and function of tissues in animals and plants. Although gene expression, including the regulation of transcription, is taught in most introductory cell biology courses, the relationship between differential gene expression and the formation of specialized cell types is challenging to understand for even upper-level life science students. In order to decrease this learning gap, I have developed a suite of in-class problem-solving activities and a lab experiment on Axolotl embryos that support student learning and integration of content related to differential gene expression and cell specialization. Although axolotls are best known as a model system for tissue regeneration, recent advances in genomic and molecular tools has increased their application as a model for studying gene expression during embryonic development as well. I tested the activities in an upper-level undergraduate course and found an increase in student understanding of the importance of differential gene expression during cell specialization processes, and the techniques used to study these processes, particularly Real Time quantitative PCR (RTqPCR). Teachers can examine student understanding of techniques and concepts using in-class assignments, exam questions, homework assignments and laboratory notebook assignments. Importantly, by analyzing a specific gene associated with a specialized cell type during different axolotl embryonic stages, students connect and integrate molecular, cellular and organismal level concepts of differential gene expression and cell specialization. This engagement deepens their understanding of the gene expression processes involved in cell specialization and of the role of model systems in biological research.
Citation
Eastman D. 2020. Investigating gene expression and cell specialization in Axolotl embryos. CourseSource. https://doi.org/10.24918/cs.2020.14
Society Learning Goals
Developmental Biology
- Gene Networks
- How do differences in regulation of gene expression explain the different cell types?
- Experimental Approaches
- How do different organisms help us understand development? And what are their strengths and limitations?
Lesson Learning Goals
From Developmental Biology Learning Framework:
- How do differences in regulation of gene expression explain the different cell types?
- How do different organisms help us understand development? And what are their strengths and limitations?
Lesson Learning Objectives
Students will be able to:
- identify characteristics of each stage of axolotl embryonic development.
- understand the importance of model organisms in the study of biological processes.
- compare strengths and limitations of axolotls as model organisms.
- understand the concepts related to differential gene expression and cell specialization.
- integrate their understanding of differential expression and cell specialization.
- explain the process and purpose of PCR, qPCR, and reverse transcriptase.
- calculate expression levels from raw qPCR results.
- analyze gene expression levels in embryos at different stages of development.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
In both plants and animals, embryonic development involves a complex set of intercellular interactions and molecular outcomes that result in cells with specialized functions. Differential gene expression is critical to this specialization of cells and is a central concept in cell, molecular, and developmental biology (1,2).
Although undergraduate students in many introductory cell, molecular, and/or developmental biology courses generally gain a basic understanding of gene expression and cell specialization, the ability to integrate these concepts and apply them at the organismal level is often a great challenge for them, even as they progress to upper-level biology courses (3). Recent studies on theory of knowledge integration in molecular and cell biology have shown that understanding the connection of molecular mechanisms to their roles in cells is challenging for students at all levels (4). Addressing this learning objective requires the development of applied experiences for student integrative thinking and intentional awareness of these difficulties by faculty. There are excellent cell biology and developmental biology text books that provide clear explanations of both gene expression regulation and cell specialization (1,2). These texts typically provide a paragraph on the relationship between differential gene expression and cell specialization; however, the concepts are detailed in two separate chapters and are not deeply integrated. Similarly, there are pedagogy resources for gene expression activities; however, examples of these are limited and are more focused on examining genotype-phenotype relationships in single cell organisms (5) or on transcriptome approaches in specific cell types. Although transcriptome methods such as RNAseq or Microarrays can potentially provide students with understanding of differential gene expression and cell specialization, these experiments tend to take many weeks, if not all of the semester to pursue (6). A recent article by Shifley (7) provides an excellent approach for students to study differential gene expression in frog embryos using in situ hybridization; however, again, there is not an emphasis on integration of differential gene expression and cell specialization concepts. Examining the differential expression of a gene associated with a specialized cell type in different stages of an embryo could provide students with a framework to integrate their understanding of basic concepts.
I have taken advantage of the Ambystoma mexicanum (axolotl) embryo system to develop students' abilities to integrate concepts and techniques related to differential gene expression and cell specialization. Although not as well-known as flies, worms, fish, or frogs, axolotls provide an excellent model system to study regeneration and development, both for research and for teaching. Axolotls are particularly accessible for undergraduate biology laboratory experiments, given their large embryo size and inexpensive availability from the Ambystoma Genetic Stock Center (https://ambystoma.uky.edu/genetic-stock-center/). They develop from fertilized egg to hatched larvae in 10 days in a simple salts solution, and the timing can be manipulated by incubating at lower temperatures without affecting developmental outcomes (8). Although molecular tools are not yet as readily available for axolotls as they are for other model organisms, the genome has recently been sequenced (9), and there are several transcriptome assemblies (10) available for obtaining sequence information (https://www.axolotl-omics.org/; https://ambystoma.uky.edu/genome-resources). The new availability of these tools allows for a discussion with students about why the study of multiple model organisms is important, why different organisms are used to study specific processes, and how an organism becomes a model system. Importantly, cell specialization and differential gene expression have been studied in axolotl embryos (11,12). One particular study by Banfi et al. (13) focused on muscle cell development in different stages of axolotl embryos and determined that myf5, an important conserved muscle development gene (14), is differentially expressed during this process.
