Understanding Host-Pathogen Interactions With the Use of Galleria mellonella
Editor: Rachel Horak
Published online:
Abstract
The Galleria mellonella; the larvae of the Greater Wax Moth, is a safe and effective means to study host-pathogen interactions in the undergraduate setting. The use of animal models in the teaching classroom provides an opportunity to discuss proper animal use and the benefits of alternative model systems in research. In this lesson, we developed a laboratory experience for students to learn about the use of G. mellonella as a model to study bacterial virulence. Students evaluate the virulence of a suite of microbes by injecting larvae and analyzing their survival over time. Students gain the opportunity to reinforce basic microbiology techniques such as aseptic technique and bacterial enumeration, while learning about new methods involving animal use in research. Students learn how to report their data using survival plots. More importantly, this lesson gives students a hands-on experience working with an in vivo system and complements discussions of innate immunity and bacterial virulence by providing a visual and quantitative approach to the study of virulence.
Primary image: Uninfected Greater wax moth larvae (Galleria mellonella) on petri dishes.
Citation
Santiago-Narvaez B. 2021. Understanding host-pathogen interactions with the use of Galleria mellonella. CourseSource. https://doi.org/10.24918/cs.2021.1
Society Learning Goals
Microbiology
- Cell Structure and Function
- How do unique bacterial cell structures make them targets for antibiotics, immunity, and phage infection?
- Systems
- How do microorganisms, cellular and viral, interact with both human and non-human hosts in beneficial, neutral, or detrimental ways?
Lesson Learning Goals
Students will:
- Learn about the use of invertebrate animal models in microbiology research.
- Compare invertebrate/vertebrate animal models.
- Identify at least two innate immune responses used by Galleria mellonella.
- Describe how innate immune responses are used to combat bacterial infections.
- Know how to use dilution plating to estimate bacterial load.
- Learn to collect and interpret virulence data using the G. mellonella larval model.
- Graph survival data to create a Kaplan-Meier Plot.
Lesson Learning Objectives
Students will be able to:
- Describe at least two innate physical defenses in the human body that are used to fend off an infection.
- Describe how bacterial structures stimulate a non-specific immune response.
- Describe innate immune responses used against bacterial pathogens.
- Describe how animal models can be used in the study of bacterial virulence.
- Identify innate immune responses in G. mellonella and how these can be used for virulence studies.
- Define pathogenic behavior.
- Compare and contrast the virulence of various microbes.
- Collect and accurately report survival data using a Kaplan-Meier plot.
- Evaluate survival data to determine virulence properties of a given microbe.
Article Context
Course
Article Type
Course Level
Bloom's Cognitive Level
Vision and Change Core Competencies
Vision and Change Core Concepts
Class Type
Class Size
Audience
Lesson Length
Pedagogical Approaches
Principles of How People Learn
Assessment Type
INTRODUCTION
Microbial virulence, the ability of the microbe to cause disease in an infected host, is a fascinating area of microbiology. The immune response of the host together with the pathogenic toolset of the microbe represent a complex interplay of virulence that while intellectually engaging, can be difficult to demonstrate to students in vivo. The study of microbial virulence is often limited by the requirement of cell lines or the use of highly-regulated and costly animal models that for the most part are not well suited for the undergraduate laboratory. Galleria mellonella, or the Greater Wax Moth, is an invertebrate animal model that is well-established and validated in microbiology for the study of human pathogens (1,2). G. mellonella has been used to study a variety of microbial pathogens including fungi, yeast, viruses and bacteria (3). G. mellonella is a useful model to study virulence because of its high correlation with virulence of microbes in mice (4). Recently, it has been increasingly used not only for the study of microbial host interactions, but also in the study of novel antimicrobials and antibiotic susceptibility testing (2,5).
G. mellonella's immune system shares similarities to that of mammalian innate immunity. G. mellonella contain the presence of a cuticle (skin barrier) and specifically, hemolymph containing hemocytes, which help distinguish self from non-self, a similar function to that of immune cells. These hemocytes act as patrolling immune cells that detect and destroy the presence of foreign entities, such as bacteria (6). The preferential form of killing for these immune cells is by encapsulation, melanization, and phagocytosis. Once microbes are engulfed by these cells an oxidative burst mediates killing through reactive oxygen species (7). Disease progression in G. mellonella can be visually assessed because the presence of microbes triggers the production of melanin. G. mellonella immune cells code for prophenoloxidases (PPO) which are an important part of its immune response. Upon development of a microbial infection, serine proteases activate the PPO cascade which consequently will oxidize phenolic substances and produce melanin (8,9). The production of melanin is thought to aid in the process of encapsulation and phagocytosis and is equivalent to that of our immune response during chronic infections that lead to abscess formation. Bacterial infections have been shown to trigger melanization in G. mellonella larvae, making it an easy trait to use for microbial virulence studies. Correlation between higher virulence and higher melanin production support the use of this model as a valid means to assess bacterial virulence. Melanization can be used to visually track microbial infections in the larvae where the level of melanization serves as an indicator of health (10).
This unique melanization in response to the presence of an invading microbe provides a visual assessment tool for students to learn about bacterial virulence in the microbiology laboratory. When students study how the immune system targets and eliminates bacterial infections, it is often times limited to images and visuals in a textbook. With this model, students can combine their study of the immune system with the study of bacterial virulence to have a more comprehensive view on what these host-pathogen interactions are like in vivo. Combined with the use of commonly used microbiology techniques such as serial dilution plating, students can use quantitative means to assess the virulence of a microbe. G. mellonella is especially useful in the undergraduate laboratory setting as it is easy to use, does not require any specialized equipment to maintain nor does it require IACUC (Institutional Animal Care and Use Committee) approval for its use. In its larval stage, G. mellonella can be injected with a microbial strain to study disease progression. The larvae can survive in temperature ranges from 25- 37°C, which are optimal growth conditions for the tested microbes, specifically when studying model organisms of human pathogens (11,12).
