A CRISPR/Cas Guide RNA Design In Silico Activity
Editor: Megan K. Barker
Published online:
Abstract
CRISPR biotechnologies inspired by the Clustered Regularly Interspaced Short Palindromic Repeat RNA-guided nuclease adaptive bacterial immune system have revolutionized biology research and become ubiquitous tools for hypothesis and discovery-driven research. Though properly a collection of technologies, today "CRISPR" is synonymous with CRISPR/Cas9 genome targeting. CRISPR is an important interdisciplinary tool and a modern topic to include in the undergraduate biology curriculum. To achieve gene targeting, Cas9 forms a complex with a guideRNA (gRNA). The complex scans double-stranded DNA for a NGG Protospacer Adjacent Motif (PAM) and interrogates complementarity of the adjacent DNA with the 20-nt spacer of the gRNA. If there is a perfect match, Cas9 cleaves the DNA. Resulting repair can yield genome variants: cells repair the damage erroneously to cause indels, and edits can be introduced by leveraging homology-directed repair. In this in silico lab activity, students synthesize and apply their knowledge of gene expression and CRISPR/Cas9 gene-targeting to design an optimized CRISPR/Cas9 gRNA. Students complete a pre-lab quiz then select one of three authentic research scenarios for their lab. The activity guides students through the process of navigating multiple bioinformatics tools, including a genome browser, DNA sequence annotation software, and a browser-based gRNA prediction tool, to complete a guided note sheet and select a suitable gRNA for their chosen scenario. Students report their design and justify their selection in an experiment summary lab report. This interdisciplinary lesson is developed for a special topics CRISPR course and is suitable for students in biochemistry/molecular biology; bioinformatics; cell, plant, animal and developmental biology; microbiology, and especially genetics.
Primary image: Schematic overview of the CRISPR/Cas gRNA design in silico lab activity. Image made in ©BioRender - biorender.com.
Citation
Samsa LA, Anderson L, Groth A, Goller C. 2020. A CRISPR/Cas guide RNA design in silico activity. CourseSource. https://doi.org/10.24918/cs.2020.46
Society Learning Goals
Bioinformatics
- Computation in the life sciences
- What is the role of computation in hypothesis-driven discovery processes within the life sciences?
- DNA - Information Storage [GENOMICS]
- Where are data about the genome found (e.g., nucleotide sequence, epigenomics) and how are they stored and accessed?
- How can bioinformatics tools be employed to analyze genetic information?
- Computational Skills
- How do biologists employ software development as part of the scientific discovery process?
- What higher-level computational skills can be used in bioinformatics research?
Cell Biology
- Methods & Tools of Cell Biology
- How do the methods and tools of cell biology enable and limit our understanding of the cell?
Genetics
- Genetic variation
- How do different types of mutations affect genes and the corresponding mRNAs and proteins?
- Genetics of model organisms
- How do the results of molecular genetic studies in model organisms help us understand aspects of human genetics and genetic diseases?
Lesson Learning Goals
By the end of this lesson, students will...
- understand how to use existing bioinformatic tools to design molecular tools for CRISPR/Cas9 reverse genetics experiments.
- gain a deeper knowledge or mastery of the following learning goals.
From the Genetics Core Competencies:
- Students should be able to tap into the interdisciplinary nature of science.
- Students should be able to communicate experimental results effectively, including writing research papers and giving presentations.
Lesson Learning Objectives
After the lesson, students should be able to:
- Explain why each of the following are required for CRISPR/Cas9 genome engineering: (1) gene and genome sequences, (2) gRNA targeting the genome, (3) source of Cas9, and (4) endogenous DNA repair machinery.
- Devise a strategy to approach a genome-editing goal.
- Use genome databases to identify the genome sequence of a target gene.
- Read and interpret the graphical summary of a gene in a genome browser.
- Explain how the CRISPOR algorithm identifies optimal gRNA target sites in a sequence by predicting on- and off-target effects.
- Justify selection of a CRISPR/Cas9 target sequence based on gene structure and gRNA characteristics.
- Diagram gene structure, target region and gRNA target site.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
CRISPR/Cas is a revolutionary and now essential tool for any biotechnological application that involves genome targeting. Yet, the NSF Workshop for Undergraduate Faculty notes that, "Despite its [CRISPR's] newfound mainstream presence in research, it has not yet become a standard offering in undergraduate biology courses." (1). Instructor-friendly resources have demystified this new technology and provided much-needed materials for integration of the tool into lecture materials (2). And, multi-session lab activities are available in published literature for CRISPR experiments in many experimental and model systems including cell-free transcription-translation (3), C. elegans (4), E. coli (5-7) Saccharomyces cerevisiae (8-11), Arabidopsis thaliana (12), Drosophila (13), and Danio rerio (14). A typical CRISPR experiment workflow starts with a design phase in which the gene is analyzed and a target selected that aligns with the experimental goal (1,4). Three steps follow, in which appropriate tools for the model system are synthesized, the experiment is performed, and on-target activity is validated (1). Of these steps, the first phase (design) is arguably the most important because failing to design an appropriate gRNA undermines the entire experiment. This design phase is so important that it has spawned a whole new field of research focused on developing bioinformatics tools to predict optimal target sites (15-17). The gRNA prediction tools are continuously improving; CRISPOR and GuideScan are considered the most sensitive and are preferred by many researchers (16-18).
Although most of the currently available lab activities include a design phase with variable levels of student choice in selecting the gRNA, most utilize other gRNA design tools and the design phase is a de-emphasized aspect of the lab activity. This is understandable considering the complications and risks of lab activity failure associated with using untested guides. With the growing need for lessons that can be delivered for online-learning and students' intense interest in CRISPR technologies, we created an in silico lab activity that isolates and explores the design phase of a CRISPR experiment workflow.
CRISPOR was selected for this activity in large part because the same factors that lend it utility to researchers also provide good teaching points. It has the largest selection of reference genomes (558 genomes) of any tool we have encountered, and these reference genomes include multiple builds and strains for some species, opening up questions about what a reference genome is, what strains are, and how genomes are named. CRISPOR has a large selection of protospacer adjacent motifs (PAMs) that correspond to Cas9 variants and other Cas effectors such as Cas12a and Cpf1. This invites students to explore why there are so may variants. Further, for each gRNA predicted by GuideScan, CRISPOR outputs a convenient summary of the commonly used gRNA prediction scores. In this regard, it is score-agnostic, allowing the researcher/user to account for the peculiarities of their specific experiment when selecting appropriate guides. CRISPOR also provides outputs that aid researchers in the second step of a CRISPR experiment - tool generation. In our activity, students learn about gRNAs by exploring these scores and putting themselves in the researcher's shoes to select the "best" gRNA. When students encounter these many options, it piques their curiosity, creating an experience where they learn by exploring or yielding a potential teaching moment. Additionally, although the user manual is comprehensive, it is written with novice-friendly language, which supports the student to investigate and answer their own questions. In this activity, students select one of three authentic research scenarios for their lab and navigate multiple bioinformatics tools, including a genome browser (NCBI), DNA sequence annotation software (SnapGene/SnapGeneViewer), and a browser-based gRNA prediction tool (CRIPSOR), to complete a guided note sheet and select a suitable gRNA for their chosen scenario. Students report their design and justify their selection in an experiment summary lab report where they practice research communication skills. This activity design was based on learning outcomes (backward design), implemented, and assessed, and is adaptable to any CRISPR design phase.