Studies of gene expression regulation and differential expression rely on methods that detect RNA transcripts and protein products. Methods for examining RNA transcripts from single genes include Northern blots, in situ hybridization, and Reverse Transcriptase quantitative PCR (RTqPCR). Northern blots require radioactivity and in situ hybridization involves many steps. Thus, both are challenging to incorporate into the undergraduate laboratory. Over the past 15 years, RTqPCR has become the most commonly used method to quantify single gene transcriptional levels (15,16). RNA is isolated from cells, tissues or embryos and reverse transcriptase is used to synthesize cDNA from the isolated RNA. Primers specific to the genes of interest are used in the reaction that allows detection of the PCR product in real time. By using a reference gene known to be equivalently expressed in all cells, changes in expression levels can be identified for the target genes.
In order to develop students' ability to understand integrated differential gene expression and cell specialization concepts in an upper-level developmental biology course, I have used multiple problem-based in-class activities, a primary research article critique and discussion, and an inquiry-based two-week laboratory activity. The in-class activities encourage students to learn and apply their understanding of basic concepts and methods related to gene expression and cell specialization. Students then read and critique the primary research article by Banfi et al. (13), which focuses on muscle development in axolotl embryos and the differential expression of muscle specific genes at different developmental stages. Students learn about components and functions associated with specialized muscle tissue, as well as the conserved muscle development gene myf5, which is differentially expressed during axolotl development. In critiquing the article, they also analyze the results from in situ hybridization and RTqPCR experiments. Based on reading and analysis of the Banfi et al. article, students develop a hypothesis about the expression of myf5 in an embryonic stage not studied in the article. They then perform a RTqPCR experiment on cDNA provided from this stage, calculate myf5 expression levels in their stage sample, compare to their hypothesis, and share results with peers in the class who analyzed cDNA from different stages. Both the experimental set-up and analysis require students to apply basic quantitative skills. Students record conclusions to the experiment in their laboratory notebooks and the class then discusses their overall results, both in comparison to results in Banfi et. al. and to larger questions of differential gene expression and specialized cell development. A timeline for these activities is provided in Table 1. These and related exercises can be used in more advanced biology courses focused on cell, molecular, and/or developmental biology concepts. In addition to addressing important concepts, the activities involve problem-solving and inquiry-based experiences, which have been shown to enhance student understanding and commitment to science (14-16), and can be particularly impactful for first generation and other underrepresented students (17).
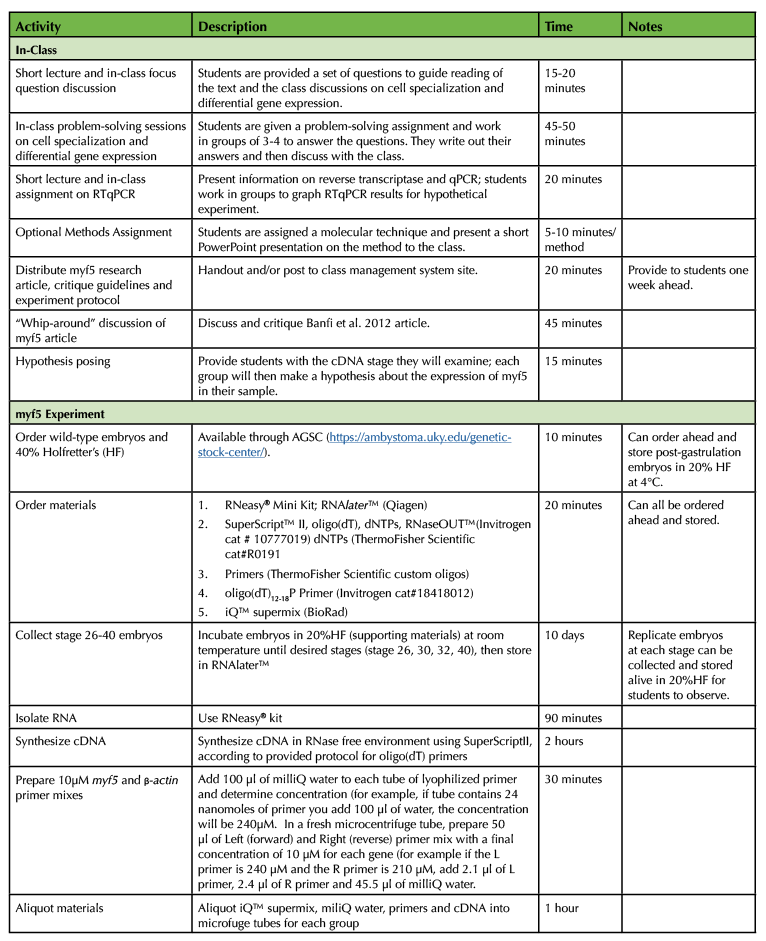
Table 1. Differential Expression in Axolotl Embryos Teaching Timeline

Table 1. Differential Expression in Axolotl Embryos Teaching Timeline (continued)
I have tested all of these activities in an upper-level undergraduate developmental biology course. They have worked very well to enhance student understanding of differential gene expression, to promote integration of concepts, and to help students understand the importance of model organisms for studying biological processes. Understanding differential gene expression and cell specialization during embryonic development is important for the study of human health, as many diseases involve these developmental processes (18).
Intended Audience
The in-class and laboratory activities were designed for undergraduate biology students in an upper-level course focused on cell, molecular, and developmental biology concepts. This course is taken by juniors and seniors majoring in Biology, Biochemistry, Cell and Molecular Biology, and Behavioral Neuroscience.