In this lab exercise, students inject G. mellonella with an assigned microbe (Biosafety Level 1 or BSL-1, following ASM Guidelines for Teaching Laboratories) to determine the virulence of their assigned microbe in comparison to that other bacterial species. Overnight cultures of the desired microbe are serially diluted and plated to determine bacterial load. After injection with the desired microbial strain, larvae are placed in an (37°C) incubator (or desired incubation temperature) for a period of 5 days. Melanization and lack of movement are used to enumerate dead vs. live larvae to generate survival curves. Bacterial virulence is determined by the analysis of survival plots comparing all bacterial strain tested in the classroom. Students can then determine which microbe of all those tested exhibits the highest virulence by determining the differences in percent survival among the groups. This exercise provides the opportunity to discuss innate immunity, host-pathogen interactions as well as introduce students to the use of animal models as tools for the study of bacterial virulence. Additionally, it provides an opportunity for students to learn quantitative approaches for pathogenic analysis.
Intended Audience
This laboratory lesson is intended for introductory to intermediate bioscience majors in a microbiology course. We have used this lesson with biology and marine biology majors in a small liberal arts college.
Required Learning Time
This lesson was taught as part of a microbiology laboratory course for two class periods of 75 minutes in length each. The discussion of the data was completed in an additional 50-minute lecture period. This lesson was taught in conjunction with lecture material.
Prerequisite Student Knowledge
Students participating in this lesson should also be familiar with basic concepts of immunity such as innate and adaptive immune responses. Students should also be familiar with aseptic technique, dilution plating and bacterial enumeration (CFU/ml).
Prerequisite Teacher Knowledge
Instructors should be familiar with microbiology techniques such as aseptic technique, dilution plating and colony counts including adjusting OD to achieve specific CFU/ml. The instructor should understand innate and adaptive immune responses. They should also be familiar with survival curves (Kaplan-Meier Plots) to assess bacterial virulence, and the use of spreadsheet or statistical software to create those curves. The G. mellonella model can be easily learned to instruct the class from reading the widely available literature on the subject (3,13,14). Specifically, Pereira et. al (6) and Ramarao et. al (10) provide the necessary background to instruct this exercise.
SCIENTIFIC TEACHING THEMES
Active Learning
Outside of class, students are expected to complete assigned reading material from both textbook and the laboratory handout. In the lecture presentations, application style questions are included for students to test their understanding of the concepts discussed. To practice interpreting and analyzing Kaplan-Meier plots, graphs are included in the lesson. Students get the opportunity to practice making these graphs in class following a video tutorial. Students work in pairs for the completion of this laboratory exercise. In all exercises the instructor is readily available to answer questions and provide feedback.
Assessment
Short lectures introducing the laboratory exercise are accompanied by application style questions for students to test their understanding of the topic. To practice making survival curves in class, students complete a video tutorial using provided hypothetical data. To practice the analysis and interpretation of survival plots, students are presented with survival curves from published work. Students demonstrate their ability to properly report their data by generating a survival curve using collective class data. Upon completion of the laboratory, students demonstrate their ability to interpret and identify bacterial virulence by creating a figure with a title sentence. Figures are assessed by the presence of proper variables (x and y-axis), a proper conclusion statement sentence that correctly describes what the experiment is demonstrating, a complete figure legend briefly summarizing the methods as well as their professional appearance.
Inclusive Teaching
All students can benefit from this lesson as microorganisms impact human life in a variety of ways. The principles behind animal models and their use is an important lesson that students from various backgrounds can benefit from, especially due to its ethical implications. This exercise gives students the opportunity to share their thoughts and ideas about animal research with their peers. The format of the exercises encourages students to work as a team and share ideas with their peers thus being exposed to a variety of ideas and perspectives. This lesson allows students to experience working with animal models to study bacterial virulence. The low cost of the materials required for the exercise makes it readily accessible for resource-limited institutions. In addition, the nature of the exercise and its adaptability allows for the instructor to adapt and modify the exercise based on student learner's needs and the goals of the specific course where the lesson is being used.
LESSON PLAN
This lesson is intended to be used in conjunction with a microbiology lecture discussing the immune system and bacterial virulence. Students should be familiar with the concept of host- pathogen interactions and should understand the interplay between a microbe and its host.
The goal of the experiment is to have students test the virulence of various bacterial strains by injecting them into G. mellonella larvae (15). Based on the survival outcomes of the different injection groups and the interpretation of the collected class data, students determine which microbe(s) are the most or least virulent. The lesson is broken down into three parts. An introduction to the G. mellonella model and methodology, the practical laboratory exercise, and the data collection and analysis. The lesson is split into three lessons. It is designed to encourage collaboration between the students and their instructor. Student pairs work autonomously to break down tasks and work together to complete the exercise. The instructor provides hands on experience and feedback for the students as the procedure is being completed in the laboratory. Collaboration between all students in the classroom is also required, as their final assignment is dependent on the collective class data. Table 1 provides details for this lesson.