Notably, this in silico lab activity is similar to the dry bioinformatics module of a previously reported lesson (11) but is distinct in three ways. 1) Students are provided web tool-specific instructions where they use the authentic tools available to researchers. 2) Students are not given the option to manually design gRNAs. 3) The lab activity emphasizes student decision making and interpretation of on- and off-target prediction metrics. And 4) an experiment summary lab report directs student effort away from organizing information and towards interpreting and communicating their findings.
Using CRISPR/Cas bioinformatic design tools is a directly transferable skill in biotechnology and integrates an understanding of concepts across many areas of the life sciences. In particular, CRISPR/Cas gRNA design requires students to master and apply concepts in gene structure, information flow, genetics, and model systems. Proper CRISPR/Cas tool design requires students to consider where the gene should be targeted, the intended consequence of the targeting, what mechanisms are available to introduce foreign agents into the model system, and other considerations that focus attention on the details of how biology works.
Additionally, this activity supports student learning of bioinformatics skills. Sequence databases have become powerful tools in modern biological sciences research; there is widespread agreement over the importance of integrating bioinformatics competencies into the undergraduate biology major (19). These repositories may be curated (managed and maintained by bioinformaticians) or available as simple repositories (not curated by professionals) and are used as a central, publicly available access point to sequence data. Sequence databases can be used for simple tasks that do not require programming skills (such as obtaining a gene sequence) or more complex tasks that may require programming skills (such as genome-wide data-mining).
This lesson uses the National Center for Biotechnology Information (NCBI) Gene database (GenBank®), a portal that aggregates gene-specific information into a central page (20). Students access the database to explore the gene structure in the genome browser section and locate the chromosome region that contains their gene of interest. Then, students import the sequence into an easy-to-use sequence management, annotation and cloning tool, SnapGene (paid version) or SnapGene Viewer (free version) (21). In doing so, they notice that the chromosome region is pre-annotated with relevant information from the NCBI gene database (https://www.ncbi.nlm.nih.gov/gene/) and appreciate the value of central sequence repository connections. Then, students use CRISPOR - a comprehensive, browser-based tool (http://crispor.tefor.net) that enables researchers to perform a complex, genome-wide comparison task without additional programming skills (16). Students define the DNA sequence, reference genome, and Cas protein (for the PAM sequence). The CRISPOR algorithm then identifies all the potential gRNA sequences found within the user-defined DNA sequence, compares those sequences to the reference genome, and reports only the unique sequences. Further, it applies multiple, previously published algorithms to report multiple metrics of predicted on- and off-target effects for the gRNA.
Combining authentic research scenarios with the tools and workflow that researchers are currently using, students develop discipline-specific research skills that are highly transferable to any cell and molecular research laboratory.
Intended Audience
The intended audience is upper-level undergraduate and graduate students from diverse fields of study in or related to genetics; cell biology; cell, plant, animal and developmental biology; biochemistry/molecular biology; microbiology; biotechnology; and/or biomolecular engineering. This lesson is also designed to provide students who plan to apply CRISPR technologies in their field of study direct, practical experience in designing a critical component of the engineered CRISPR/Cas system, the gRNA.
Required Learning Time
This lesson is designed to be taught in one ~90-120 minute class period. This activity requires ~45 minutes for students to read the lab and complete a pre-lab quiz before class, 90-120 minutes of time for the in silico lab, and ~2-4 hours of out of class time for students to write the experiment summary lab report and respond to open-ended assessment questions. The 90-120 minute allocation includes time for students to explore and familiarize themselves with CRISPOR and SnapGene. Allow an additional 30 minutes each for the in silico activity if students have not used a genome browser in previous courses or lessons or if class members are known to have extra challenges learning new software.
Prerequisite Student Knowledge
Students should have a very good understanding of the central dogma of molecular biology, gene structure, gene expression, and general molecular biology/biochemistry. It is preferred for students to have previous exposure to genome browsers and SnapGene; students who do not have prior exposure may require extra support during the lab activity and additional time for completion. Tutorials and user manuals are readily available through the developers' websites for the software and browser applications (e.g., SnapGene). Additionally, students may find it helpful to have additional knowledge of CRISPR/Cas9 from lectures earlier in the course sequence. However, this level of background knowledge of CRISPR/Cas9 is reviewed in the Background section of the student-facing lab materials and includes concepts in how CRISPR/Cas9 works as a biotechnology, comparative properties of Cas9 and dCas9 (catalytically dead Cas9), and how bioinformatics algorithms are used to predict target sites and their properties.
Prerequisite Teacher Knowledge
In addition to the prerequisite student knowledge, instructors and teaching assistants should have sufficient background knowledge to 1) assist students in selecting an appropriate region of a gene to achieve gene knockout, repression, and editing goals outlined in the lab; 2) explain the relationship between PAM sequence and Cas effector variant to help students select CRISPOR inputs; 3) guide students through the decision-making process of selecting and justifying a gRNA target sequence; and 4) assist students in navigating software and in-browser apps used in this lesson. We recommend that teachers read and understand the CRISPOR user manual http://crispor.tefor.net/manual/manual.pdf pages 1-8 to learn how to use CRISPOR and interpret outputs. For the lecture portion of the lesson, the instructor should have the knowledge and skills to explain CRISPR-mediated approaches to reverse engineering (see ref. (2)).
SCIENTIFIC TEACHING THEMES
Active Learning
Each phase of this lesson has active learning components to support student engagement throughout the in- and out-of-class activities.
The pre-lab quiz prompts students to interact with the lab background materials, recall concepts from previous courses, and plan their experiment summary lab report. The in silico lab activity leverages authentic, impactful research scenarios, problem-based learning and student choice to promote active engagement. Students select one of three research scenarios that provide a rough outline of how a researcher is planning to use CRISPR/Cas9 in a model system to study a real-world human disease. Students get to put themselves in the position of the scientist studying these diseases using cutting edge-CRISPR techniques by designing the gRNA target sequence. The experiment summary lab report is a structured, data-centric writing activity where students revisit their lab activity results to summarize and report their findings. In the report, students explain why they selected their particular gRNA and practice making discipline-appropriate figures. The writing process requires students to revisit their work and, in doing so, reinforces their learning. Following the activity and report, free-response questions prompt students to apply their knowledge to a new scenario and to reflect their experience with the lab activity.
Assessment
The instructors measure learning primarily through an experiment summary lab report. Leading up to that summative assessment are several formative assessments including a pre-lab quiz and short answer prompts throughout the lab activity. After the experiment summary, student learning is further evaluated with low-stakes, post-activity free response questions. Each of these assessments is described further below.
Pre-lab quiz:
Students read the background information provided with the lab activity, read the lab protocol, and are prompted to answer open-note comprehension questions (Supporting File S1. In silico CRISPR gRNA design – Pre-lab Quiz Questions and Key, Supporting File S2. In silico CRISPR gRNA design – Lab Activity). With these questions, students apply concepts learned in previous, foundational courses in molecular biology and similar subjects to this new context, demonstrate understanding of the lab protocol, and plan for their experiment summary report. The pre-lab is completed independently by each student. The instructor grades and provides feedback on the pre-lab quiz by addressing frequently missed questions at the beginning of the in silico lab.