Required Learning Time
Short in-class lectures and discussion of focus questions require approximately 15-20 minutes and in-class problem solving assignments approximately 45-50 minutes. A "whip-around" discussion of the primary research article takes approximately 40 minutes. Hypothesis development and myf5 RT-qPCR set-up takes approximately 1.5 hours. After qPCR results are obtained, I reserve one hour to analyze and discuss results (normally during the following lab period). Advanced investigative projects require 4-5 weeks, depending on the specific experiments that students pursue. A timeline that includes estimated time allotments for each of the activities is provided in Table 1.
Prerequisite Student Knowledge and Skills
Prior to performing the experiment, students should be introduced to basic concepts of embryonic development and eukaryotic gene expression as part of classroom discussions. This includes the processes of transcription, translation, and regulation of transcription, as well as the basic molecules involved. They should also be familiar with traditional PCR and have experience with using micropipettors to set up small-volume reactions.
Prerequisite Teacher Knowledge
Instructions for all of the related activities are included and do not require any prerequisite specific teacher knowledge beyond what is described for student knowledge. Since the teacher will need to isolate RNA, make cDNA and set-up qPCR experiments, teacher familiarity with molecular biology assays using DNA and/or RNA would be beneficial, but not necessary. Care and staging of axolotl embryos is relatively easy, as both the embryos and solutions can be purchased inexpensively from the Ambystoma Genetic Stock Center (https://ambystoma.uky.edu/genetic-stock-center/), and detailed drawings of the different stages are accessible on-line through Sal-Site™ (https://ambystoma.uky.edu/education1/embryo-staging-series) (19).
SCIENTIFIC TEACHING THEMES
Active Learning
The activities for this lesson involve multiple active learning approaches. For the weeks prior to and during the laboratory exercises, pre-class focus questions, in-class short lectures, and in-class problem-solving assignments on cell specialization and gene expression regulation concepts (Supporting File S1. Axolotl gene expression and specialization – Class assignments and activities) are provided. For the in-class problem solving assignments, students work in groups, write up their answers and report out to the rest of the class. Class discussions and problem-solving assignments engage students in the basic concepts, molecules, processes and methods related to cell specialization and regulation of gene expression. Students can then learn about different techniques used to study molecular processes during development through peer presentations on different methods, including methods to study gene expression products (Supporting File S1. Axolotl gene expression and specialization – Class assignments and activities). As part of the methods presentations, a short lecture on RTqPCR is provided, followed by an in-class problem solving assignment that asks students to translate results from a Northern Blot to raw RTqPCR results (Supporting File S1. Axolotl gene expression and specialization – Class assignments and activities). Students are then assigned the Banfi et al. article on muscle development in axolotl embryos, which they read and critique using a set of assigned questions (Supporting File S2. Axolotl gene expression and specialization – Lab assignments and activities). The article is discussed class-wide during a lab sessions, ending with an in-depth discussion of Figure 7 (Supporting File S2. Axolotl gene expression and specialization – Lab assignments and activities). Each lab group is assigned a developmental stage not studied by Banfi et al. and formulates a hypothesis for the expression of the muscle development gene, myf5, in their assigned stage. Students work collaboratively to perform a myf5 RTqPCR experiment and calculate expression levels (Supporting File S2. Axolotl gene expression and specialization – Laboratory assignments and activities). As a group, they compare their results to their hypothesis. They then share out results in a class-wide discussion in which all students participate. At the end of this discussion students are asked to discuss and write in their notebooks how the class-wide and Banfi et al. results for myf5 differential expression might relate to muscle cell specialization in axolotls. All aspects of these assignments require that students take responsibility for their learning and engage fully.
Assessment
Different levels of student learning were assessed using a variety of approaches. Student understanding of differential expression and cell specialization was assessed multiple times throughout the semester: during in-class learning of concepts and methods (Supporting File S1. Axolotl gene expression and specialization – Class assignments and activities) through answers to problem-solving assignments (Supporting File S1. Axolotl gene expression and specialization – Class assignments and activities), during the laboratory experiments through recordings in their notebook (Supporting File S2. Axolotl gene expression and specialization – Laboratory assignments and activities), several weeks after performing the experiment using an in-class closed-book exam question, later in the semester as part of a graded homework assignment, and at the end of the semester with graded take-home final exam questions (Supporting File S3. Axolotl gene expression and specialization – Assessments).
Inclusive Teaching
The activities for this Lesson are intentionally designed to include participation from all students. Throughout the activities, students work in small assigned groups of three to four. Assignment of these groups is based on student self-identified asset maps (20). This is an approach that has been shown to enhance equity and inclusion in group work (20). They are encouraged to share their strengths and challenges regarding the material and communicate how they will work as a group to achieve full participation for all group members. Students discuss the research article using a "whip-around," which is an inclusive pedagogy approach that has been shown to enhance student engagement and cultivate equity in the classroom (21,22). This requires each student to share their understanding of background information on muscle development and of the results for myf5 expression in axolotl embryos within a supportive environment. The use of focus questions to structure text reading and multiple active learning approaches provides opportunities for all students to be supported in their learning. Since no prior knowledge of muscle development or myf5 is required, all students are able to participate in developing a hypothesis, performing the experiment and analyzing results. Throughout the exercise I encourage students to share ideas and questions in order to learn from one another, since each student has different areas of expertise, both knowledge and skill-wise, to contribute.