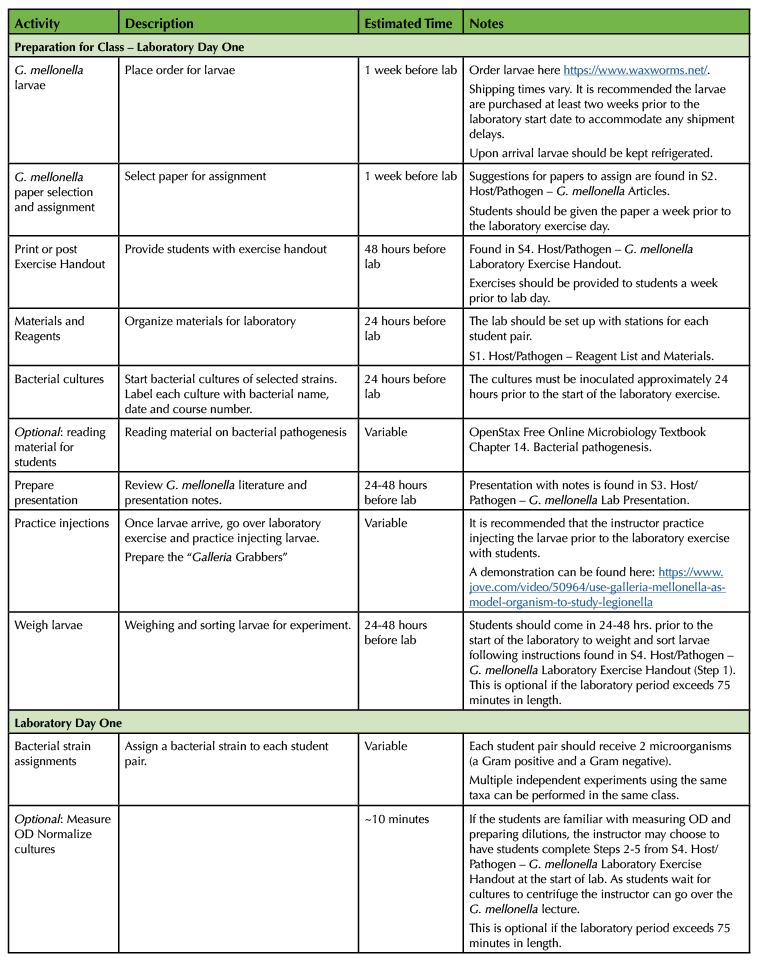
Table 1. Lesson plan timeline for G. mellonella pathogenesis laboratory.

Table 1. Lesson plan timeline for G. mellonella pathogenesis laboratory (continued).

Table 1. Lesson plan timeline for G. mellonella pathogenesis laboratory (continued).
Classroom Environment
This lesson was taught in a microbiology laboratory with 16 students working in pairs. Student pairs were assigned at the start of the semester. The laboratory sessions were 75 minutes in length, meeting twice a week. Students were required to come back to collect results outside of class time for a period of five days. The lecture session for discussion of data was 50 minutes in length.
Safety Procedures
Due to the use of live bacterial cultures for injection, students are required to wear Personal Protective Equipment (PPE) at all times. The use of a needle resistant glove is recommended. An inexpensive device called the "Galleria Grabber" designed to minimize self-injection can also be used (16). The "Galleria Grabber" can easily be made using readily available materials. Materials are listed in Supporting File S1. Host/Pathogen – Reagent List and Materials. Microorganisms used in this laboratory are non-pathogenic, BSL-1, and approved for use within the undergraduate microbiology lab setting (17). However, the use of a syringe presents certain risks if students are not properly trained for their use. It is recommended that students practice injecting the larvae with water or PBS before using live cultures. Hamilton syringes require no recapping which minimizes the risks of accidental injection. To facilitate the process of injection, larvae are kept in the refrigerator until right before injecting. A cold surface (such as a small cutting board kept refrigerated) is recommended to keep them dormant and prevent them from moving during the injection process. The use of the "Galleria Grabber" greatly minimizes any risks and facilitates the procedure (16). Students should be properly informed of the risks of the experiment prior to its execution.
Instructor Preparation for the Lesson
A list of all laboratory reagents and materials are found in Supporting File S1. Host/Pathogen – Reagent List and Materials. This document includes the protocol with instructor notes. This laboratory requires the purchase of live organisms for its execution. Larvae can be purchased from the following vendor https://www.waxworms.net/. The larvae should be purchased with sufficient time to account for shipping prior to the start of the laboratory exercise. As students should already be familiar with host-immune responses, it is recommended for the instructor to select and assign a scientific paper describing G. mellonella as a model system for bacterial pathogenesis prior to the laboratory session. Students should be given the opportunity to research and become familiar with this model organism independently. Suggested articles to assign are found in Supporting File S2. Host/Pathogen – G. mellonella Articles. The instructor is tasked with preparing the presentation materials used in the lesson. A PowerPoint presentation was used by this instructor found in Supporting File S3. Host/Pathogen – G. mellonella Lab Presentationn. The presentation was used for both laboratory periods. Bacterial cultures used for the lesson should be inoculated 24 hours prior to the laboratory session.
Student Preparation for the Lesson
Students should be familiar with the topics of immunity and bacterial virulence or are in a course where these topics are concurrently being discussed. Students can be assigned to read background material in these topics found in OpenStax Free Online Microbiology Textbook (18) as part of their preparation for the laboratory. Students should read the assigned G. mellonella paper prior to attending the laboratory and go over the Laboratory Exercise handout found in Supporting File S4. Host/Pathogen – G. mellonella Laboratory Exercise Handout. For graphing survival curves, students should have access to a computer or laptop with Microsoft Excel and have online access to view the tutorial video.
Laboratory Day One
G. mellonella model
To begin the lesson, the instructor starts with a PowerPoint presentation (Supporting File S3. Host/Pathogen – G. mellonella Lab Presentation). The instructor should begin by asking the class why animal models are important in microbiology research. They also will ask students to think about why animals are necessary in virulence studies. Students are given the time to share their answers with the class. Following this discussion, the instructor introduces the use of G. mellonella as an invertebrate animal model for research. The instructor briefly explains G. mellonella's innate immune response of melanization. It is important that the instructor emphasizes how these specific characteristics will be used in their experiment. The presentation should include specific questions about G. mellonella to assess the students understanding of its use to measure bacterial virulence. If the instructor assigns a specific paper, questions should also be included to verify student understanding of the assigned reading.
The instructor should then explain the procedure for the G. mellonella laboratory experiment. Using the visuals provided in the presentation (Supporting File S3. Host/Pathogen – G. mellonella Lab Presentation), the instructor should walk students through the experimental design described in the Bacterial Virulence Lab exercise (Supporting File S4. Host/Pathogen – G. mellonella Laboratory Exercise Handout). The PowerPoint presentation includes notes to facilitate the explanation of the experiment. The instructor can also use any available online videos that demonstrate the procedure (19). The Ramarao et. al paper (10) provides a downloadable video explaining the procedure. Following the explanation of the methods, the instructor should allow students to ask questions regarding the protocol. Students should clearly understand how they can quantify virulent behavior with the use of the larvae. To end the presentation, the instructor should briefly show how the data is reported by including an image of a survival curve. Students do not have to be taught how to graph the data at this point. In the next laboratory session, as they are still in the process of collecting their data, they can use the lab period to practice graphing their data and the instructor can explain how to accurately interpret survival curves.