Lab activity:
Students are prompted to fill in provided notes that serve two purposes (Supporting File S2. In silico CRISPR gRNA design – Lab Activity). The notes help students collect the information they will need to report in the experiment summary, and they provide formative feedback on student comprehension of the subject matter. The instructor circulates throughout the classroom as students are working on the lab activity, thus allowing ample opportunity to observe and correct misconceptions and for students to ask questions in an informal setting.
Experiment summary lab report:
This modified lab report is modeled after a mini-manuscript (Supporting File S3. In silico CRISPR gRNA design – Experiment Summary Lab Report Instructions and Rubric, Supporting File S4. In silico CRISPR gRNA design – Experiment Summary Lab Report Instructions Writing Guidelines). Students write a brief summary of their methods and generate well-labeled, captioned, and multi-part figures that communicate their approach and findings to the standards of the field. Students are provided with an experiment summary grading rubric and outline tailored for this specific lab activity and a writing guidelines document. The outline includes guiding questions and suggested figure layout. As students write the document, they use the rubric, outline and guiding questions to self-evaluate their progress and understanding. The instructor evaluates the final piece of writing using a rubric heavily weighted towards clear, concise, and accurate communication of data figures, results, and discussion.
Free response questions:
After the lab activity, students compose free responses to two questions (Supporting File S5. In silico CRISPR gRNA design – Open-ended Assessment Questions with Key and Sample Answers). One prompts students to summarize their knowledge. The other prompts students to reflect on their experience, describe their experience and provide feedback on the challenges they experienced.
Inclusive Teaching
This lesson involves inclusive teaching practices in several ways. It incorporates realistic human disease research scenarios where all students can visualize themselves as the scientist doing meaningful work. The format incorporates universal design for learning principles that improve access and learning opportunities for all students. It uses (or can be rapidly adapted to use) only materials and software that are available to students at no cost, which lowers access barriers for resource-limited students. Electronic materials can be read with a screen reader and the display modified. The structured lab notes guide students through expert thinking processes and resource management. The experiment summary lab report provides a suggested structured outline to guide information processing and offers opportunity for creativity in data presentation. The post-activity free response questions and conclusion section of the experiment summary lab report gives each student the opportunity for individual reflection, which supports metacognition and self-monitoring of learning. Additionally, the lesson is readily adaptable to asynchronous and online teaching using supporting materials resources provided.
LESSON PLAN
Summary
There are four components to this lesson - (1) a pre-lab quiz, (2) the in silico lab activity, (3) an experiment summary lab report assessment and (4) post-lab free response questions (Figure 1, Table 1, Supporting Files S1-5). The lesson is completed within one ~90-120 minute class period and requires student work outside of class before and after the lab activity (~45 minutes and ~2-4 hours, respectively). The 90-120 minute allocation includes time for students explore and familiarize themselves with CRISPOR and SnapGene, Allow an additional 30 minutes each for the in silico activity if students have not used a genome browser in previous courses or lessons or if members of the class are known to have extra challenges learning new software.
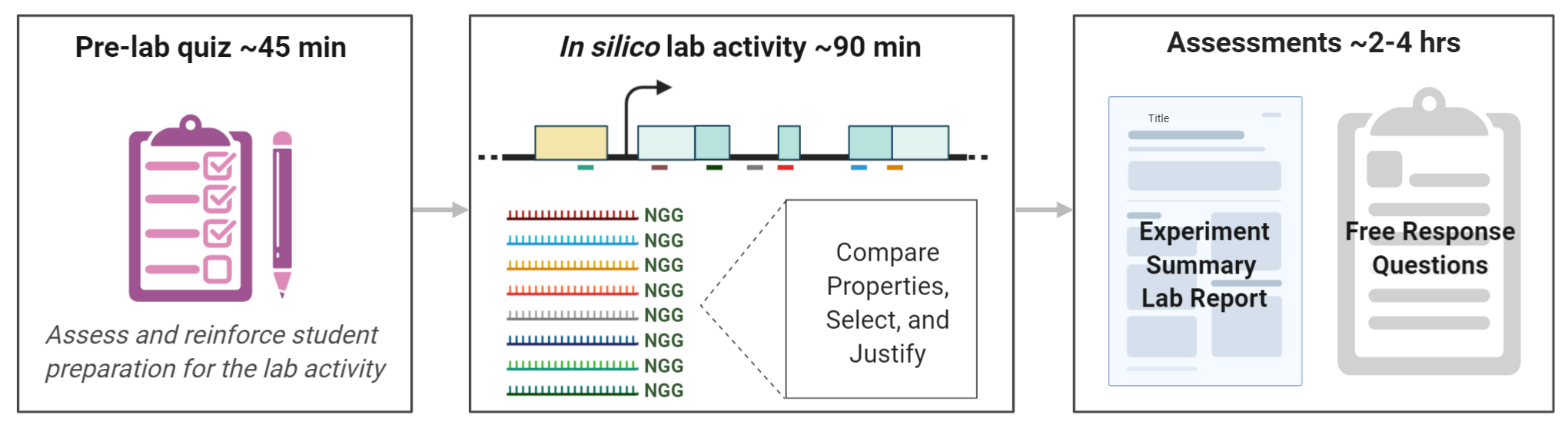
Figure 1: Lesson summary schematic. Students complete a pre-lab quiz before class. In class, they complete the in silico lab activity where they identify an appropriate region of interest for their target gene and all the potential CRISPR/Cas9 gRNA spacers (shown as multi-colored protospacers and Cas9 PAM NGG) for that region. They compare properties of those potential gRNA spacers, select a “best” one and justify their selection based on these metrics. Students document their activity in a summative experiment summary lab report, then reflect on their experience and apply their knowledge to a new problem with free response questions. Image made in ©BioRender - biorender.com
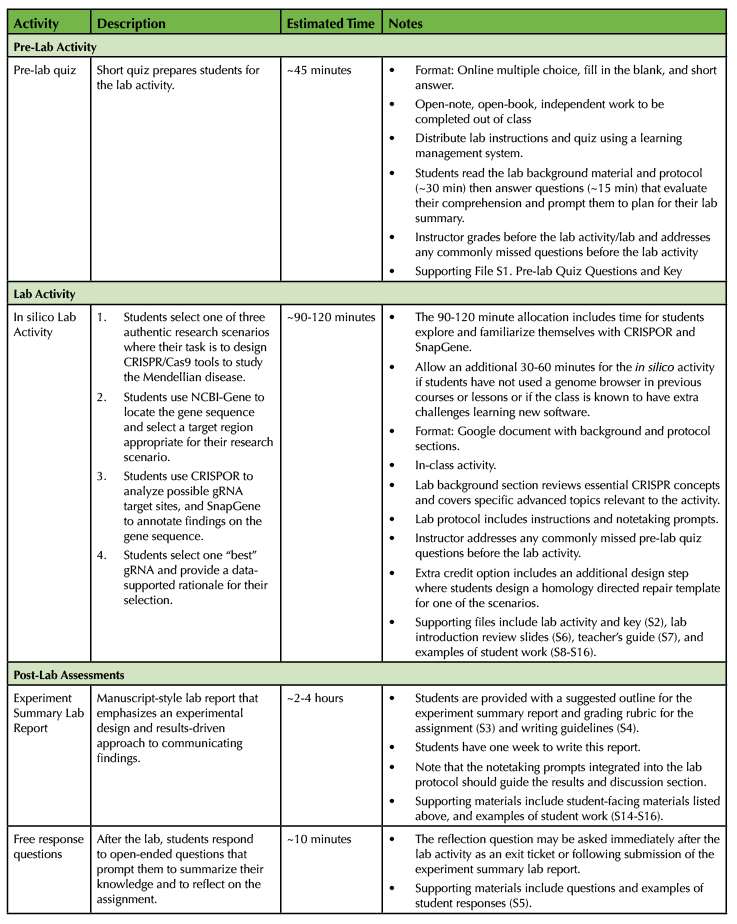
Table 1. Lesson summary and timeline.