LESSON PLAN
Pre-class Preparation
Textbook page numbers, a pre-class video (15 minutes maximum time), and focus questions (Supporting File S1. Axolotl gene expression and specialization – Class assignments and activities) for class discussions on differential gene expression and on cell specialization are provided to students at least one-week prior to class time. Optional methods assignment (Supporting File S2. Axolotl gene expression and specialization – Lab assignments and activities), Banfi et al. article (https://onlinelibrary.wiley.com/doi/full/10.1111/j.1440-169X.2012.01338.x) and critique questions (Supporting File S2. Axolotl gene expression and specialization – Lab assignments and activities) are provided two weeks prior to class time.
Pre-lab Preparation
Materials
Embryos: Early stage wild-type axolotl embryos can be ordered from the Ambystoma Genetic Stock Center (AGSC; https://ambystoma.uky.edu/genetic-stock-center/). They are shipped overnight and upon arrival are normally in the cleavage stage. Information about axolotl embryo husbandry, staging, and other educational materials are available at Sal-Site™ (18). For some experiments, the jelly coat should be removed at the beginning of the experiment using two #5 forceps). The use of animals throughout the lesson should be in compliance with local Institutional Animal Care and Use Committee (IACUC) guidelines. Normally, the use of axolotl embryos before hatching (stage 43) does not require an IACUC protocol. Embryos from stages 26, 30, 32 and 40 should be used for RNA isolations and for student viewing during lab. These stages were not part of the Banfi et al. study, so student results will be novel.
Solutions and conditions for axolotl embryos: Order embryos a week prior to use from AGSC and incubate in 20% HF (40% HF is available from AGSC) in glass finger bowls at room temperature. They will continue to develop if incubated at lower temperatures; however, they should not be incubated at temperatures lower than 10°C until after gastrulation is complete (stage 13). Details on identifying embryo stages are available through Sal-Site™(https:// ambystoma.uky.edu/education1/embryo-staging-series). For embryos used for RNA isolation, once at the desired stage, anesthetize them in 0.1% Tricaine methane sulfonate solution (Sigma, Cat#A5040-250) for 5 minutes and then store them in RNAlater™ (an RNA stabilizing reagent, ThermoFisher Cat#AM7020) until all samples are collected. For live embryos used during class for students to observe, collect embryos at desired stages and store them at 4°C for use within the following week.
Reagents for isolation of RNA and cDNA: Order an RNeasy® Mini Kit (QIAGEN, Cat#74104). The protocol is provided with the kit and is straightforward. Briefly, embryos are homogenized with a Dounce homogenizer in Buffer RLT (part of RNeasy® Kit). The QIAGEN RNeasy® kit's reagents are then used to isolate RNA from the homogenized sample. Superscript™ II (ThermoFisher, Cat#18064014) and oligo(dT)12-18 Primer (ThermoFisher, Cat#18418012) are used to synthesize cDNA from the isolated total RNA. The protocol for cDNA synthesis is provided by ThermoFisher along with the Superscript™ II enzyme. Follow these directions using oligo(dT) primers.
RNAaseOUT™ (ThermoFisher Cat#10777-019) is a reagent that can be used to protect RNA during the isolation and synthesis processes.
PCR primers: The sequences for myf5 and actin primers are:
- Axo Myf5F 5' CCCTGCCCGGCCAGCACTGC 3'
- Axo Myf5R 5' GGGTGTTGATTTTGTCTGTGGGGTAAA 3'
- Axo B-actinF 5' CTGAACCCCAAAGCCAACAGAGAAAA 3'
- Axo B-actinR 5' GCGTAAAGGGACAGCACAGCTTGAAT 3'
Primers for additional muscle-specific (and other) genes can be identified by typing the gene name into the "Search for Genes in assembly V.0" box. The sequence can then be used to identify qPCR primers using primer3 (http://bioinfo.ut.ee/primer3/) by pasting the sequence into the primer3 source sequence box and selecting for products that are 100-200bp. (Supporting File S7. Axolotl gene expression and specialization – Adapting Activities).
Oligonucelotide primers can be ordered from Thermofisher as standard oligos. Oligonucleotide primers arrive in powder form and must be rehydrated and then diluted to a final working concentration of 10 µM. Briefly, add 100µl milliQ water to the tube of dried oligonucleotide (generally arrive with 10-30 nmol; for example: if the forward primer is 17 nmol and the reverse primer is 24 nmol, if 100 µl of milliQ water is added, the concentrations will be 170µM and 240 µM, respectively). Then make a 100µM stock that contains both primers: for a 50µl stock of 10µM, add 2.94 µl of forward primer, 2.1 µl of reverse primer and 44.96 µl milliQ water. You will need to calculate the exact volumes for each primer based on the nanomole amount of primer in each tube.
Specific pre-lab preparation for Myf5 experiment: For the myf5 experiment, students are provided cDNA from different stages of embryos. Thus, stage embryos, remove the jelly coat, isolate RNA, and synthesize cDNA for each stage before the laboratory exercise. Order both myf5 and actin primers/oligonucleotides, and prepare 10µM stocks containing forward and reverse primers for each gene.
In-Class Facilitation
For concept classes, students should be expected to have read the text, watched the pre-class video and considered the focus questions. Encourage them to write answers to the focus questions prior to class. Organize the class around the focus questions: ask students to discuss each focus question with a group of three to four peers and then agree on a brief answer. Provide a short (5-10 minutes) lecture on the focus question, ask students to discuss again, and have groups report out a more detailed response to the focus question. After the focus questions have been discussed, provide the in-class problem-solving assignment on gene expression regulation or cell specialization (Supporting File S1. Axolotl gene expression and specialization – Class assignments and activities). Students then discuss and work on the problem as a group, but each student hands in their own written answers to the problems.