G. mellonella lab exercise
The exercise requires the use of G. mellonella larvae. Each student pair will need approximately 50 larvae. These are shipped with food/wood chips and require no feeding during the storage process. Upon arrival, the larvae can be stored in a refrigerator (5°C) for one week. Larvae will start to pupate if they are left out at room temperature, therefore these should be used as quickly as possible and not stored for more than two weeks in the refrigerator (5°C). An image of the different stages of larval development can be found in Singkum et al. (3).
To begin the exercise, students need to weigh the larvae. The larvae used in the experiment should all be of similar weight and size for consistency. This allows students to understand how variables (differences in weight) can affect the outcome of their experiment. Larvae weight should be between 200 and 300 milligrams. Any larvae that are immobile, desiccated or appear dark in color should be discarded. This is a good opportunity for the instructor to point out larvae that show melanization to the students.
After weighing the larvae, these are sorted in groups of 10 and placed into empty petri dishes with wood chips. To save time, students may choose to weigh the larvae prior to the laboratory session. The larvae are kept refrigerated until they are ready to be used in the experiment. Each student pair should be provided with the materials for the experiment listed in Supporting File S1. Host/Pathogen – Reagent List and Materials and a working space for injection and dilution plating. Each student should be assigned a specific microbe to test. Students pairs inject two microorganism total, one Gram positive and one Gram negative microbe.
Following the provided protocol (Supporting File S4. Host/Pathogen – G. mellonella Laboratory Exercise Handout) students will normalize their bacterial cultures as described in their lesson before proceeding with the injection. Normalizing the cultures allows for all cultures to have a similar bacterial density based on optical density (OD). If the cultures are not normalized, the larvae will be injected with varying amounts of bacteria and the results will be inconclusive. The instructor may choose to ask students why this matters. It helps to highlight the importance of each group being exactly the same (host weight, bacterial load, etc.) with the exception of the taxa of bacteria used. In our experiment, we normalized to an OD of 0.6. For recommended bacterial loads to use and the expected kill curves see Jia et al. (2).
After diluting and washing the culture, the bacterial pellet is resuspended in sterile PBS. This step allows for the removal of media and any bacterial byproducts secreted into the medium during cultivation. This resuspension will be used for the injection. Students should be reminded not to discard their resuspended culture after injecting. They will be using the remaining culture for their serial dilutions. Students should only work with one group of larvae at a time. Larvae should be maintained inside the refrigerator until right before the injection procedure.
Before the start of the experiment, students should be given the opportunity to practice injecting the larvae using saline. By injecting 5-10 larvae as practice, the instructor can provide feedback on injection technique. These larvae will not be used in the experiment. Depending on the laboratory period length, students can be given additional time to practice injecting larvae before they start the lab exercise. Students should begin by injecting the saline control group. As this does not involve the use of live cultures, it is an opportunity for them to become familiar with the procedure. It is extremely important that students are properly trained to do this step as they will be working with live cultures in their experimental group injections. The injection site is located is the right-hand side last pro-leg of the larvae. They should be able to see the pro-legs when the larvae are flipped upside down. There are multiple options to facilitate injection and increase safety. Larvae can be held down belly side up against the cutting board using the index and middle finger while using a needle resistant glove. An alternative to this protocol is to use a serological pipette. The larvae can be held against the pipette which curves the body of the larvae and makes injecting easier (20). Finally, students can use the device called the "Galleria Grabber" (16). This decreases the chances of accidental injection as the individual does not have to hold the larvae for injection. Students should never hold the larvae in their hand while attempting an injection. If the students are injecting the larvae correctly, they should be able to lift the larvae from the cutting board surface or "grabber" while the syringe is still inside the larvae. To remove the larvae simply slide it off the syringe tip using the middle and index finger. The process is demonstrated in this video (20).
After injection the larvae should be placed in their corresponding petri dish and set aside. Continue the injection process with the experimental group. This is the group that will be injected with bacteria. Students need to proceed with the injection cautiously as they are working with live cultures. After the experimental group has been injected with the assigned bacterial strain, the larvae should be placed inside their corresponding petri dish. All petri dishes should be placed inside a tray and transferred to the incubator. The larvae should be incubated for a period of five days. Students should come in and record the number of live larvae every 24 hours. To determine if larvae are alive or dead, students should observe the coloration and appearance of the larvae (Figure 1). They can use a P1000 micropipette tip to prod the larvae. If the larva is unmoving and appears dark or black in color, it is dead (Figure 1B).