Preparation
To teach this lesson, first confirm that the classroom has reliable internet access and that all computers have SnapGene (or SnapGene Viewer) and a suitable web browser (Chrome, Firefox, or Safari) installed. Post the pre-lab quiz questions (Supporting File S1. In silico CRISPR gRNA design – Pre-lab quiz Questions and Key) to your learning management system (LMS) and distribute an electronic copy of the in silico lab activity document to students (Supporting File S2. In silico CRISPR gRNA design – Lab activity). Distribute electronic copies of the Experiment Summary Report Instructions and Rubric (Supporting File S3) and the Experiment Summary Lab Report Writing Guidelines (Supporting File S4). Grade student responses to the pre-lab quiz before class and be prepared to address any commonly missed questions.
Lab activity
Start the in silico lab class period by reviewing commonly missed questions from the pre-lab. Then, use the provided slides to review the central dogma of biology with emphasis on how nucleotide sequence dictates protein sequence and to introduce the concept of CRISPR/Cas9 reverse genetics (Supporting File S6. In silico CRISPR gRNA design – Review slides). The presentation helps get students thinking about CRISPR/Cas9 as a genetic engineering tool, which is important for contextualizing the lab activity, and should take ~10 minutes.
For the remainder of the class period, students work on the in silico lab activity by following the protocol and filling in the associated guided notes.
For the lab activity, students first select from one of three research scenarios where they will design a CRISPR/Cas9 gRNA to use as a reverse genetics tool to study a Mendelian disease (Figure 2, top). Students select from Gaucher Disease (knockout out Gba in mouse cells), Polycystic Kidney Disease (repress PKD1 in human kidney organoids), and Progeria (allelic modification of LMNA in human fibroblasts) (Figure 2, top). After selecting a scenario, students find the reference sequence of the gene using NCBI's Gene database and genome browser. They import the reference sequence into SnapGene using genome where it can be more easily visualized and annotated. Applying their understanding of genetic engineering approaches and the experimental goal of their selected scenario, students identify a suitable target region of the gene (Figure 2, middle). They enter the sequence of their selected region into the CRISPOR (http://crispor.tefor.net/) gRNA design tool where they select design parameters including the genome and Cas9 variant and interpret the output (Figure 2, bottom). Students select the single "best" gRNA spacer sequence from the CRISPOR output and justify their selection based on the predicted gRNA structure, on-target scores (6 scores), and predicted off-target sites (Figure 2, bottom). CRISPOR outputs are simple to interpret - scores predict specificity, efficiency, or outcome and are scaled 0-100 (high scores are favorable). Off-targets counts are provided for 0, 1, 2, 3, and 4 mismatches. Based on their experiment's needs, students weigh the relative importance of each of these factors to select a guide sequence. For additional guidance, see Supporting File S7. In silico CRISPR gRNA design – Teacher Guide). Students annotate their SnapGene sequence with their target site, potential gRNAs and selected gRNA. Guiding questions and instructions in the lab protocol prompt students to collect information and images to use in their experiment summary report. Sample student notes (filled in protocols) for each scenario are provided in Supporting Files S8-S10 and annotated SnapGene files in Supporting Files S11-13.
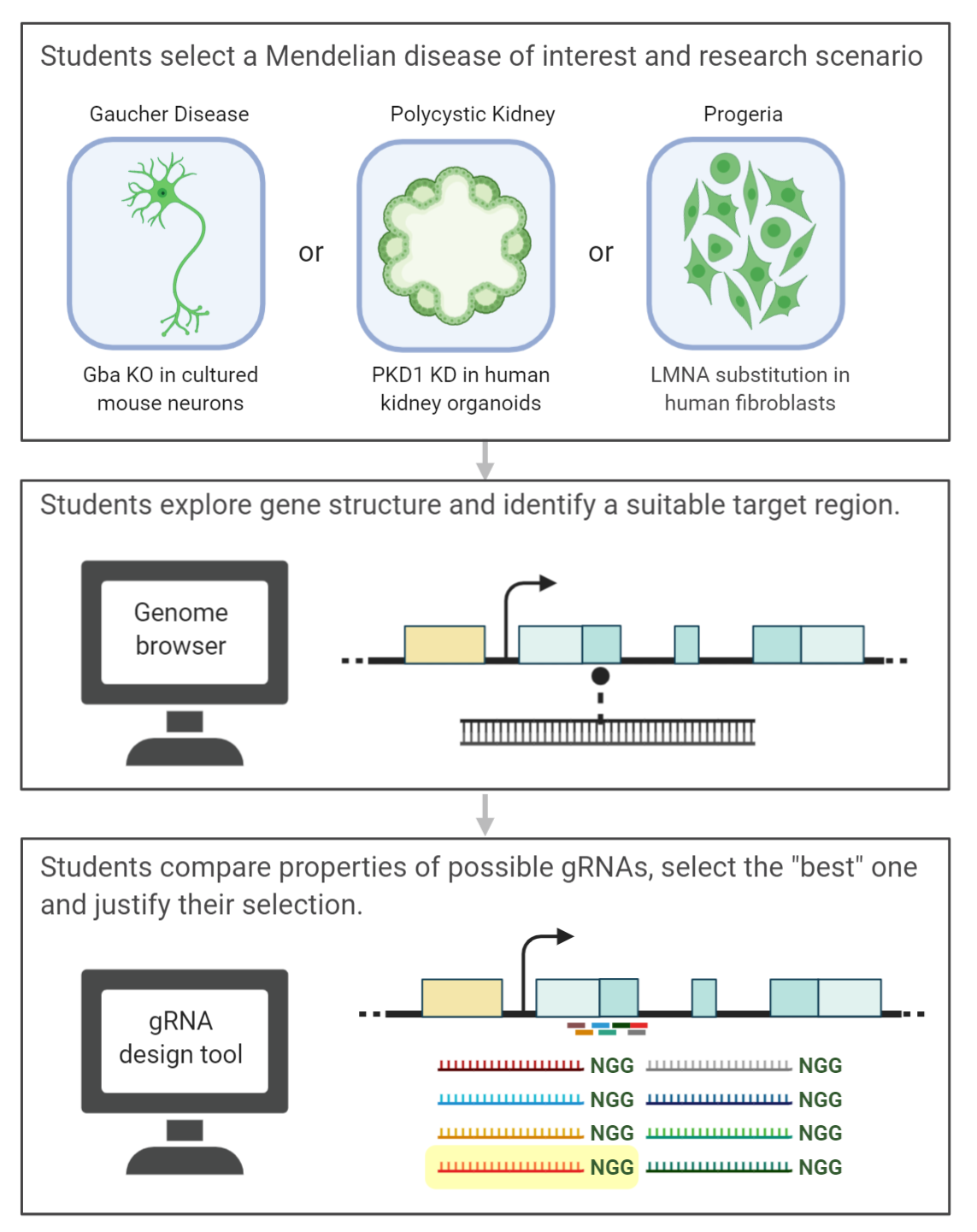
Figure 2: Schematic overview of the in silico lab activity. Image made in ©BioRender - biorender.com
Tips for lab activity instruction
Instruct students to work individually but to feel free to collaborate with one another to complete the protocol portion of the lab activity (Supporting file S2. In silico CRISPR gRNA design – Lab activity). While students are working, circulate throughout the room, answer questions, and guide students. If students encounter common sticking points, take advantage of peer teaching techniques and direct students who have the answer to help out those around them. Common sticking points and solutions are summarized in Table 2, below. Students who have a minimal level of familiarity with GenBank, reviewing sequence data, or Excel may need additional support to complete this activity.