For the differential gene expression methods assignment, introduce Northern blot and RTqPCR as methods for studying levels of RNA in cells and tissues, using a short lecture and video during class. Organize students in small groups of 3-4 and hand out the in-class problem-solving assignment on qPCR (Supporting File S1. Axolotl gene expression and specialization – Class assignments and activities). Allow groups to work on the problem for 10-15 minutes. Each student writes their own answers that are collected. Draw a graph on the board with "cycle number" on the x-axis and "expression level" on the y-axis. Have each group draw results for one of the tissue types, and discuss student answers to confirm understanding of 1) why lines are different between some of the samples and 2) how the results relate to cell functions in the different tissues. For the optional additional methods assignment, students give a 5-10 minute PowerPoint presentation on a molecular method prior to the instructor introducing Northern blots and RTqPCR.
In-Lab Facilitation
Two weeks prior to the scheduled lab, provide a copy of the article by Banfi et al., critique guidelines and the myf5 experiment handout, which has stepwise instructions for the laboratory protocol. Students are required to submit answers to critique questions prior to the start of the laboratory period. Each of the critique questions is discussed class-wide using a "whip-around," and students use learning from their critique along with the discussion to formulate their hypotheses for the investigative experiment that examines expression of a muscle development gene, myf5, in embryo stages not yet examined in published studies. Students work collaboratively to perform and analyze results from the myf5 experiment. As a group, they compare their results to their hypothesis. They then share out results in a class-wide discussion in which all students participate. At the end of this discussion students are asked to discuss and write in their notebooks how the class-wide and Banfi results for myf5 differential expression might relate to muscle cell specialization in axolotls.
Step-by-Step Guide for the Activities
Early in the semester
Using an approach developed by Wobbe and Stoddard (20) students fill out an asset map in which they write down their strengths in areas such as team skills, creativity, related courses, research experiences, passions and interests, quantitative skills, and personal background, as well as three areas in which they would like to grow. Assign lab groups based on these asset maps to create groups with members who have different areas of strength, which they can share with group members. During a lab meeting members share their asset maps with the rest of the group and together they build a team asset map.
Week 1-2
- Day 1 in-class short-lectures and discussion on focus questions followed by problem-solving on cell specialization.
- Day 2 in-class short-lectures and discussion on focus questions focused on problem-solving on differential gene expression.
- Day 3 in-class short-lecture and problem-solving on RTqPCR and other methods to study transcription products; (optional methods presentations assignment prior to RTqPCR).
Week 2
- Collect critiques and discuss the Banfi et al. article, using a "whip-around" and the article critique questions. Discuss each figure in detail, specifically focusing on Figure 7 in the article.
- Provide each lab group with axolotl embryos from multiple stages and cDNA from a different stage of development not studied in the Banfi article, as well as qPCR reagents (thawed and kept on ice). Students should first observe the embryos under a dissecting scope and identify the stage of embryo that they have cDNA from. Students record their observations in their laboratory notebook.
- Ask each group to write a hypothesis in their notebooks for what they expect to observe with their results.
- Each group sets up three qPCR reactions for myf5 and three for β-actin using the steps provided in the hand-out.
- Members of the group pipette the triplicate reaction into wells of a 96-well plate for qPCR or into individual PCR tubes for traditional PCR.
- The qPCR cycle program settings should be:
- 10 minutes 95°C
- 40 cycles :95°C for 15 seconds
- 60°C for 60 seconds
- Run the qPCR reactions in an iCycler or other qPCR machine.
Week 3
- Send students raw Cycle Threshold (Ct) results and a model Excel file to assist with calculations (Figure 3; Supporting Material S3. Axolotl gene expression and specialization – Assessments). Students can determine the normalized Ct (∆Ct) by subtracting the average actin Ct from the average myf5 Ct. They can then determine ∆Ct expression by using the calculation 2∆Ct, (16,23). Students then normalize expression of their stage to stage 40 by calculating ∆∆Ct and 2-∆∆Ct. Details of how to perform the calculations are found in Supporting File S5. Axolotl gene expression and specialization – Model Excel file for myf5 experiment).
- If students performed traditional PCR, they will need to run a 2% gel to visualize their results and determine whether there is a band for each of the stages.
- Discuss the results and have students compare their results to their original hypothesis.
Expected Results for myf5 Experiment
Students should observe PCR product in all stages for both myf5 and β–actin. The lowest level of expression (highest ∆cT values) should be observed in stage 40 (Figure 1; Supporting File S4. Axolotl gene expression and specialization – Raw sample data in Excel; Supporting File S5. Axolotl gene expression and specialization – Model Excel file for myf5 experiment; Supporting File S6. Axolotl gene expression and specialization – Sample results Excel file). The highest levels of expression should be seen in stage 30, which corresponds to the stage when cells within the forming somites are differentiating into muscle. Lowest levels of expression should be observed in stages 24 and 40, before and after muscle differentiation is occurs, respectively.