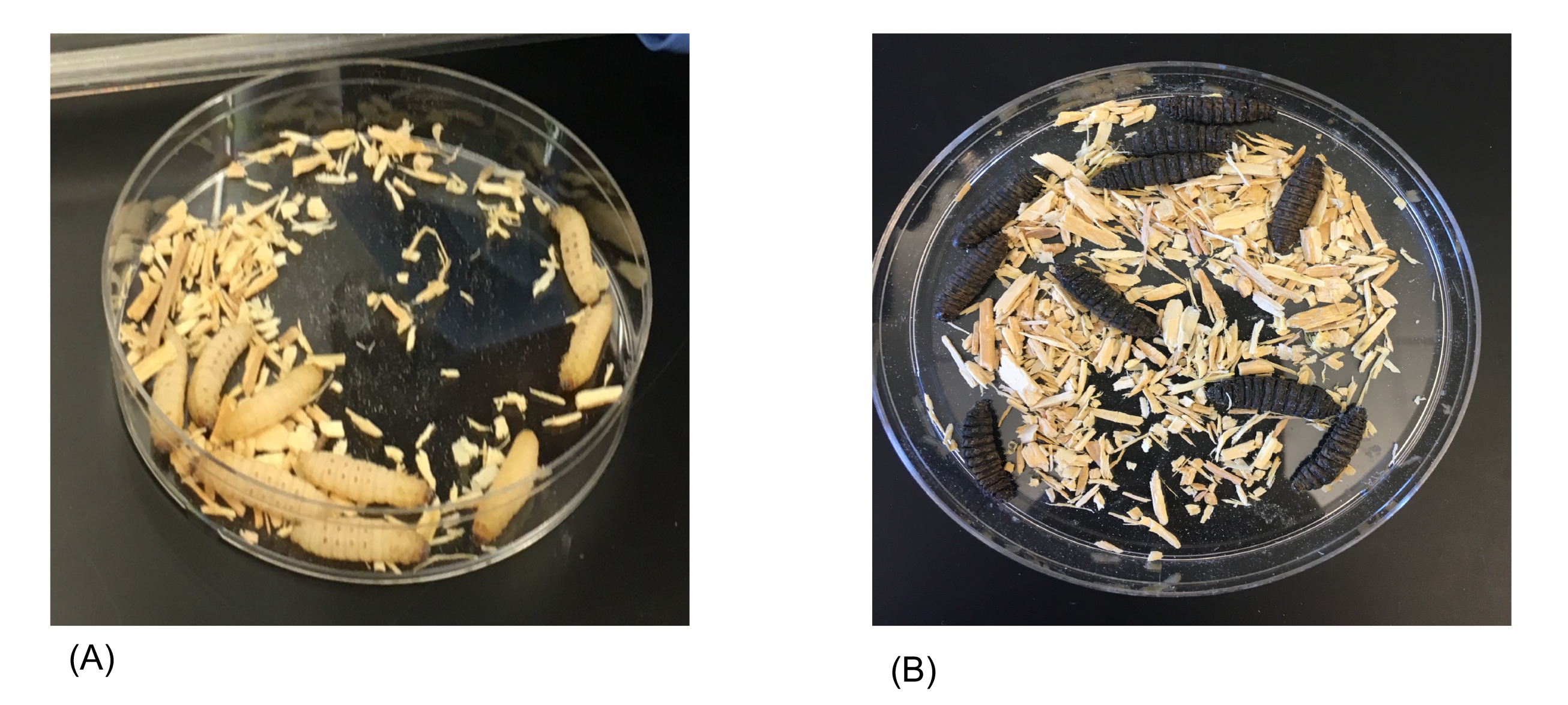
Figure 1. Visual assessment of Galleria mellonella larvae before and after bacterial infection. Galleria mellonella larvae are typical beige or yellow colored as shown in our control plate (A). They do not have any blue or dark spots present when they are healthy. After injection with bacteria followed by incubation, melanization occurs as evidenced by dark, black coloration of the larvae (B). Larvae are immobile, indicating they are dead.
Upon completion of the experiment all larvae (injected and untreated - including unused larvae) should be placed in a biohazard bag and placed inside a (- 80°C) freezer for 24 hours. This bag should then be autoclaved. The greater wax moth can infest beehives and should never be released. Remind students that they will be using the entire class's data for their figure assignment. The class can work with a Google Docs to facilitate collaboration and sharing of class data (Supporting File S9. Host/Pathogen – Class Data Google Doc Template). This class data should also include their CFU/ larva (bacterial load). Students should take a photo of their plates at the end of the experiment, before they discard their larvae (Figure 1).
Laboratory Day Two
Data Collection: Survival Curves
The instructor begins the discussion with a brief introduction explaining the use of Kaplan-Meier plots to report survival data. The instructor explains how collecting the data (number of live larvae over time in a table) allows the investigator to determine the virulence of a single microbe. Since we are testing a variety of microbes, the survival curve allows us to compare the virulence of all groups to each other to determine if there are any differences between the groups. The presentation should include an example of a survival curve from published work to demonstrate what the graphs look like. The instructor can use the image later in the presentation to explain how these graphs are interpreted. Students will have the opportunity to practice graphing their data in class following a video tutorial found on https://www.vialogues.com/vialogues/play/19756 . This video can be posted online for students to watch in their own time before lab or can be shown to the class during the lab period. Students can follow the tutorial using the practice data posted in Supporting File S5. Host/Pathogen – Excel Spreadsheet for Survival Curves. This file contains hypothetical data from a G. mellonella experiment.
After completing the tutorial, the instructor should check the students graphs to make sure they have successfully graphed their data. Supporting File S5. Host/Pathogen – Excel Spreadsheet for Survival Curves includes a key for the instructor to use (Sheet 2). Students graph their data as percent survival over time (days). The instructor answers any questions the students have about how to graph the data. Using this same graph or a graph from a publication, the instructor will explain how to interpret a survival curve. The presentation (Supporting File S3. Host/Pathogen – G. mellonella Lab Presentation, Day two) contains detailed notes to prompt the discussion of the interpretation. After this explanation, students are given the chance to interpret a survival curve on their own. After completing the exercise, students continue to record their survival data for the remaining days left leading up to the five days. After collecting their data and that of their classmates they should prepare and submit their figure (Figure 2). Figure assignment details can be found in Supporting File S6. Host/Pathogen – Figure Assignment Instructions. The final figure is graded using the provided rubric (Supporting File S7. Host/Pathogen – Figure Rubric).
Laboratory or Lecture Day Three
Data Analysis Class Discussion
After students submit their figures, the instructor discusses the class data with students. Talking points for the interpretation of survival curves can be found on Supporting File S8. Host/Pathogen – Survival Curve Analysis. Students have the chance to interpret the data and share their ideas with the class regarding their experiment. The instructor starts by asking the students "what is the goal of the experiment?" This is an opportunity for students to articulate the purpose of the exercise. Most students at this point are able to mention that the purpose was to determine the virulence properties for a variety of microbes by looking at the end point readout or death of the larvae over time. Some may also comment on "learning how to use animals in research." The instructor should ask why the experiment was done using G. mellonella. Students can explain why this model system is useful and why it is a good system for the experiment the class performed. A figure with the class data should be projected for the students to view. The instructor should go over the features of the graph, emphasizing what each feature is for (axis, controls, groups-experimental, etc.). Students are given time to share their thoughts on the data and how they interpreted it. Students pairs can also be assigned to present their individual data to the class as part of the class discussion.