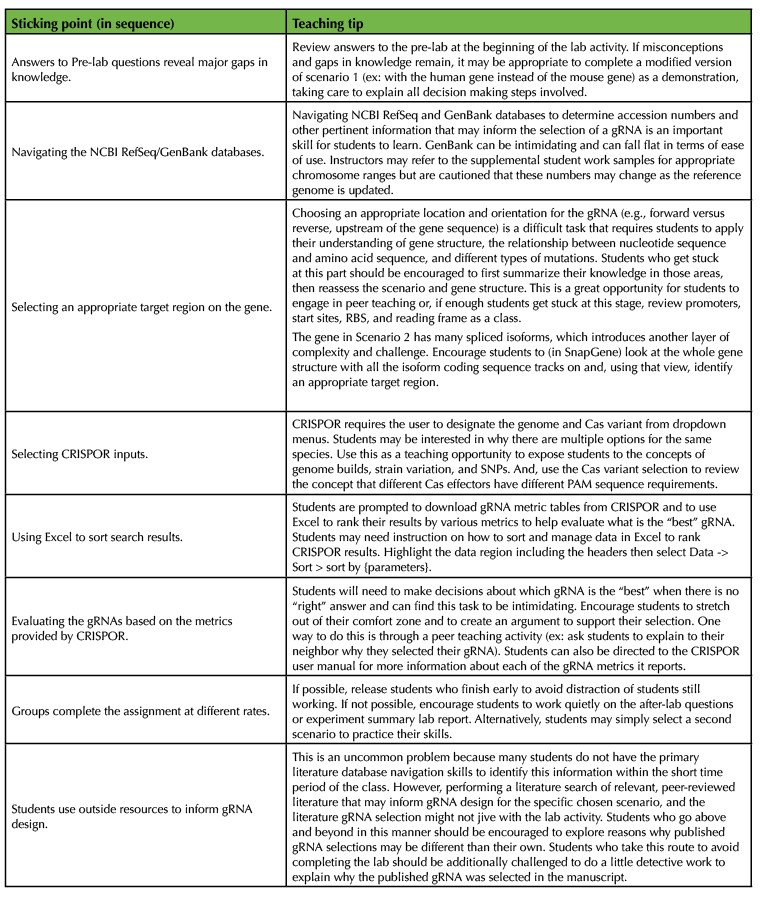
Table 2. Tips for lab activity instruction.
Experiment Summary Lab Report
Assign and collect
Students have one week to complete the experiment summary lab report. If students follow the writing guidelines, it should take ~2-4 hours to write the report. The experiment summary lab report assignment is modeled after primary molecular biology manuscripts.
Tips for experiment summary evaluation
The Experiment Summary Report Instructions and Rubric (Supporting File S3) and the Experiment Summary Lab Report Writing Guidelines (Supporting File S4) should be referred to while evaluating experiment summaries. The provided rubric (Supporting File S3) includes notes on items that should be included to receive a maximum score on the individual components (sections) of the written assignment. Experiment summaries are designed to be graded as individual assignments rather than graded on a curve or to the highest score.
Scientific writing is a challenge for many students. Direct students to the experiment summary report writing guidelines (Supporting File S4. In silico CRISPR gRNA design – Experiment Summary Lab Report Writing Guidelines). Additionally, the provided student work samples (Supporting Files S14-16. In silico CRISPR gRNA design – Lab Activity Case Option [1-3] experiment summary lab report) can be modified to use as an example lab report with the caveat that students will copy the structure. If students struggle to create the data figures, consider recording a tutorial video using data from a modified version of scenario 1 (ex: with the human gene instead of the mouse gene).
Free Response Questions
Assign and collect
Post the free response questions to your learning management system (LMS) as a quiz (Supporting File S5. In silico CRISPR gRNA design – Open-ended Assessment Questions with Key and Sample Answers). Question 1 asks, "In CRISPR-Cas9 gene editing of a eukaryotic gene, what factors do you consider to justify the selection of a specific gRNA and DNA target sequence?" Free response essay questions yield rich data on an upper level Bloom's task of summarizing knowledge and provide a metric for the learning objective "Devise a strategy to approach a genome-editing goal." Question 2 asks, "What was the most challenging part(s) of Lab 1?" which prompts students to reflect on their experience; note: students also reflect on the lab in the conclusion section of the experiment summary. Give students 10-15 minutes to complete the quiz during a class period after the experiment summary is due.
TEACHING DISCUSSION
Effectiveness at achieving the stated learning goals and objectives
Analysis of student work for data as research was approved by NC State University IRB # 20522.
This lesson was developed for an upper-level, Special Topics in Biotechnology laboratory course focused exclusively on CRISPR Technologies. Student data from two sections of the course, one in Fall 2019 and the other Spring 2020 were combined for analysis (N=29). Both sections were ~50% upper-level undergraduate and graduate students, and this lesson was taught in face-to-face format. Learning objectives were assessed as described in Table 3 (below) and student performance on graded assessments is summarized in Figure 3 with additional details for the Experiment Summary Lab Report provided in Supporting File S17.
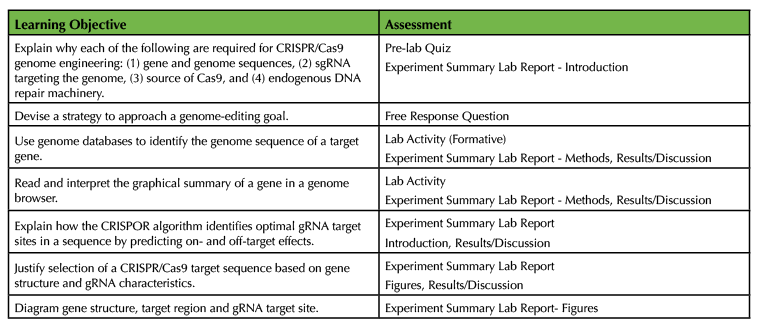
Table 3. Learning Objective Assessment.