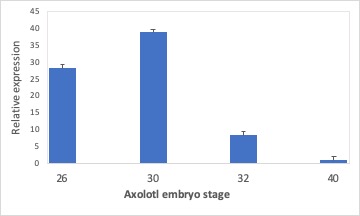
Figure 1. Sample results for myf5 differential expression experiment. Relative expression determined by the ratio of ∆Ct for each stage compared to the ∆Ct for stage 40 is presented in the graph. As expected, the highest level of myf5 expression occurs in stages 26 and 30, when muscle development is occurring in axolotl somites.
TEACHING DISCUSSION
The suite of class and lab activities described in this Lesson address two main learning goals: for students to understand 1) the importance of differential gene expression to the process of cell specialization and 2) the usefulness of model organisms in biological studies. The concepts of differential gene expression and cell specialization, as well as the techniques used to analyze these processes, are central to the laboratory activities, and student understanding of the relationship between these two concepts was enhanced by engagement in the activities. The in-class problem-solving activities required students to apply their knowledge and deepened their understanding of the molecular aspects of the concepts. The article discussion and myf5 experiment then provided an opportunity for them to actively integrate the concepts. Focus questions and pre-class videos encouraged students to come to class prepared to engage in discussions and problem-solving activities. The concepts for cell specialization were introduced first, and the problem-solving activity required that students read sections of a review article and use the online database NCBI to identify proteins and relate their functions and expression patterns in different cell types. This allowed students to apply their understanding of proteins as functional molecules to the concept of cell specialization. By the end of class, all students understood that the presence of different protein products in different cell types allows these cells to specialize and have different functions. The problem-solving assignment for differential gene expression requires that students apply their understanding of molecular mechanisms for gene expression regulation including binding of regulatory transcription factors to enhancer elements to promote transcription. By drawing and discussing the molecular events that occur at a muscle-specific and a neuronal-specific gene within specialized cell types, they are able to integrate differential expression and cell specialization. Since RTqPCR has many applications in a wide-array of biological, environmental and medical fields, it is an important technique for undergraduate students to understand. The in-class assignment on RTqPCR provided an introduction to the technique (Supporting File S1. Axolotl gene expression and specialization – Class assignments and activities) and allowed students to immediately apply their learning. When first working on the in-class assignment, many students struggle with understanding why real time qPCR, but not traditional PCR, allows for quantitative analysis. In order to enhance student understanding, I re-engage the class and draw a graph with two samples, one that produces product at an early cycle and one that produces product at a later cycle. I then re-emphasize the basic concept of product amplification during PCR reactions pointing out that based on this amplification, if there is more starting DNA template in one sample then it takes fewer cycles to produce a specific amount of product than a sample with lower amounts of starting DNA. By identifying the difference in cycle numbers needed to produce this product amount in real time, we can quantify the difference. After this discussion, all student groups were able to correctly draw predicted results for the hypothetical qPCR experiment. Students were assessed individually for their understanding of this concept on a take-home final exam (Supporting File S3. Axolotl gene expression and specialization – Assessment). At least 84% of developmental biology students were able to demonstrate their understanding of the technique by answering this question correctly.
The article discussion and myf5 experiment provide a simplified research experience in which students are able to engage in reading sections of a primary research article, make a hypothesis, perform qPCR, analyze results and relate conclusions to published research. Laboratory notebook entries, an in-class exam question and a take-home writing assignment (Supporting File S2. Axolotl gene expression and specialization – Lab assignments and activities; Supporting File S3. Axolotl gene expression and specialization – Assessments) served as assessment tools to determine students' understanding of the concepts of and relationship between differential expression and cell specialization. All students participated in the discussion of the primary research article and worked with their laboratory group to develop a hypothesis, which they wrote in their notebook. This required students to understand the results in the article and apply them to their experiment. Students worked in groups to perform the experiment and to analyze the results. Quantitative skill development was an intentional goal for these activities, and students were required to calculate specific amounts of reagents for the qPCR reactions as well as analyze quantitative results. Students worked in groups to perform this analysis during lab time. Approximately half of the students were able to easily do and understand all of the calculations; however, some students struggled with understanding the purpose of the reference gene and how to do the expression calculations. I encouraged them to perform the calculations using Excel by providing a model file (Supporting File S5. Axolotl expression and specialization – Model Excel file for myf5 experiment); however, some students preferred to calculate and graph directly in their notebooks. By the end of the laboratory discussion, all students were able to perform the calculations. Analysis of laboratory notebook entries showed that almost all the students understood the RTqPCR results, specifically that lower ∆Ct values represented higher levels of expression in samples. All students related these results to their hypotheses, and 74% of students related these results to differential gene expression and muscle cell specialization in their laboratory notebooks. Student understanding of differential gene expression and cell specialization was further assessed on an in-class closed-book exam (two weeks after the myf5 lab experiments) and near the end of the semester as part of a take-home assignment (Supporting File S3. Axolotl gene expression and specialization – Assessments). For the in-class exam question, 95% of students understood the basic concepts of differential gene expression and cell specialization and 79% of students understood the concepts as well as the relationship between them.
Importantly, the activities reported here require students to consider the organismal outcomes of the molecular events that occur during differential gene expression and cell specialization. By viewing axolotl embryos at different stages, reading a primary research article on a specific tissue as it develops in the embryo, and performing gene expression analysis on a specific gene, students were able to expand their understanding beyond cell-type specializations to the level of tissue development and functions at the organismal level. In addition, analysis of experiments in axolotls in comparison to other vertebrates, including humans, allowed students to understand the importance of studying model organisms and how different organisms become model systems for studying different biological processes.