Important aspects of the discussion revolve around students considering bacterial loads, traits indicative of virulence, and the effectiveness of controls in the experiment. Students oftentimes ignore bacterial loads when making their interpretations. It is important to remind students that the bacterial load influences the outcome. Based on the specific results obtained by the class (which can vary), students should share their explanation of the data. They are encouraged to use their understanding of immunology and bacterial virulence to support their answers. To end the discussion the instructor should provide the overall conclusion of the class experiment and answer any remaining questions on the topic.
TEACHING DISCUSSION
Bacterial virulence is a challenging concept to demonstrate in an undergraduate laboratory setting. The use of living organisms to teach bacterial virulence can be challenging due to the ethical limitations and the costs presented by animal research. Invertebrate animal models such as G. mellonella are highly popular in the study of virulence, and as such provide a unique, inexpensive way to teach bacterial virulence in undergraduate microbiology courses. This model, which has been validated and is consistently used in research, is a great tool to demonstrate how animals are used to study pathogens (2). This exercise provides students with a visual assessment of the interactions between a host and the pathogen. In addition, the topics of immunology with microbiology can be discussed, providing a unique opportunity for students to see the interconnection of these processes. As ASM Curriculum Guidelines highlight the importance of students learning about systems (which encompasses host-microbe interactions) this exercise can serve as a link for students to understand how microbes and hosts interact (21).
With this laboratory exercise, students use their understanding of a variety of concepts relevant to microbiology and infectious diseases to determine the virulence of microbes. As this exercise is designed to be taught in conjunction with a microbiology course, students first learn about immune responses and bacterial pathogenesis from a theoretical perspective. This may not be as engaging as seeing the process happen in real life. Students can then reinforce these concepts by identifying characteristics that support virulent behavior within an in vivo setting. By injecting the larvae with a variety of microbes, students can compare and contrast the virulence of the microbes tested in the class by measuring larval survival over time. By researching the tested microbes, students can determine their virulence properties and mechanisms of infection in order to support the observed virulence traits from their experiment. They can also make predictions on which microbes are the most virulent simply by observing the appearance of the groups at different time points of the experiment. By collecting and graphing their data, students are able to use quantitative reasoning to decipher the characteristics of the tested microbes.
Although this exercise is not intended to be a biostatistics lesson, the simplified use of survival curves is sufficient for students to grasp how data can be processed and used to reach conclusions. Survival curves are widely used in medical research, ecology, and engineering. The exercise is an opportunity for students to learn how to graph and interpret data that is clinically relevant.
The use of animals in research is controversial. Undergraduates should be exposed to course content that allows them to understand why animals are used in research. A recent study by Sandgren et al. (22) looked at how undergraduates view and understand the use of animals in research. In the majority of the cases, students expressed the need for more animal research-related course content as a means to strengthen their confidence in the topic. The authors highlighted how course-based content on animal research should be part of a college's best practices. This lesson provides instructors with an opportunity to teach undergraduates about invertebrate animal models and their use in microbiology related research (10). Alternative animal models for research are especially welcome as they minimize many of the challenges and ethical concerns regarding the topic, making them even more accessible to undergraduates. Lastly, the experimental design of the exercise allows students to understand the steps of the scientific process and the use of an invertebrate model to answer a question. In vitro assays are not always sufficient to mimic the complexity behind microbe-host interactions. With this laboratory students are encouraged to consider the different approaches used to conduct research.
After the completion of this exercise students were able to successfully create survival plots to report their data (Figure 2). In their figures, students were able to accurately describe the outcome of the experiment, supporting their ability to analyze and interpret the collective class data. Students successfully determined bacterial loads with the use colony counts and were able to identify this as a variable affecting the experiment's outcome. Students were also able to identify and differentiate the virulence properties among the microbes through the assessment of G. mellonella survival curves. During the class discussion, they analyzed the collective class data, and were able to provide plausible explanations in support of their observations. Finally, the exercise provided students with the opportunity to understand the use and the benefits of alternative animal models in microbiology studies.