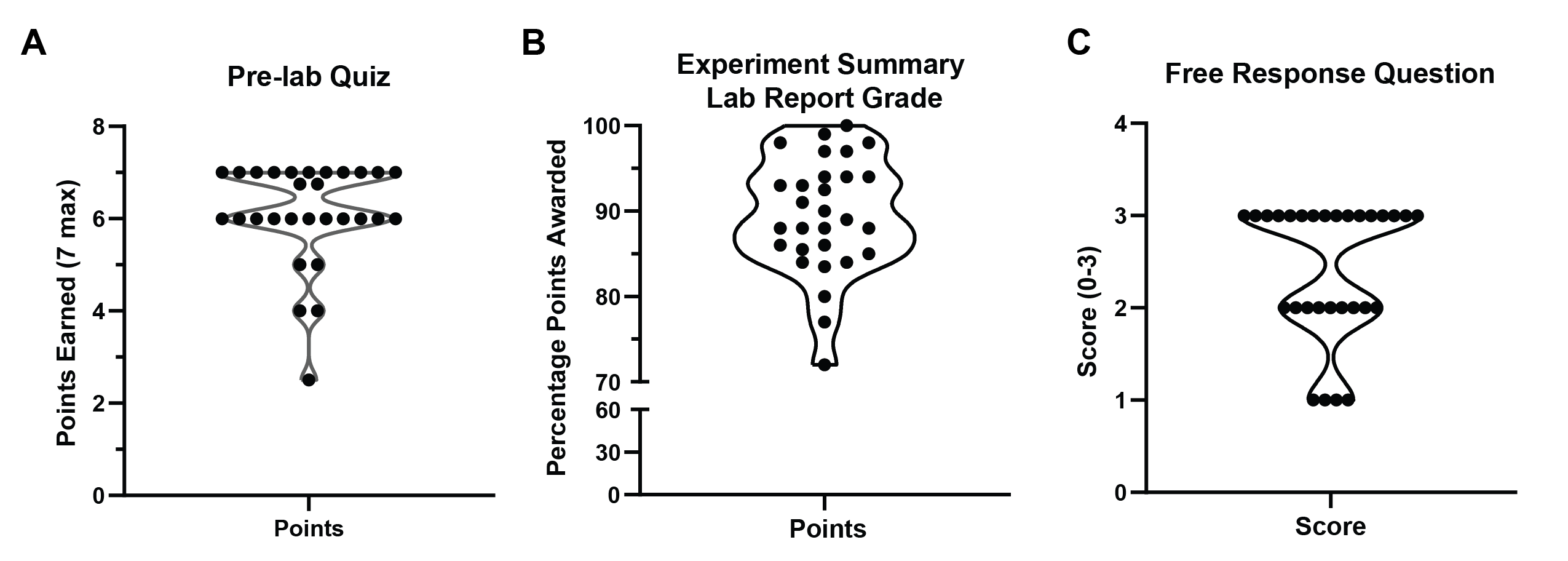
Figure 3: Student performance. Distribution of performance on (A) Pre-lab Quiz, (B) Experiment Summary Lab Report, and (C) a post-activity free response question. Student data was collected and pooled from two sections of BIT 495/595-005 CRISPR Technology enrolled in the Fall 2019 (N=15) and Spring 2020 (N=14). Data plotted with GraphPad Prism 8.
All students completed the pre-lab quiz, and most missed zero or one question (Figure 3A). For the lab activity, 12/29 students selected Scenario 1 (Gaucher's disease), 6/29 students selected Scenario 2 (Polycystic Kidney Disease), and 11/29 students selected scenario 3 (Progeria). Of those who selected Scenario 3, 5/11 completed a bonus assignment where they designed an HDR template to accompany their selected gRNA. Students scored between 72.0-100.0% (excluding bonus points) (mean = 89.4, median = 89.0%) on the experiment summary points (Figure 3B). The most common reasons for point deductions were failure to include content in a section (ex: leaving out the Conclusions section), omitting details in the methods or figures, and failing to address each piece of data in the results section. The free response question asking students to summarize their understanding was administered as a very-low stakes exit ticket and responses were scored on a scale of 0-3 (mean = 2.4, median = 3) (Figure 3C). We suggest that, taken in the context that all students demonstrated material mastery in the introduction section of the summative writing assignment, the presence of some "developing" scores (3/29) in the distribution of the free response question is likely more a reflection of how the assignment was administered rather than of student learning. Indeed, most students (26/29) produced "good" or "excellent" responses. Together, these assessments indicate that the lesson is effective at achieving the learning objectives.
Student reactions to the lesson
Based on the student responses to the reflection prompt and experiment summary, this lesson provides an engaging educational activity for students new and experienced with CRISPR technologies. New students were exposed to the basic programs and requirements for gRNA design. More experienced students expanded their knowledge of design and challenged themselves through the extra credit opportunity. The lesson also showcased the difficulty of choosing an optimal gRNA location. Students were surprised to learn there is not an 'ideal' gRNA location based on sequence alone. After the lesson was complete, there was a desire to test the gRNAs generated to verify if the gRNA chosen would be functional or ideal.
Students generally liked and enjoyed the lab:
"I enjoyed completing the lab summary one. It took me a lot longer than I expected but that was because I decided to redo the whole thing using a different option. I thought it was a great first lab because we went through how to conduct a real experiment using the actual software that is used to create the best guide RNA."
"The experiment performed in class was very helpful, since the design of gRNAs may be a little confusing sometimes. I really enjoyed how we enter deeply into each predicted result from CRISPOR, considering all parameters before designing a gRNA (target region location, specificity, efficiency, etc)."
Though most students (>50%) reported that learning to use different software was the most challenging part of the lab, all students were able to complete the lab and some reported that after the lesson they were more familiar with the programs needed to do basic gRNA design (NCBI Genbank, CRISPOR, and SnapGene). We interpret this student feedback optimistically - CRISPR and gRNA design concepts were not the most difficult part - and in our experience the software used in this lab are intuitive and user-friendly. This was reported from students who were new to gRNA design and those with previous experience, the latter of which expressed appreciation for this laboratory helping develop a greater depth of understanding for using these tools:
"The most challenging aspect of lab 1 was the novelty of the software. I had used SnapGene briefly about a year ago, but some refreshing on the software was needed. Also, since CRISPOR itself was new and the categorization of the scores was new, it took some time to understand what the numbers meant and how to utilize that information to pick the best gRNA, especially since a clear winner in each category was not evident."
Importantly, students valued the structure of the assignment and found the lab activity to be relevant to their current and future research:
"[...] Thoughts: (1) I really, really appreciate that the labs are provided with questions to answer that effectively fill in our lab summaries for us. That is awesome. (2) I provided my lab with the template for a lab summary because we have had some in-house issues with recording all of the data as it is collected... so thank you for that as well!"
"Since gene-editing is something I am interested in improving for use in model system genetic research of cancers, gRNA design is imperative to learn with excellent proficiency. This lab taught me that many options for design will be available and that further testing will be needed to pick a "best" candidate for each experiment. I am more familiar with the concepts of gRNA and where they bind after this lab. I am also more confident using SnapGene, CRISPOR, and RefWorks software which will be beneficial in future career fields. [...] I mostly learned that of the gRNA possibilities, one clear winner may not prevail and that it will take much more critical thinking to select the most correct gRNA for your desired project. Looking back at the gRNAs after lab, I realized that there may have been a better gRNA to pick or one that had a better argument for its use than the one I originally selected."
Notably, this lab does not include experimental validation of student gRNA designs.
Anecdotally and based on our conversations with students, some students find it uncomfortable to learn that they will not experimentally validate their gRNA design - they may never know if it works! We encourage students to redirect this energy into questions like, "How would I go about testing if my design works?" and "How could I compare efficacy between several gRNA candidates?" A small number of our students have used this lab as a launching point for independent research projects.