Adapting Activities for Introductory Courses
Although these activities were tested in an upper-level course, they can be adapted for an introductory cell or molecular biology course to engage in another core concept, genomic equivalence. Before students can appreciate differential gene expression mechanisms and how they relate to cell specialization, they must understand that the genome is the same within nearly every cell of a multicellular organism. In-class activities can focus on the evidence for genomic equivalence including somatic cell nuclear cloning, as well as how qPCR can be used to quantify differences in genomic DNA between samples. For the laboratory component, students quantify the levels of a small region of the β-actin gene (actin) from embryos at different stages of development (Supporting File S7. Axolotl expression and specialization – Adapting activities). Pre-class preparation involves genomic DNA isolation (Quiagen kit) and DNA concentration measurements for different samples. Students perform qPCR reactions and analyze results using standard curves. The results show students that the same amount of a specific sequence of DNA is present in cells of different embryo stages, supporting genomic equivalence.
Adapting Activities for Advanced Course-Based Research Experiences
The simplified myf5 experiment can be modified into a multi-week investigative laboratory experience for advanced students. Students work in groups to identify possible questions, signaling pathways, tissues, cell types and genes of interest to study in different stages of axolotl embryos. This allows them to engage relevant content related to their project as well as important steps of experimental design. Students then propose a hypothesis based on their literature research and develop a list of genes and axolotl embryo stages. They design oligonucleotide primers for quantitative PCR (Supporting File S7. Axolotl expression and specialization – Adapting activities), isolate RNA from specified axolotl embryo stages, and synthesize cDNA from these RNA samples (as described in pre-lab preparation). For the investigative projects, students meet as groups to first discuss their roles and then to perform the experiment, analyze results, and present conclusions. Students work together to first search for and read primary literature in order to develop their research question and identify the genes they propose to study. Each week, students share informal progress reports on their projects with the rest of the class. For the informal project reports, I use a think-pair-share approach to enhance the depth and participation during the discussion. At the end of the project each group develops a poster that they then present in a class-wide poster session. I have tested this in my upper-level developmental biology course and it has been very successful.
Adapting Activities to Use Traditional PCR
If a qPCR machine is not available, the myf5 and genomic equivalence experiments can be simplified to a semi-quantitative experiment using a OneTaq® One-Step RT-PCR kit (NEB cat#E5315S) and gel electrophoresis. The same in-class discussions and problem solving assignments can be done along with the research article discussion and hypothesis posing described in this Lesson. Students are provided RNA from different stages and perform reverse transcriptase and PCR reactions using One-Step Reaction Mix. The same primers and annealing temperatures provided for the qPCR reactions can be used for the traditional PCR reactions. In order to visualize PCR products, students run PCR products on a 2% agarose gel and determine which lanes have bands corresponding to PCR product. Details for gel electrophoresis are provided in section III of the Genomic Equivalence handout (Supporting File S7. Axolotl expression and specialization – Adapting activities). In order to see differences in myf5 expression students should analyze stages where myf5 is either not expressed or has very low expression (stages 10-15 or stages 41-42).
Conclusions
In summary, this set of activities may enhance student understanding of the relationship between differential gene expression and cell specialization as well as the importance of model organisms in biology. Both the in-class and lab activities are useful for an upper-level undergraduate course.
SUPPORTING MATERIALS
- S1. Axolotl gene expression and specialization – Class assignments and activities
- S2. Axolotl gene expression and specialization – Lab assignments and activities
- S3. Axolotl gene expression and specialization – Assessments
- S4. Axolotl gene expression and specialization – Raw sample data in Excel
- S5. Axolotl gene expression and specialization – Model Excel file for myf5 experiment
- S6. Axolotl gene expression and specialization – Sample results Excel file
- S7. Axolotl gene expression and specialization – Adapting activities
ACKNOWLEDGMENTS
Many thanks to Connecticut College students in Bio 302 Fall 2015, 2017 and 2018 for their work on the differential expression activities and for their willingness to share responses for this study. I also appreciate the engagement by students in Bio 110, where I tested the genomic equivalence laboratory activity. The Ambystoma Genetic Stock center is supported by NIH P40-OD019794.
References
- Baressi, M, Gilbert, S. 2019. Developmental Biology. Oxford University Press, Sunderland (MA).
- Alberts, B, Johnson, AD, Lewis, J, Morgan, D, Raff, M, Roberts, K, Walter, P. 2015. Molecular Biology of the Cell. Garland Science.
- Reinagel, A, Bray Speth, E. 2016. Beyond the Central Dogma: Model-Based Learning of How Genes Determine Phenotypes. CBE Life. Sci. Educ. 15:ar4-0105. doi: 10.1187/cbe.15-04-0105.
- Southard, K, Wince, T, Meddleton, S, Bolger, MS. 2016. Features of Knowledge Building in Biology: Understanding Undergraduate Students' Ideas about Molecular Mechanisms. CBE Life. Sci. Educ. 15:ar7-0114. doi: 10.1187/cbe.15-05-0114.
- Aronson, BD, Silveira, LA. 2009. From genes to proteins to behavior: a laboratory project that enhances student understanding in cell and molecular biology. CBE Life. Sci. Educ. 8:291-308. doi: 10.1187/cbe.09-07-0048.
- Procko, C, Morrison, S, Dunar, C, Mills, S, Maldonado, B, Cockrum, C, Peters, NE, Huang, SC, Chory, J. 2019. Big Data to the Bench: Transcriptome Analysis for Undergraduates. CBE Life. Sci. Educ. 18:ar19-0161. doi: 10.1187/cbe.18-08-0161.