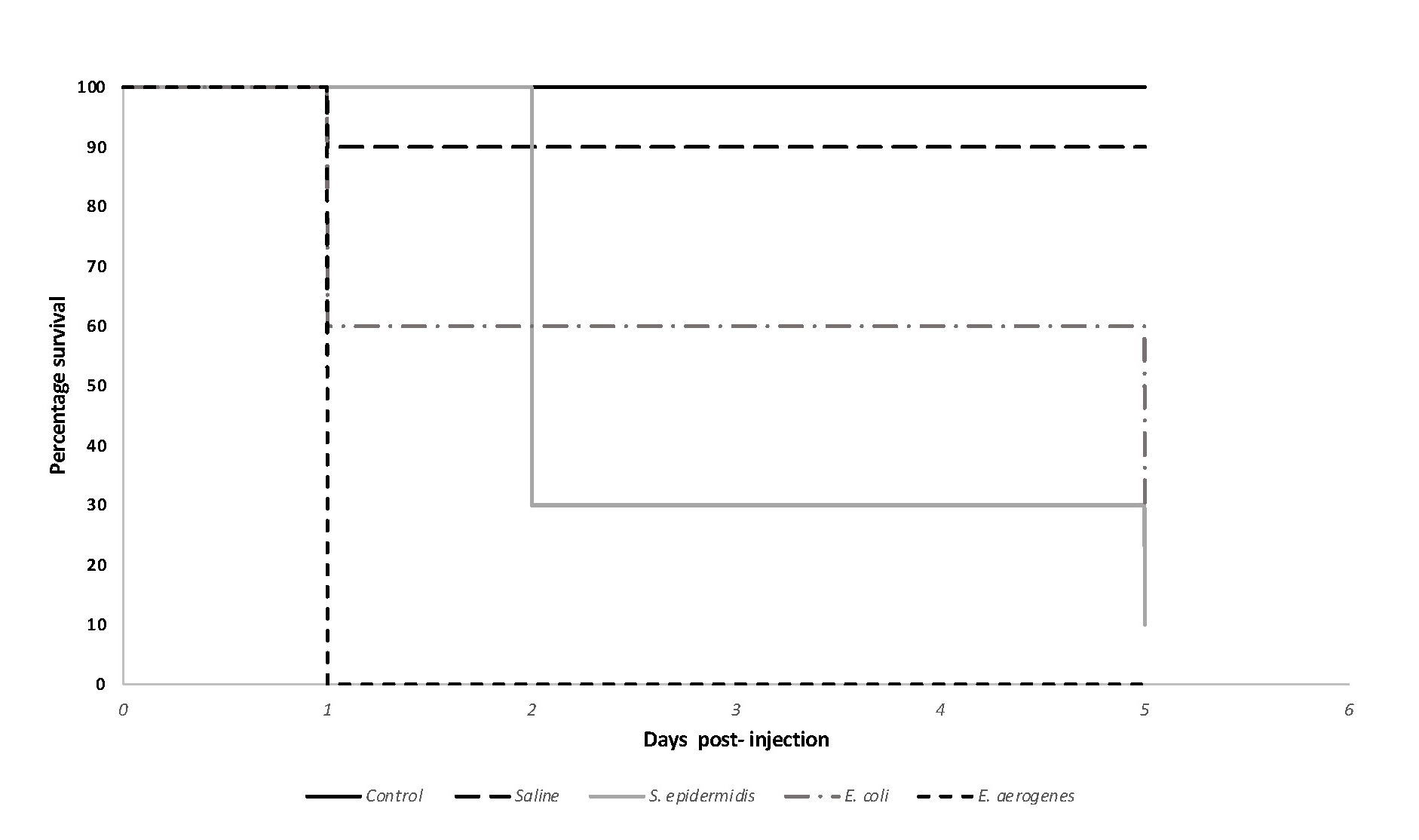
Figure 2. Survival curve for Galleria mellonella larvae injected with various bacteria. After normalization, students injected larvae (n =10/group) with 10 uls of their assigned microbe (approx. 1 X 106 CFU/ml as confirmed by colony counts). Control (no treatment) and saline control groups were also included. In this sample image, 1 larva died from saline control group. Larval survival was tracked for a total of 5 days (approximately 120 hours) where the number of live larvae were recorded daily. Kaplan-Meier plots were graphed using Microsoft Excel. Survival curve differences were statistically significant based on p < 0.0001.
Modifications
For laboratories with longer class periods (three hours in length or more), the experiment and the survival curve tutorial exercise can be taught together and would not require two separate laboratory sessions. As a supplement to this lab exercise, students can test the effect of bacterial load on virulence and survival by repeating the procedure using a variety of bacterial loads for a single microbe. Recommended bacterial loads are described in the literature (2). As a complement to the study of antibacterials in a microbiology course, disruption of disease progression using a therapeutic can be also tested by treating infected larvae with antibiotics after injection with a bacterium (23). Larvae are first injected with microbes, incubated for 1-2 hrs. to allow bacterial infection to develop followed by antibiotic injection in the final left-side proleg. Recommended antibiotic concentrations can be determined by selecting levels within known MIC values for the desired microbe to be tested. Amount of antibiotic used is determined per kg of larval weight. Approximately 5 mg/ kg of body weight is recommended (23,24). An antibiotic or treatment control group should be included if antibiotics or treatments are tested. The larvae are injected with antibiotic only (no pathogen) to account for any potential toxicity effects of the treatment.
As another application to the exercise, students can make observations on the degree of melanization of their infected larvae using images found in the literature (3). The characteristic melanization response in the larvae allows students to make direct observations on the innate immune responses used to fight bacterial infections. With observations on the level and degree of melanization, students can identify the success or failure of the innate immune response in clearing an infection. As a class they can decide on a severity score to assess their larvae based on published images. Students can perform the injections and collect qualitative data based on degree of melanization. For each larvae group (control, saline and experimental) students determine the average score using their index and create a graph allowing them to connect how qualitative data can be used to generate quantitative data.
Student Feedback for the Lesson
After completing the laboratory exercise students enrolled in the course were asked to share their comments on the use of animal models, and the laboratory exercise as a whole. Selected comments are shared below.
"Yes, the exercise helped me understand how physical observations of bacterial virulence and the mortality of G. mellonella can be used to study virulence."
"The use of G. mellonella helped me to understand how animal models are used in research, and also why certain animal models are chosen over others."
"The use of Kaplan-Meier plots helped me learn how to compare data that involved multiple bacterial species. Additionally, I learned how to read plots that represent data across different bacterial strains and how to interpret that information from a graph."
"I learned why animal models are necessary and useful for research because they allow the safety and effectiveness of treatments to be tested more efficiently. There are many different animal models that can be used, and it is important to select a model with many similarities to the organism that will benefit from the long-term effects of the research."
SUPPORTING MATERIALS
- Supporting File S1. Host/Pathogen – Reagent List and Materials. This file Includes a material list and protocol notes for Instructor.
- Supporting File S2. Host/Pathogen – G. mellonella Articles
- Supporting File S3. Host/Pathogen – G. mellonella Lab Presentation
- Supporting File S4. Host/Pathogen – G. mellonella Laboratory Exercise Handout
- Supporting File S5. Host/Pathogen – Excel Spreadsheet for Survival Curves
- Supporting File S6. Host/Pathogen – Figure Assignment Instructions
- Supporting File S7. Host/Pathogen – Figure Rubric
- Supporting File S8. Host/Pathogen – Survival Curve Analysis
- Supporting File S9. Host/Pathogen – Class Data Google Doc Template
ACKNOWLEDGMENTS
The author would like to thank the students enrolled in BIO229 Microbiology Course as well as their colleagues Dr. Jay Pieczynski and Dr. Sabrice Guerrier for feedback in the revision of this manuscript.