Suggestion for improvement and adaptations to different courses or student populations
Though originally designed as a stand-alone, modular lab in an upper-level biotechnology course, this lesson is adaptable across the undergraduate biology curriculum where it can be used to introduce students to CRISPR with a biotechnology and human-health lens. Additionally, simply by removing the scenarios and adjusting the embedded questions in the procedure, the process can be adjusted to serve as the bioinformatics gRNA design phase for almost any CRISPR workflow. To complement this modification, we recommend adding a full gRNA assembly step towards the end of the lab activity - students could be provided with gRNA scaffold sequences for the selected Cas protein or could be promoted to locate this information from extant literature.
This lab could be readily adapted as the beginning of a course based undergraduate research experience. A natural extension of this activity is to allow students to test their gRNA using gRNA validation labs activities. These could be relatively simple in vitro activities using synthesized DNA and RNA and purified Cas proteins or more involved experiments using mouse and human cell lines. In practice, gRNA validation labs are resource-intensive and highly dependent on the model system, time, and available resources. As an alternative, one of the reviewers of this manuscript provided an excellent suggestion for an in silico validation activity--we would like to share this suggestion as a possible extension for advanced-level students: Students may identify original research papers in which these genes are edited using the CRISPR technology. Then, they can work backwards and use CRISPOR to identify the scores of those gRNAs that have been shown to work in these published papers. With this approach, students may compare known, functional gRNAs in these published papers to their selected gRNAs to learn how the in silico selection is effective. This may provide a validation component of this dry lab class and deepen students' understanding of the in silico design and selection of gRNAs. We envisioned the challenges that an instructor might face in adapting this lesson to their specific classroom and learning environment and summarized them in Table 4, below. We encourage instructors to engage in creative improvements and adaptations to use this lesson in their courses. For example, instructors can swap out the scenarios for new ones that better fit their course research objective. Or, this lesson can be adapted into a case study or problem-based-learning activity by adjusting the case scenario and relaxing the rigidity of the procedure instructions.
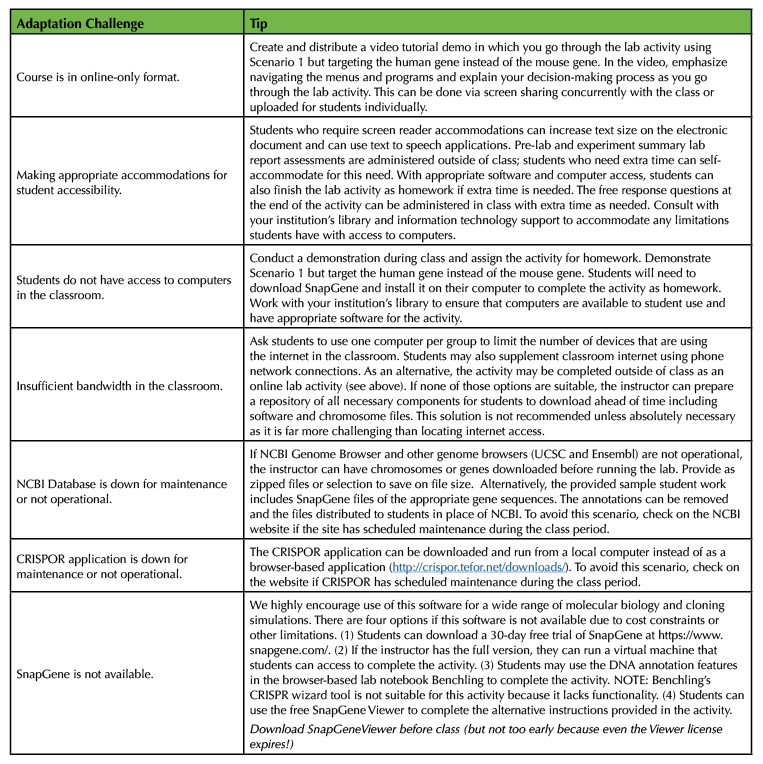
Table 4. Suggested solutions to adaptation challenges.
SUPPORTING MATERIALS
- S1. In silico CRISPR gRNA design – Pre-lab Quiz Questions and Key
- S2. In silico CRISPR gRNA design – Lab Activity
- S3. In silico CRISPR gRNA design – Experiment Summary Lab Report Instructions and Rubric
- S4. In silico CRISPR gRNA design – Experiment Summary Lab Report Writing Guidelines
- S5. In silico CRISPR gRNA design – Open-ended Assessment Questions with Key and Sample Answers
- S6. In silico CRISPR gRNA design – Review Slides
- S7. In silico CRISPR gRNA design – Teacher Guide
- S8. In silico CRISPR gRNA design – Sample work - Lab Activity Case Option 1 lab notes
- S9. In silico CRISPR gRNA design – Sample work - Lab Activity Case Option 2 lab notes
- S10. In silico CRISPR gRNA design – Sample work - Lab Activity Case Option 3 lab notes
- S11. In silico CRISPR gRNA design – Sample work - Lab Activity Case Option 1 SnapGene file
- S12. In silico CRISPR gRNA design – Sample work - Lab Activity Case Option 2 SnapGene file
- S13. In silico CRISPR gRNA design – Sample work - Lab Activity Case Option 3 SnapGene file
- S14. In silico CRISPR gRNA design – Sample work - Lab Activity Case Option 1 experiment summary lab report
- S15. In silico CRISPR gRNA design – Sample work - Lab Activity Case Option 2 experiment summary lab report
- S16. In silico CRISPR gRNA design – Sample work - Lab Activity Case Option 3 experiment summary lab report
- S17. In silico CRISPR gRNA design – Experiment Summary performance by assignment section
ACKNOWLEDGMENTS
We are grateful to the BIT program for support and discussion. We thank all the students in Biotechnology course BIT 495/595 CRISPR Technologies at NC State University who completed this lab activity and provided candid feedback on their experience. We thank Dr. Chase Beisel and Dr. Eric Lazear for sharing previous BIT 495/595 CRISPR Technologies course materials including documents that formed the foundation of the activity described in this document. The authors thank BIT-SURE student John Britt for his diligent work in piloting the activity and Stephen Serrano for critical reading of the manuscript. Use of deidentified student data for this study is approved by NC State IRB Protocol #20522. The authors declare that they have no conflicts of interest.
References
- Wolyniak MJ, Austin S, Bloodworth LF, Carter D, Harrison SH, Hoage T, Hollis-Brown L, Jefferson F, Krufka A, Safadi-Chamberlin F, Santisteban MS, Soneral P, VanWinkle B, Challa AK. 2019. Integrating CRISPR-Cas9 Technology into Undergraduate Courses: Perspectives from a National Science Foundation (NSF) Workshop for Undergraduate Faculty, June 2018. J Microbiol Biol Educ. 20(1). doi: 10.1128/jmbe.v20i1.1702.
- Thurtle-Schmidt D, Lo T. 2018. Molecular biology at the cutting edge: A review on CRISPR/CAS9 gene editing for undergraduates. Biochem Mol Biol Educ. 46(2):195-205. doi: 10.1002/bmb.21108.
- Collias D, Marshall R, Collins SP, Beisel CL, Noireaux V. 2019. An educational module to explore CRISPR technologies with a cell-free transcription-translation system. Synth Biol. 4(1):ysz005. doi: 10.1093/synbio/ysz005.