- Emily T. Shifley. 2019. Differential gene expression during Xenopus laevis development. 06:. doi: 10.24918/cs.2019.32.
- Khattak, S, Murawala, P, Andreas, H, Kappert, V, Schuez, M, Sandoval-Guzman, T, Crawford, K, Tanaka, EM. 2014. Optimized axolotl (Ambystoma mexicanum) husbandry, breeding, metamorphosis, transgenesis and tamoxifen-mediated recombination. Nat. Protoc. 9:529-540. doi: 10.1038/nprot.2014.040.
- Nowoshilow, S, Schloissnig, S, Fei, JF, Dahl, A, Pang, AWC, Pippel, M, Winkler, S, Hastie, AR, Young, G, Roscito, JG, Falcon, F, Knapp, D, Powell, S, Cruz, A, Cao, H, Habermann, B, Hiller, M, Tanaka, EM, Myers, EW. 2018. The axolotl genome and the evolution of key tissue formation regulators. Nature. 554:50-55. doi: 10.1038/nature25458.
- Voss, SR, Athippozhy, A, Woodcock, MR. 2015. Transcriptomics using axolotls. Methods Mol. Biol. 1290:309-319. doi: 10.1007/978-1-4939-2495-0_24.
- Jiang, P, Nelson, JD, Leng, N, Collins, M, Swanson, S, Dewey, CN, Thomson, JA, Stewart, R. 2017. Analysis of embryonic development in the unsequenced axolotl: Waves of transcriptomic upheaval and stability. Dev. Biol. 426:143-154. doi: S0012-1606(15)30261-X.
- Zajdel, RW, McLean, MD, Dube, S, Dube, DK. 2013. Expression of tropomyosin in relation to myofibrillogenesis in axolotl hearts. Regen. Med. Res. 1:8-8. eCollection 2013 Dec. doi: 10.1186/2050-490X-1-8.
- Banfi, S, Monti, L, Acquati, F, Tettamanti, G, de Eguileor, M, Grimaldi, A. 2012. Muscle development and differentiation in the urodele Ambystoma mexicanum. Dev. Growth Differ. 54:489-502. doi: 10.1111/j.1440-169X.2012.01338.x.
- Hernandez-Hernandez, JM, Garcia-Gonzalez, EG, Brun, CE, Rudnicki, MA. 2017. The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. Semin. Cell Dev. Biol. 72:10-18. doi: S1084-9521(17)30353-1.
- Wagner, EM. 2013. Monitoring gene expression: quantitative real-time rt-PCR. Methods Mol. Biol. 1027:19-45. doi: 10.1007/978-1-60327-369-5_2.
- Jones, NL. 2002. PCR. Principles, procedures, and parameters. Methods Mol. Biol. 187:37-46. doi: 10.1385/1-59259-273-2:037
- Rodenbusch, SE, Hernandez, PR, Simmons, SL, Dolan, EL. 2016. Early Engagement in Course-Based Research Increases Graduation Rates and Completion of Science, Engineering, and Mathematics Degrees. CBE Life. Sci. Educ. 15:10.1187/cbe.16-0117.
- Singh, AJ, Ramsey, SA, Filtz, TM, Kioussi, C. 2018. Differential gene regulatory networks in development and disease. Cell Mol. Life Sci. 75:1013-1025. doi: 10.1007/s00018-017-2679-6.
- Smith, JJ, Putta, S, Walker, JA, Kump, DK, Samuels, AK, Monaghan, JR, Weisrock, DW, Staben, C, Voss, SR. 2005. Sal-Site: integrating new and existing ambystomatid salamander research and informational resources. BMC Genomics. 6:181. doi: 10.1186/1471-2164-6-181.
- Wobbe, K, Stoddard, E, Bass, R. 2019. Project-based learning in the first year: beyond all expectations. Stylus.
- Tanner, KD. 2013. Structure matters: twenty-one teaching strategies to promote student engagement and cultivate classroom equity. CBE Life. Sci. Educ. 12:322-331. doi: 10.1187/cbe.13-06-0115.
- Gehring, KM, Eastman, DA. 2008. Information fluency for undergraduate biology majors: applications of inquiry-based learning in a developmental biology course. CBE Life. Sci. Educ. 7:54-63. doi: 10.1187/cbe.07-10-0091.
- Pfaffl, MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45 [doi].
Article Files
Login to access supporting documents
Investigating Gene Expression and Cell Specialization in Axolotl Embryos(PDF | 204 KB)
S1. Axolotl gene expression and specialization- Class assignments and activities.docx(DOCX | 47 KB)
S2. Axolotl gene expression and cell specialization-Lab assignments and activities.docx(DOCX | 15 KB)
S3. Axolotl gene expression and cell specialization- Assessments .docx(DOCX | 32 KB)
S4. Axolotl gene expression and cell specialization-Model for myf5 experiment .xlsx(XLSX | 11 KB)
S5. Axolotl gene expression and cell specialization-Raw sample data.xlsx(XLSX | 13 KB)
S6. Axolotl gene expression and cell specialization-Sample results.xlsx(XLSX | 13 KB)
S7. Axolotl gene expression and cell specialization- Adapting activities.docx(DOCX | 35 KB)
- License terms

Comments
Comments
There are no comments on this resource.