References
- Cook SM, McArthur JD. 2013. Developing Galleria mellonella as a model host for human pathogens. Virulence 4:350-353.
- Jia C, Tsai -Yun, Mei J, Loh S, Proft T. 2016. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7:214-229.
- Singkum P, Suwanmanee S, Pumeesat P, Luplertlop N. 2019. A powerful in vivo alternative model in scientific research: Galleria mellonella. Acta Microbiol Immunol Hung 66:31-55.
- Jander G, Rahme LG, Ausubel FM. 2000. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J Bacteriol.18(13): 3843-3845
- Cutuli MA, Petronio Petronio G, Vergalito F, Magnifico I, Pietrangelo L, Venditti N, Di Marco R. 2019. Galleria mellonella as a consolidated in vivo model hosts: New developments in antibacterial strategies and novel drug testing. Virulence 10:527-541.
- Pereira T, de Barros P, Fugisaki L, Rossoni R, Ribeiro F, de Menezes R, Junqueira J, Scorzoni L. 2018. Recent advances in the use of Galleria mellonella model to study immune responses against human pathogens. J Fungi 4:128.
- Bergin D, Reeves EP, Renwick J, Wientjes FB, Kavanagh K. 2005. Superoxide production in Galleria mellonella hemocytes: Identification of proteins homologous to the NADPH oxidase complex of human neutrophils. Infect Immun 73:4161-4170.
- Leonard C, Ratcliffe NA, Rowley AF. 1985. The role of prophenoloxidase activation in non-self recognition and phagocytosis by insect blood cells. J Insect Physiol 31:789-799.
- Schmit AR, Rowley AF, Ratcliffe NA. 1977. The role of Galleria mellonella hemocytes in melanin formation. J Invertebr Pathol 29:232-234.
- Ramarao N, Nielsen-Leroux C, Lereclus D. 2012. The insect Galleria mellonella as a powerful infection model to investigate bacterial pathogenesis. J Vis Exp 70.
- Fuchs BB, O'Brien E, Khoury JB El, Mylonakis E. 2010. Methods for using Galleria mellonella as a model host to study fungal pathogenesis. Virulence 1:475-482.
- Desbois AP, Coote PJ. 2012. Utility of greater wax moth larva (Galleria mellonella) for evaluating the toxicity and efficacy of new antimicrobial agents. Adv Appl Microbiol 78:25-53.
- Mikulak E, Gliniewicz A, Przygodzka M, Solecka J. 2018. Galleria mellonella as a model organism used in biomedical and other studies. Przegl Epidemiol 72:57-73.
- Velikova N, Kavanagh K, Wells JM. 2016. Evaluation of Galleria mellonella larvae for studying the virulence of Streptococcus suis. BMC Microbiol 16:1-9.
- Junqueira JC. 2012. Galleria mellonella as a model host for human pathogens. Virulence.
- Dalton JP, Uy B, Swift S, Wiles S. 2017. A novel restraint device for injection of Galleria mellonella larvae that minimizes the risk of accidental operator needle stick injury. Front Cell Infect Microbiol 7:99
- Byrd JJ, Emmert E, Maxwell R, Townsend H. 2019. Guidelines for biosafety in teaching laboratories version 2.0: A revised and updated manual for 2019. J Microbiol Biol Educ. 20(3):57
- Parker N, Schneegurt M, Thi Tu A-H, Lister P, Forster BM. 2016. Microbiology. OpenStax, Houston.
- Dunn MJ, Woodruff AL, Anderson MZ. 2018. The Galleria mellonella waxworm infection model for disseminated candidiasis. J Vis Exp 141.
- Harding CR, Schroeder GN, Collins JW, Frankel G. 2013. Use of Galleria mellonella as a Model Organism to Study Legionella pneumophila Infection. J Vis Exp 81:50964.
- Merkel S. 2012. The development of curricular guidelines for introductory microbiology that focus on understanding. J Microbiol Biol Educ 13:32-38.
- Sandgren EP, Streiffer R, Dykema J, Assad N, Moberg J. 2019. Assessing undergraduate student and faculty views on animal research: What do they know, whom do they trust, and how much do they care? PLoS One 14:e0223375.
- Desbois AP, Coote PJ. 2011. Wax moth larva (Galleria mellonella): An in vivo model for assessing the efficacy of antistaphylococcal agents. J Antimicrob Chemother. 66(8 ):1785-1790
- Ignasiak K, Maxwell A. 2017. Galleria mellonella (greater wax moth) larvae as a model for antibiotic susceptibility testing and acute toxicity trials. BMC Res Notes. 10: 428
Article Files
Login to access supporting documents
Understanding Host-Pathogen Interactions With the Use of Galleria mellonella(PDF | 840 KB)
S1. Host Pathogen - Reagent List and Materials.docx(DOCX | 39 KB)
S2. Host Pathogen - G. mellonella Articles.docx(DOCX | 25 KB)
S3. Host Pathogen - G. mellonella Lab Presentation.pptx(PPTX | 11 MB)
S4. Host Pathogen - G. mellonella Laboratory Exercise Handout.docx(DOCX | 40 KB)
S5. Host Pathogen - Excel Spreadsheet for Survival Curves.xlsx(XLSX | 31 KB)
S6. Host Pathogen - Figure Assignment Instructions.docx(DOCX | 20 KB)
S7. Host Pathogen - Figure Rubric.docx(DOCX | 23 KB)
S8. Host Pathogen - Survival Curve Analysis.docx(DOCX | 30 KB)
S9. Host Pathogen - Class Data Google Doc Template.docx(DOCX | 16 KB)
- License terms

Comments
Comments
There are no comments on this resource.