- Kee HL, Kushner JK, Deuchler CP, Becker AK, Clarke DG, Pieczynski JN. 2019. Using CRISPR-Cas9 to teach the fundamentals of molecular biology and experimental design. CourseSource. doi: 10.24918/cs.2019.21.
- Militello KT, Lazatin JC. 2017. Discovery of Escherichia coli CRISPR sequences in an undergraduate laboratory. Biochem Mol Biol Educ.;45(3):262-9. doi: 10.1002/bmb.21025.
- Pieczynski JN, Deets A, McDuffee A, Lynn Kee H. 2019. An undergraduate laboratory experience using CRISPR-cas9 technology to deactivate green fluorescent protein expression in Escherichia coli. Biochem Mol Biol Educ. 47(2):145-55. doi: 10.1002/bmb.21206.
- Ziegler H, Nellen W. 2020. CRISPR-Cas experiments for schools and the public. Methods. 172:86-94. doi: 10.1016/j.ymeth.2019.08.009.
- de Waal E, Tran T, Abbondanza D, Dey A, Peterson C. 2019. An undergraduate laboratory module that uses the CRISPR/Cas9 system to generate frameshift mutations in yeast. Biochem Mol Biol Educ. 47(5):573-80. doi: 10.1002/bmb.21280.
- Sehgal N, Sylves ME, Sahoo A, Chow J, Walker SE, Cullen PJ, et al. 2018. CRISPR Gene Editing in Yeast: An Experimental Protocol for an Upper-Division Undergraduate Laboratory Course. Biochem Mol Biol Educ. 46(6):592-601. doi: 10.1002/bmb.21175.
- Vyas VK, Bernstein DA. 2019. An Introduction to CRISPR-Mediated Genome Editing in Fungi. J Microbiol Biol Educ. 20(3). doi: 10.1128/jmbe.v20i3.1809.
- Ulbricht RJ. 2019. CRISPR/Cas9 in yeast: a multi-week laboratory exercise for undergraduate students. CourseSource. doi: 10.24918/cs.2019.19.
- Ruppel NJ, Estell LE, Jackson RI, Wolyniak MJ. 2019. An Undergraduate Research Project Utilizing CRISPR-Cas9 Gene Editing Technology to Study Gene Function in Arabidopsis thaliana. J Microbiol Biol Educ. 20(2). doi: 10.1128/jmbe.v20i2.1666.
- Adame V, Chapapas H, Cisneros M, Deaton C, Deichmann S, Gadek C, Lovato TL, Chechenova MB, Guerin P, Cripps RM. 2016. An undergraduate laboratory class using CRISPR/Cas9 technology to mutate drosophila genes. Biochem Mol Biol Educ. 44(3):263-75. doi: 10.1002/bmb.20950.
- Bhatt JM, Challa AK. 2018. First Year Course-Based Undergraduate Research Experience (CURE) Using the CRISPR/Cas9 Genome Engineering Technology in Zebrafish. J Microbiol Biol Educ. 19(1). doi: 10.1128/jmbe.v19i1.1245.
- Klein M, Eslami-Mossallam B, Arroyo DG, Depken M. 2018. Hybridization Kinetics Explains CRISPR-Cas Off-Targeting Rules. Cell Rep. 22(6):1413-23. doi: 10.1016/j.celrep.2018.01.045.
- Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud J, Schmeider-Maunoury SS, Shkumatava A, Teboul L, Kent J, Joly SP, Concordet JP. 2016. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17(1):148. doi: 10.1186/s13059-016-1012-2.
- Alkhnbashi OS, Meier T, Mitrofanov A, Backofen R, Voss B. 2020. CRISPR-Cas bioinformatics. Methods. 172:3-11. doi: 10.1016/j.ymeth.2019.07.013.
- Perez AR, Pritykin Y, Vidigal JA, Chhangawala S, Zamparo L, Leslie CS, Ventura A. 2017. GuideScan software for improved single and paired CRISPR guide RNA design. Nat Biotechnol. 35(4):347-9. doi: 10.1038/nbt.3804.
- Wilson Sayres MA, Hauser C, Sierk M, Robic S, Rosenwald AG, Smith TM, Triplett EW, Williams JJ, Dinsdale E, Morgan WR, Burnette JM, Donovan SS, Drew JC, Elgin SCR, Fowlks ER, Galindo-Gonzalez S, Goodman AL, Grandgenett NF, Goller CC, Jungck JR, Newman JD, Pearson W, Ryder EF, Tosado-Acevedo R, Tapprich W, Tobin TC, Toro-Martínez A, Welch LR, Wright R. 2018. Bioinformatics core competencies for undergraduate life sciences education. PLoS One. 13(6):e0196878. doi: 10.1371/journal.pone.0196878.
- Gene [Internet].; 2020 [updated 2004 - [cited 6/5/]; cited 6/5/2020].
- Insightful Science. SnapGene software.
Article Files
Login to access supporting documents
A CRISPR/Cas Guide RNA Design In Silico Activity(PDF | 635 KB)
S1 In silico CRISPR gRNA design - Pre-lab Quiz Questions and Key.docx(DOCX | 20 KB)
S2 In silico CRISPR gRNA design - Lab Activity.docx(DOCX | 356 KB)
S3 In silico CRISPR gRNA design - Experiment Summary Lab Report Instructions and Rubric.docx(DOCX | 23 KB)
S4 In silico CRISPR gRNA design - Experiment Summary Lab Report Writing Guidelines.docx(DOCX | 23 KB)
S5 In silico CRISPR gRNA design - Open-ended Assessment Questions with Key and Sample Answers.docx(DOCX | 22 KB)
S6 In silico CRISPR gRNA design - Review Slides.pptx(PPTX | 523 KB)
S7 In silico CRISPR gRNA design Teacher Guide.pptx(PPTX | 1006 KB)
S8 In silico CRISPR gRNA design - Sample work - Lab Activity Case Option 1 lab notes.docx(DOCX | 29 KB)
S9 In silico CRISPR gRNA design - Sample work - Lab Activity Case Option 2 lab notes.docx(DOCX | 29 KB)
S10 In silico CRISPR gRNA design - Sample work - Lab Activity Case Option 3 lab notes.docx(DOCX | 53 KB)
S11 In silico CRISPR gRNA design - Sample work - Lab Activity Case Option 1 SnapGene file.txt(TXT | 69 KB)
S12 In silico CRISPR gRNA design - Sample work - Lab Activity Case Option 2 SnapGene file.txt(TXT | 285 KB)
S13 In silico CRISPR gRNA design - Sample work - Lab Activity Case Option 3 SnapGene file.txt(TXT | 285 KB)
S14 In silico CRISPR gRNA design - Sample work - Lab Activity Case Option 1 experiment summary lab report.docx(DOCX | 138 KB)
S15 In silico CRISPR gRNA design - Sample work - Lab Activity Case Option 2 experiment summary lab report.docx(DOCX | 127 KB)
S16 In silico CRISPR gRNA design - Sample work - Lab Activity Case Option 3 experiment summary lab report.docx(DOCX | 53 KB)
S17 In silico CRISPR gRNA design - Experiment Summary performance by assignment section.docx(DOCX | 295 KB)
- License terms

Comments
Comments
There are no comments on this resource.